SUMMARY
The mitotic checkpoint acts to maintain chromosome content by generation of a diffusible anaphase inhibitor. Unattached kinetochores catalyze a conformational shift in Mad2, converting an inactive open form into a closed one that can capture Cdc20, the mitotic activator of the APC/C ubiquitin ligase. Mad2 binding is now shown to promote a functional switch in Cdc20, exposing a previously inaccessible site for binding to BubR1’s conserved Mad3 homology domain. BubR1, but not Mad2, binding to APC/CCdc20 is demonstrated to inhibit ubiquitination of cyclin B. Closed Mad2 is further shown to catalytically amplify production of BubR1-Cdc20 without necessarily being part of the complex. Thus, the mitotic checkpoint is produced by a cascade of two catalytic steps, an initial one acting at unattached kinetochores to produce a diffusible Mad2-Cdc20 intermediate and a diffusible one in which that intermediate amplifies production of BubR1-Cdc20, the inhibitor of cyclin B ubiquitination by APC/CCdc20.
INTRODUCTION
The mitotic checkpoint (also known as the spindle assembly checkpoint) delays the irreversible transition into anaphase until kinetochores on all chromosomes successfully attach to spindle microtubules. Each unattached kinetochore produces an inhibitory signal(s) that blocks ubiquitination of cyclin B and securin by inhibiting Cdc20, the pre-anaphase activator of the E3 ubiquitin ligase, the anaphase-promoting complex or cyclosome (APC/C) (reviewed in (Lara-Gonzalez et al., 2012; Peters, 2006)). Anaphase onset is triggered by silencing the checkpoint-derived inhibitor and the subsequent degradation of cyclin B and securin. Critical components of the mitotic checkpoint include Bub1, Bub3, Mad1, Mad2, Mad3 (known as BubR1 outside of yeast), and Mps1 (Hoyt et al., 1991; Li and Murray, 1991; Weiss and Winey, 1996).
Mad2 is an essential mitotic checkpoint protein that has been shown to directly bind to Cdc20 (Fang et al., 1998). Structural studies have revealed that Mad2 undergoes a major conformational change from an open to a closed form when it binds to Cdc20 or Mad1 (Luo et al., 2000; Luo et al., 2002; Luo et al., 2004; Mapelli et al., 2007; Sironi et al., 2002; Sironi et al., 2001). In vivo and in vitro fluorescence recovery after photobleaching has led to a model that open Mad2 is converted to a closed form by recruitment to a stably-bound Mad1/Mad2 complex at each unattached kinetochore (Shah et al., 2004; Vink et al., 2006). This second molecule of Mad2 then quickly cycles on and off unattached kinetochores (De Antoni et al., 2005; Mapelli et al., 2006; Nezi et al., 2006; Shah et al., 2004; Vink et al., 2006). In vitro reconstitution has established that unattached kinetochores with stably bound Mad1 serve as “Mad2 templates” to catalyze the production of a diffusible Cdc20 inhibitor (Kulukian et al., 2009).
BubR1 has also been shown to associate with Cdc20 (Wu et al., 2000) and can inhibit Cdc20 activation of APC/C either alone (Tang et al., 2001) or in combination with Mad2 (Fang, 2002; Kulukian et al., 2009; Lara-Gonzalez et al., 2011). Two Cdc20 binding domains have been identified in BubR1: one within the N-terminal Mad3 homology domain and a second at an internal site (Davenport et al., 2006). Mad2 can enhance BubR1 association with Cdc20 (Davenport et al., 2006; Kulukian et al., 2009), but the mechanistic contribution(s) to mitotic checkpoint signaling of the two Cdc20 binding domains in BubR1 remains to be established.
Attempts have been made to identify the mitotic checkpoint inhibitor produced by unattached kinetochores (Chao et al., 2012; Kulukian et al., 2009; Nilsson et al., 2008; Sudakin et al., 2001). While BubR1, Bub3, and Mad2 have all been found to be complexed with Cdc20 during mitotic checkpoint-mediated arrest, there are major discrepancies in the reported level of Mad2 in the inhibitory complex, spanning from equimolar (Chao et al., 2012; Sudakin et al., 2001) to negligible (Nilsson et al., 2008) levels compared to Cdc20. Correspondingly, multiple models have been put forward for how APC/CCdc20 is inhibited. One proposal, based on recent evidence in budding yeast, is that the function of Mad3 (i.e., BubR1) is to promote the inhibition of Cdc20 by Mad2 (Lau and Murray, 2011). An alternative model for Cdc20 inhibition relies on cooperative binding of Mad2 and Mad3 using evidence from a crystal structure of a fission yeast mutant of Mad2 locked in the closed conformation and complexed with portions of Mad3 and Cdc20. Consistent with this latter model, activity of APC/CCdc20 has been proposed to be influenced by the sequestration by Mad2 of Cdc20’s N-terminal APC/C recognition motifs (Chao et al., 2012; Izawa and Pines, 2012). Lastly, Mad3/BubR1 may act as a ‘pseudosubstrate’ inhibitor, where it competes with substrates for Cdc20 binding in a KEN-box dependent manner (Burton and Solomon, 2007).
In all of the proposed models it remains unclear how a single unattached kinetochore can act to generate global inhibition of APC/CCdc20 activity. Using a combination of purified components and with cultured cells, we now identify two sequential, interlocking catalytic steps in the production of the mitotic checkpoint inhibitor in which BubR1, not Mad2, blocks the activity of APC/CCdc20. In an initial catalytic step, the Mad1/Mad2 complex acts locally at unattached kinetochores to catalyze the production of closed Mad2 bound to Cdc20. Subsequently, closed Mad2 then functions catalytically as a diffusible loader for generating BubR1 binding to a previously inaccessible site in Cdc20. This produces Cdc20 bound to BubR1 as the functional mitotic checkpoint-derived inhibitor that selectively blocks ubiquitination of cyclin B and securin by APC/CCdc20.
RESULTS
Inhibition of APC/CCdc20 by Mad2 and the conserved “Mad3” Cdc20 binding site of BubR1 is essential for the mitotic checkpoint
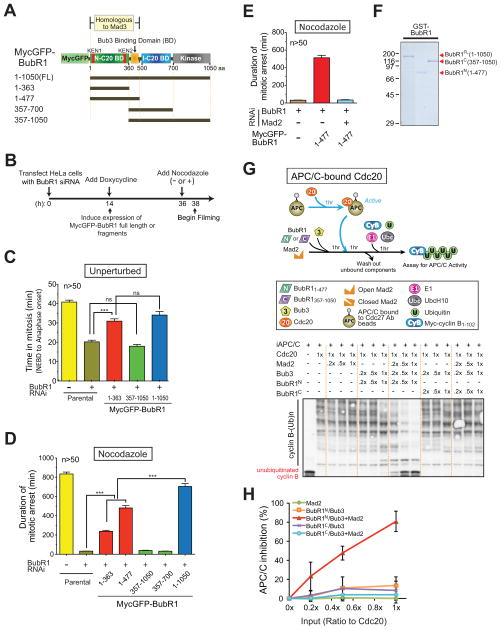
To determine the contribution of the Cdc20 binding sites in BubR1 to mitotic checkpoint signaling, endogenous BubR1 was replaced by a similar level (Fig. 1A,B; Fig. S1A–C) of MycGFP-BubR1 variants containing either or both Cdc20 binding domains. As expected, suppression of endogenous BubR1 resulted in premature mitotic exit from an unperturbed mitosis and this was rescued by expression of full-length BubR1 (BubR11-1050) or a variant (BubR11-363) that included the N-terminal, Mad3 homology domain Cdc20 binding site, but which neither bound Bub3 nor kinetochores (Fig. 1C; Fig. S1D). Similarly, both full length (BubR11-1050) and BubR1 variants with the N-terminal Cdc20 binding site (BubR11-363 or BubR11-477) sustained long term nocodazole-induced mitotic arrest, while BubR1 depleted cells exited mitosis within 35 min. (Fig. 1D).
Figure 1. Inhibition of APC/CCdc20 by Mad2 and BubR1N is critical for the mitotic checkpoint.
(A) Schematic of the MycGFP-BubR1 transgenes stably expressed in HeLa cells with functional domains of BubR1 highlighted. (B) Schematic of the protocol used for replacement of endogenous BubR1 with various BubR1 transgenes. (C,D) Time-lapse microscopy was used to determine mitotic timing in (C) an unperturbed mitosis and (D) in the presence of nocodazole (100 ng/ml). (E) Time-lapse microscopy was used to determine mitotic timing in nocodazole after replacing endogenous BubR1 with MycGFP-BubR1N. Mad2 was co-depleted where indicated using siRNA. Bars in (C–E) represent the mean of at least fifty cells from two independent experiments. Error bars represent SEM. ***=P<0.0001. (F) Purified recombinant BubR1 fragments, assessed by Coomassie staining. (G–H) In vitro generation of an inhibitor of preactivated APC/CCdc20 for ubiquitination of cyclin B1-102. Inhibitory activity was measured in (H) by depletion of unubiquitinated cyclin B1-102. (Data represent mean ± SEM). (see also Fig. S1).
The BubR1 N-terminal-mediated mitotic checkpoint arrest was dependent on Mad2, as it was completely eliminated by siRNA-mediated reduction in Mad2 (Fig. 1E). In contrast, replacement of endogenous BubR1 with variants deleted in the Cdc20 binding site within the Mad3-homology domain were unable to mediate checkpoint signaling in both unperturbed mitosis (Fig. 1C; Fig. S1D) and nocodazole treated cells (Fig. 1D). This effect did not depend on whether the BubR1 fragments contained (BubR1357-1050) or lacked (BubR1357-700) the BubR1 kinase domain. Full-length BubR1 mediated longer nocodazole-induced mitotic arrest than BubR1 variants with only the N-terminal Cdc20 binding site, consistent with a possible role of the internal Cdc20 binding site in maintaining chronic mitotic checkpoint signaling.
To determine how BubR1 generated a mitotic checkpoint inhibitor together with Mad2, full-length recombinant BubR1 and BubR1 fragments containing either Cdc20 binding domain were expressed, purified (Fig. 1F; Fig. S1E), and incubated with Cdc20 along with Bub3 in the presence or absence of Mad2. Inactive APC/C was added, incubated, and the APC/C subsequently recovered (with an antibody to Cdc27). Finally, cyclin B was added along with E1, the E2 UbcH10, and ubiquitin. APC/CCdc20 ubiquitination activity was then measured by the increase of multiubiquitinated cyclin B or by the reduction of unubiquitinated cyclin B (Fig. S1F,G). While full length wild type Mad2 alone produced no inhibition of Cdc20’s ability to activate APC/C (Fig. S1G), addition of Bub3 and full length BubR1 (hereafter BubR1FL) along with Cdc20 significantly reduced subsequent APC/C-mediated cyclin B ubiquitination in a dose dependent manner (Fig. S1G top, lanes 3–6; Fig. S1H, red triangles).
Synergistic inhibition of Cdc20 activation of APC/C was produced by co-addition of BubR1FL-Bub3 and Mad2 (Fig. S1G top, lanes 11–14; Fig. S1H, blue squares). Assay of Bub3 and a BubR1 variant containing only BubR1’s internal Cdc20 binding site (BubR1357-1050, to be referred to hereafter as BubR1C) produced a similar level of inhibition of cyclin B ubiquitination as did a comparable amount of Bub3-BubR1FL, but in the absence of the Mad3-homology region Cdc20 binding site inhibition was not enhanced by co-incubation with Mad2 (Fig. S1G bottom; Fig. S1J). By contrast, BubR11-477 (carrying the Mad3-homology region Cdc20 binding domain and to be referred to hereafter as BubR1N) had no APC/CCdc20 inhibitory activity when complexed with Bub3 (Fig. S1G middle, lanes 3–6; Fig. S1I, red triangles), but inhibition was strongly enhanced by addition of an equivalent amount of Mad2 (Fig. S1G middle, lanes 11–14; Fig. 1I, blue squares), which again had no inhibitory activity by itself. Moreover, BubR1N-Bub3 almost completely inhibited already active APC/CCdc20 when Mad2 was present (Fig. 1G). In contrast, BubR1C-Bub3 produced minimal APC/CCdc20 inhibition regardless of the presence of Mad2 (Fig. 1G,H). Thus, the N-terminal, but not the internal, Cdc20 binding domain of BubR1 can mediate Mad2-dependent inhibition of APC/CCdc20, reproducing the in vivo situation (Fig. 1C,D).
A conformational change in Mad2 is rate limiting for inhibition of APC/CCdc20 by Mad2 and BubR1N
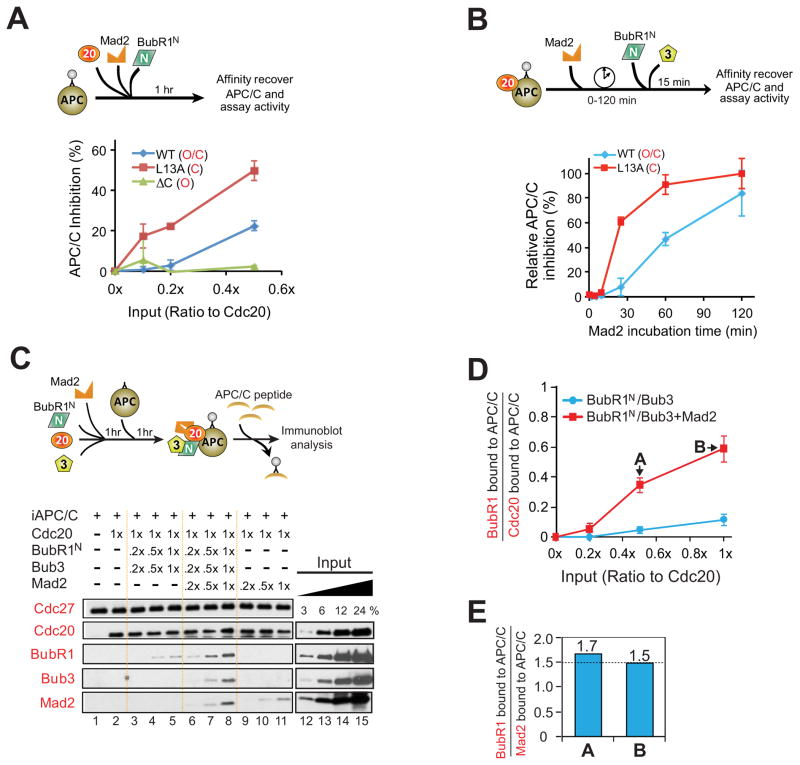
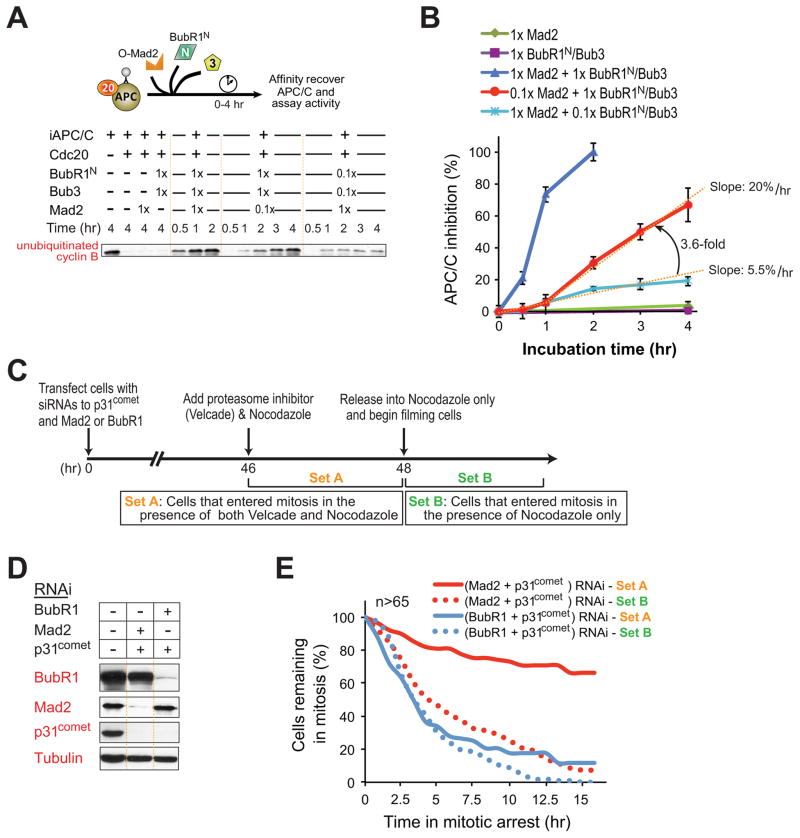
To determine how Mad2 facilitated BubR1N-dependent inhibition of APC/CCdc20, we compared inhibition produced by wild type Mad2, which exists in an open conformation that only slowly spontaneously converts to a closed form (Yang et al., 2008), with two Mad2 mutants: closed Mad2 (Mad2L13A) (Yang et al., 2007), and open Mad2 (Mad2 C) (Luo et al., 2000) (Fig. S2A). While closed Mad2L13A alone (Fig. S2D,E) had no APC/C inhibitory activity when used at equal stoichiometry with Cdc20, it was much more potent than wild type Mad2 in promoting BubR1N-mediated APC/C inhibition, producing comparable inhibition at concentrations 5 times lower than needed for wild type Mad2 (Fig. 2A; Fig. S2B). In contrast, BubR1N produced no inhibition of APC/C in the presence of open Mad2 C (Fig. 2A; Fig. S2B).
Figure 2. A conformational change in Mad2 facilitates BubR1N’s ability to inhibit Cdc20-mediated activation of APC/C.
(A) (Top) Increasing amounts of BubR1N, and Mad2 (L13A, WT, and ΔC) were incubated with Cdc20 and APC/C. APC/C was affinity-purified and assayed for activity. (Bottom) Plot of percent APC/C inhibition. Error bars represent SEM, n=3. (B) Testing how Mad2 conformation affects inhibition of preactivated APC/CCdc20 by BubR1N and wild type Mad2 or a Mad2 variant locked in the closed conformation (Mad2L13A). Data represent mean ± SEM (n=3). (C) (Top) Schematic for complexes bound to APC/CCdc20. (Bottom) Mad2 dependence for BubR1N binding to APC/CCdc20. (D) The molar ratio of BubR1N bound to APC/C compared with Cdc20 bound APC/C from lanes 3–8 in (C) was measured against a dilution series of purified proteins. Plot displays the mean from two independent assays. Error bars represent SEM. (E) Stoichiometry of BubR1 and Mad2 assembled into Cdc20 complexes (at the added stoichiometries denoted A and B in (D). (see also Fig. S2).
In light of our evidence for catalytic production of a Mad2-Cdc20 inhibitor in the presence unattached kinetochores containing the Mad1/Mad2 complex (Kulukian et al., 2009), we tested if addition of closed Mad2 - the proposed product of immobilized Mad1-Mad2 at kinetochores (De Antoni et al., 2005; Kulukian et al., 2009; Luo et al., 2002) - accelerated Mad2-dependent BubR1N inhibition of APC/CCdc20. In the absence of BubR1N-Bub3, even a stoichiometric level (relative to Cdc20) of closed Mad2L13A did not mediate any inhibition of cyclin B ubiquitination by APC/CCdc20 (Fig. S2D,E). Closed Mad2 (Mad2L13A) was much more potent than wild type Mad2 in promoting BubR1N-mediated APC/C inhibition, producing an initial rate of inhibition 8 times faster than wild type Mad2 and yielding much higher inhibition at early times (e.g., at 25 min. there was ~60% inhibition from Mad2L13A vs. only 8% inhibition with Mad2WT) (Fig. 2B; Fig. S2C). Thus, the rate of formation of the APC/CCdc20 inhibitor by BubR1N-Bub3 depended on the concentration of closed Mad2, consistent with kinetochore-catalyzed conformational change in Mad2 as an initiating step in mitotic checkpoint signaling.
The Mad2-dependent complex of BubR1N-APC/CCdc20 contains substoichiometric Mad2
To identify the mechanism by which Mad2 stimulated BubR1-dependent inhibition of APC/CCdc20, we affinity purified APC/C after incubation with BubR1N, Bub3, Mad2, and Cdc20. In the absence of Mad2, only a trace level of BubR1N-Bub3 associated with APC/CCdc20. This association was increased ~6 fold by co-incubation with Mad2 (yielding 0.6 molecules of BubR1 bound per molecule of Cdc20) (Fig. 2C, Fig. 2D). Under the same conditions, cyclin B ubiquitination by APC/CCdc20 was reduced by a comparable percentage (Fig. 1H), consistent with inhibition of APC/CCdc20 activity by stoichiometric binding of BubR1N. Although Mad2 facilitated BubR1N association with APC/CCdc20, the level of Mad2 bound to the produced APC/CCdc20 complex was lower than that of BubR1N (Fig. 2E).
Mad2 enables BubR1 binding to APC/CCdc20 without necessarily remaining bound
While a consensus has emerged that the initiating event in mitotic checkpoint signaling is production by unattached kinetochores of Mad2-Cdc20, widely divergent levels of Mad2 have been reported (Kulukian et al., 2009; Nilsson et al., 2008; Sudakin et al., 2001; Westhorpe et al., 2011) or predicted (Chao et al., 2012) in the mitotic checkpoint complexes, leaving the molecular identity of the checkpoint-produced inhibitor of APC/CCdc20 a central unresolved question. Moreover, although Mad2-stimulated BubR1N-Cdc20 binding was increased at higher Mad2 levels (Fig. S2F), the level of Mad2 bound to APC/CCdc20 in the final complex was lower than the level of BubR1N (Fig. 2E). Consistent with this, immunopurified Cdc20 complexes from nocodazole-arrested mitotic cells contained 3-fold more BubR1 than Mad2 (Fig. S2 G,H), suggesting dissociation of Mad2 from Cdc20 after enabling initial binding of BubR1N to Cdc20.
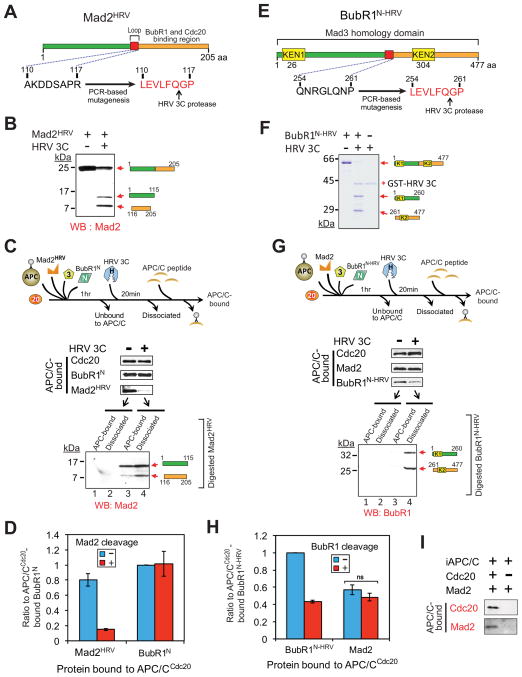
To determine if continued Mad2-Cdc20 association is required for maintaining a BubR1 association with APC/CCdc20, we created a Mad2 mutant (hereafter Mad2HRV) that can be removed from an initial APC/CCdc20 complex through cleavage by the sequence-specific HRV 3C (human rhinovirus 3C) protease (Cordingley et al., 1990) (Fig. 3A). Upon incubation with HRV 3C protease, the 24 kD Mad2HRV was cleaved into 15 kD and 9 kD fragments (Fig. 3B). The digested Mad2HRV fragments were nonfunctional as inhibitors of APC/CCdc20 (Fig. S3). APC/C was incubated with Cdc20, BubR1N, and Bub3 (at 1:1:1 stoichiometries) and a 5-fold excess of Mad2HRV, a concentration that produces similar levels of BubR1N and Mad2 bound to Cdc20 (Fig. 3D). Protease treatment reduced Mad2HRV-bound to APC/CCdc20 by 80% (Fig. 3C,D), with most of the C-terminal Mad2HRV fragment [containing residues required for binding to Cdc20 and Mad3 (the yeast homolog of BubR1) (Chao et al., 2012)] dissociated from APC/CCdc20 (Fig. 3C, lanes 3,4). Importantly, the level of BubR1N bound to Cdc20 remained almost unchanged despite a > 5 fold reduction of Cdc20-bound Mad2 (Fig. 3C,D). Taken together, these findings demonstrate that Mad2 stimulates the initial BubR1N-Cdc20 interaction, but is dispensable for maintaining this association.
Figure 3. Maintaining the association of BubR1N with Cdc20 does not require the Mad2-Cdc20 interaction.
(A) Schematic of a Mad2 variant modified to contain a HRV 3C cleavage site. (B) Immunoblot showing HRV 3C-mediated cleavage of Mad2HRV in vitro. Mad2HRV was incubated at room temperature for 20 min in the presence or absence of HRV 3C protease. (C) After incubation with BubR1N, Bub3, Mad2HRV and Cdc20, APC/C was affinity-purified to remove unbound components. Following incubation with or without HRV 3C protease, dissociated components were washed away and APC/C was peptide-eluted from beads and analyzed for associated components by immunoblotting. The supernatant from the protease digestion mixture was collected and analyzed for release of the digested Mad2HRV fragments. (D) The amount of APC/C-bound BubR1N or Mad2HRV in (C) was measured. Values are plotted as the molar ratio of APC/C-bound Mad2HRV compared with APC/C-bound BubR1N. The level of APC/C-bound BubR1N without protease treatment was set to 1. Bars represent the mean. Error bars represent SEM (n=3). (E) Schematic of an HRV 3C cleavable variant of BubR1N. (F) HRV 3C-mediated digestion of the cleavable BubR1N in vitro. Cleavable BubR1N was incubated at room temperature for 20 min in the presence or absence of HRV 3C prior to analysis. (G–H) Effect of forced BubR1N release on the association of Mad2 with APC/CCdc20. (G) Experimental procedure is the same as in (C). (H) Values were plotted as the molar ratio compared with APC/C-bound BubR1N-HRV. The level of APC/C-bound cleavable BubR1N without the protease treatment was set to 1. Bars represent the mean. Error bars represent SEM (n=3). ***=P<0.0001. (I) Assay of Mad2 binding to APC/C in the presence and absence of Cdc20 (see also Fig. S3).
BubR1, not Mad2, binds to Cdc20 in the final APC/CCdc20 inhibitor
To test if BubR1N is a bona fide APC/CCdc20 inhibitor we created a BubR1 mutant (hereafter BubR1N-HRV) that contains the recognition sequence for HRV 3C protease between amino acids 254–261. Incubation of purified BubR1N-HRV with protease produced 34 kD and 26 kD fragments, respectively (Fig. 3F). Bead-immobilized APC/C was again incubated with Cdc20, BubR1N-HRV, and Bub3 (at 1:1:1 stoichiometries) and a 5-fold excess of Mad2. Sixty percent of APC/C BubR1N-HRV was cleaved by protease addition, with all of the cleavage products released from the APC/C complexes (Fig. 3G), but this did not affect the amount of Mad2 retained (Fig. 3G, H). Additionally, since Mad2 association with APC/C was completely dependent on the presence of Cdc20 (Fig. 3I), the Mad2 remaining in the BubR1-depleted complexes must be bound to Cdc20 and the Mad2-Cdc20 interaction does not depend on the BubR1-Cdc20 interaction.
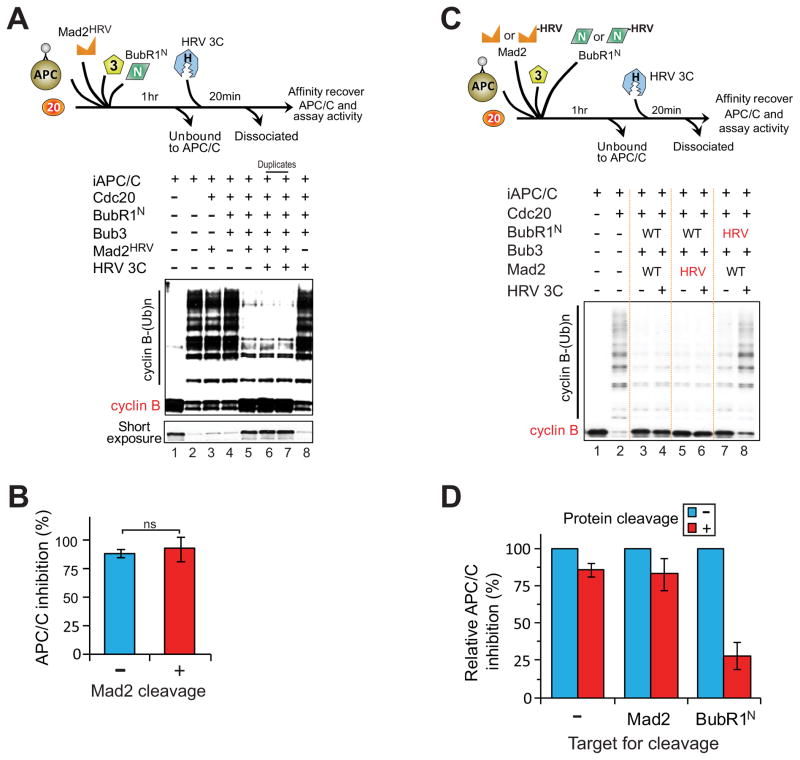
To address the individual function of Mad2 and BubR1N in inhibiting activity of APC/CCdc20, the inhibitor-bound APC/C complex containing BubR1N-HRV or Mad2HRV was incubated with the HRV 3C protease. Nearly quantitative digestion of Mad2 did not significantly alter APC/C inhibition (Fig. 4A–D). Digestion of BubR1N-HRV, on the other hand, reduced APC/C inhibition by 70% (Fig. 4C, lanes 7,8; Fig. 4D right set), with the residual 30% inhibition of APC/C attributable to the incomplete digestion of the BubR1N-HRV (Fig. 3F,G). Similarly, digestion of BubR1FL-HRV (full-length BubR1 that contains the recognition sequence for HRV 3C protease) bound to APC/CCdc20 significantly reduced APC/C inhibition (Fig. S4). Taken together, our results demonstrate that in vitro BubR1, but not Mad2, is the effective inhibitor of APC/CCdc20 and in complexes containing both BubR1 and Mad2, it is Cdc20 binding to BubR1 which inhibits recognition of cyclin B by APC/CCdc20.
Figure 4. Mad3 homology domain of BubR1, but not Mad2, is the effector for APC/CCdc20 inhibition in vitro.
(A–B) Testing if Mad2-binding to Cdc20 is required for APC/CCdc20 inhibition. (A) Bead-bound APC/C was incubated with combinations of BubR1N, Bub3, Mad2HRV, and Cdc20 followed by removal of components unbound to APC/C. Buffer either containing HRV 3C protease or not was added for 20 min prior to APC/C activity assay. (B) Graph quantifying percent inhibition of APC/C with or without protease incubation in (A). Bars represent the mean ± SEM (n=3). ns=not significant. (C) Assay to determine whether BubR1N or Mad2 is an effector for APC/CCdc20 inhibition after HRV 3C protease-mediated digestion and dissociation of BubR1N-HRV or Mad2HRV from inhibited APC/CCdc20. (D) Quantification of inhibition of APC/C activity in (C). Bars represent mean ± SEM (n=3) (see also Fig. S4).
BubR1, not Mad2, is the mitotic checkpoint inhibitor in vivo
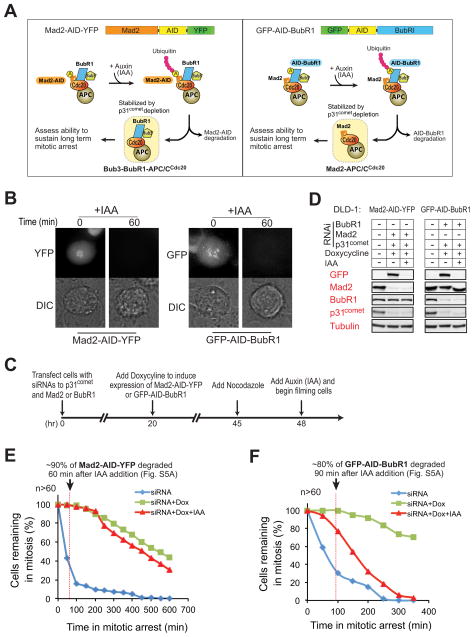
To test the in vivo requirement of BubR1 and Mad2 for inhibition of APC/CCdc20, we took advantage of a recently described auxin inducible degradation (AID) system (Holland et al., 2012; Nishimura et al., 2009). We established stable cell lines expressing doxycycline-inducible and RNAi-resistant GFP-AID-tagged BubR1 or AID-YFP-tagged Mad2 (Fig. 5A). Addition of the auxin derivative IAA to cells arrested in mitosis induced rapid destruction of both AID-tagged BubR1 or Mad2 with t1/2 of 15 and 30 min, respectively, while proteins associated with either remained stable (Figs. 5B and S5A). Suppression of endogenous BubR1 or Mad2 by transfection of appropriate siRNAs and doxycycline-induction led to their replacement with GFP-AID-BubR1 or Mad2-AID-YFP (Fig. 5D).
Figure 5. BubR1, not Mad2, is required to sustain inhibition of APC/CCdc20 in vivo.
(A) Schematic for the inducible destruction of Mad2-AID-YFP or GFP-AID-BubR1 in mitotic cells using an Auxin-inducible protein degradation system. (B) Microscopic images showing Auxin (indoleacetic acid, IAA)-induced mitotic degradation of Mad2-AID-YFP (left) or GFP-AID-BubR1 (right). (C) Schematic of the protocol used to deplete endogenous p31comet and replace Mad2 or BubR1 with an AID-tagged version. Degradation of AID-tagged proteins was induced with IAA (500 μM) and cells monitored by time-lapse microscopy. (D) Immunoblot showing the levels of various proteins before and after IAA-induced degradation of (left) Mad2-AID-YFP or (right) GFP-AID-BubR1. (E–F) Time-lapse microscopy was used to determine the duration of mitosis in nocodazole (100 ng/ml) treated cells after 1) depletion of endogenous Mad2 (E, blue) or BubR1 (F, blue), 2) replacement of endogenous Mad2 or BubR1 with Mad2-AID-YFP (E, green) or GFP-AID-BubR1 (F, green) and 3) induced destruction of Mad2-AID-YFP (E, red) or GFP-AID-BubR1 (F, red). Cells were treated with nocodazole prior to filming to facilitate accumulation of the mitotic checkpoint inhibitor(s). More than 60 cells were analyzed for each condition over three independent experiments (see also Fig. S5).
Cdc20 has been shown to undergo autoubiquitination/turnover during mitosis (King et al., 2007; Nilsson et al., 2008; Pan and Chen, 2004; Reddy et al., 2007; Varetti et al., 2011) leading to disassembly of the Cdc20-associated checkpoint inhibitor(s). Since the rapid turnover of checkpoint inhibitor(s) in vivo would preclude identifying whether Mad2 or BubR1 is the final inhibitor(s) of APC/CCdc20, we therefore co-depleted p31comet to stabilize Cdc20 inhibitor(s) (Jia et al., 2011; Mapelli et al., 2006; Reddy et al., 2007; Teichner et al., 2011; Varetti et al., 2011; Westhorpe et al., 2011). Following checkpoint activation and IAA-induced degradation of Mad2-AID-YFP or GFP-AID-BubR1, the duration of mitotic checkpoint-mediated arrest was determined by time-lapse microscopy. Addition of IAA to mitotically arrested Mad2-AID-YFP expressing cells induced destruction of 90% of Mad2-AID-YFP within 60 min, but remarkably all cells remained arrested in mitosis (Fig. 5E, red). Indeed, after Mad2-AID-YFP degradation, 60% of cells sustained a mitotic arrest for > 6 hours, similar to the duration observed in cells not treated with IAA and in which Mad2-AID-YFP remained stable (Fig. 5E, green). Mad2 was essential for initiating this sustained arrest, however, as destruction of Mad2-AID-YFP by IAA addition prior to mitotic entry in the presence of nocodazole lead to rapid mitotic exit (Fig. S5B,C). By contrast, IAA-induced destruction of GFP-AID-BubR1 not only disrupted extended mitotic arrest, but provoked mitotic exit before BubR1 was completely degraded (Fig. 5F, red). Taken together, as was seen in our in vitro observations (Fig. 4), these efforts demonstrate that under conditions where disassembly of mitotic checkpoint complexes is dampened (by depletion of p31comet), BubR1, not Mad2, is required to sustain inhibition of APC/CCdc20.
Mad2 acts catalytically to produce an APC/CCdc20 inhibitor containing BubR1, but not itself
We next tested if a single molecule of Mad2 is capable of facilitating the production of multiple BubR1-APC/CCdc20 complexes. As expected, at stoichiometric levels of Mad2, BubR1N-mediated inhibition of APC/CCdc20 increased in a time dependent manner, reaching 100% inhibition within 2 hours (Fig. 6A,B). At concentrations of Mad2 ten-fold below that of Cdc20 or BubR1, APC/CCdc20 inhibition also increased over time, but at a slower rate, and with bi-phasic kinetics, reaching ~70% inhibition by 4 hours. The initial rate of APC/CCdc20 inhibition was slow (5.5% inhibition of APC/CCdc20 per hour), but the rate of inhibitor formation in the presence of a substoichiometric level of Mad2 accelerated (by 3.6 fold) by the end of one hour (to 20% inhibition of APC/CCdc20 per hour). In contrast, addition of a substoichiometric level of BubR1N-Bub3 mediated inhibition of APC/CCdc20 activity only in proportion to the level of added BubR1, indicating the inhibition was limited by the available BubR1 (Fig. 6B, light blue line). \
Figure 6. Mad2 catalytically amplifies formation of the final BubR1-APC/CCdc20 inhibitor.
(A) The indicated amounts of Mad2, Bub3, and BubR1N were incubated with preassembled APC/CCdc20 and at various time points assayed for the inhibition of APC/C-mediated ubiquitination of cyclin B1-102. (B) APC/CCdc20 inhibition versus time for the assays denoted in (A). Data represent mean ± SEM (n=3). (C) Schematic of the protocol used to reduce endogenous p31comet and Mad2 or BubR1 levels by siRNA transfection, followed by inhibition and release with the 26S proteasome inhibitor (with 25 nM Velcade) and microtubule inhibitor nocodazole (100 ng/ml), so as to provide a temporal delay in mitotic exit and allow accumulation of mitotic checkpoint inhibitor(s), respectively. (D) Levels of depletion of p31comet, Mad2 or BubR1 by siRNA transfection. (E) Duration of mitosis determined by time-lapse microscopy for cells treated as in b. Filming began after removal of Velcade and release into nocodaozle. Cells counted in Set A entered mitosis in the presence of both nocodazole and proteasome inhibition, whereas in Set B only cells that entered mitosis after removal of the proteasome inhibitor were included. More than 65 cells were analyzed for each condition.
We further tested in an in vivo context (schematic in Fig. 6C) this model in which Mad2 and BubR1 play discrete roles as a catalytic loader and an effector for Cdc20 inhibition, respectively, with the accelerating rate of APC/CCdc20 inhibition in vitro reflecting Mad2 dissociation from Cdc20 in a conformation that can rebind another molecule of Cdc20. To do this, using siRNAs, cells were first depleted by ~90% for either BubR1 or Mad2 (Fig. 6D). To stabilize accumulated Cdc20 inhibitor(s), transfection of siRNA was used to co-deplete p31comet. Nocodazole was added to sustain activation of the mitotic checkpoint and a 26S proteasome inhibitor was added to inhibit cyclin B destruction, thereby allowing the accumulation of mitotic checkpoint inhibitor(s). After 2 hours, the proteasome inhibitor was removed and the duration of remaining mitotic arrest was determined by time-lapse microscopy.
In cells with normal levels of Mad2 but with ~10% the normal BubR1 level, washout of the proteasome inhibitor yielded mitotic exit, with >60% of cells exiting mitosis within 4 hours (Fig. 6E, blue solid line). By contrast, cells in which Mad2 levels were reduced to ~10% the normal level sustained a mitotic arrest for 16 hr after washout of the proteasome inhibitor (Fig. 6E, red solid line). The normal cellular stoichiometries of BubR1 and Cdc20 have been determined to be 1:1, with Mad2 between 1.3 (Tang et al., 2001) and 3 (Nilsson et al., 2008) fold higher. Thus, a 90% depletion of Mad2 reduces it to a level between 13 and 30% that of Cdc20. Nevertheless, our evidence demonstrates this limited level of Mad2 can sustain long-term checkpoint signaling if a sufficient time window is provided to allow a BubR1-containing mitotic-checkpoint inhibitor to accumulate, consistent with a model in which Mad2 acts catalytically to load BubR1 onto Cdc20.
Mad2 binding induces a functional switch in Cdc20 enabling BubR1 binding
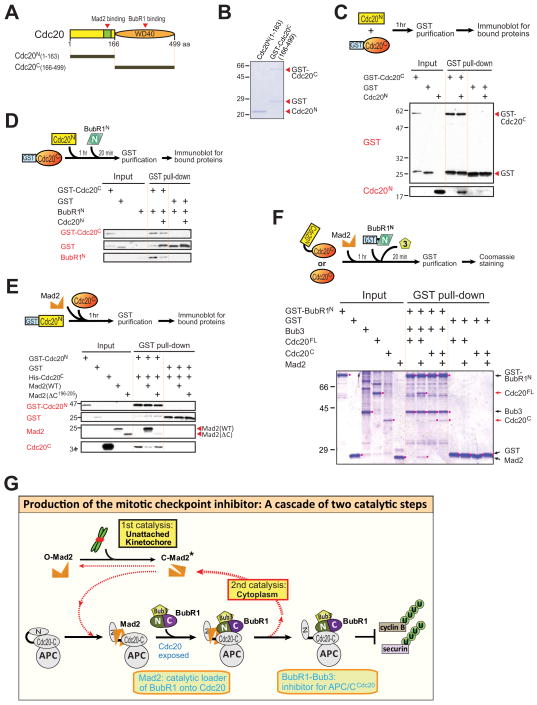
Our finding that APC/C-bound Cdc20 association with BubR1N is significantly enhanced by its prior binding to Mad2 (Figs. 2D, S2F), along with a previous yeast two-hybrid study (Davenport et al., 2006), suggested a model in which the BubR1 binding site within the C-terminal WD40 repeats of Cdc20 may be initially inaccessible as the result of self-association of the Cdc20 amino and carboxy-terminal domains. To test this directly, we expressed and purified Cdc201-163 (hereafter called Cdc20N) and GST-Cdc20166-499 (to be referred to as GST-Cdc20C) (Fig. 7A,B). After mixing Cdc20N and Cdc20C and purification by glutathione affinity, GST-Cdc20C, but not GST, co-purified Cdc20N (Fig. 7C), demonstrating a direct association between the two domains. Moreover, pre-incubation of Cdc20N with Cdc20C reduced subsequent Cdc20C association with BubR1N (Fig. 7D), consistent with blocking BubR1 binding through an intra-molecular interaction between two domains in Cdc20FL.
Figure 7. Mad2 binding unlocks Cdc20 to allow further binding of BubR1.
(A) Schematic of human Cdc20, showing the location of the N-terminal Mad2 binding domain and BubR1 binding WD40 repeat domain. (B) Purified recombinant Cdc20 fragments, assessed by Coomassie staining. (C) GST-Cdc20C or GST was affinity purified after incubation with Cdc20N for 1 hr at room temperature. GST proteins and associated Cdc20C were analyzed by immunoblotting. (D) GST-Cdc20C or GST was incubated with Cdc20N for 1 hr and then further incubated with BubR11-363 for 20 min at room temperature before affinity purification. Associated proteins were analyzed by immunoblotting. (E) GST-Cdc20N or GST was incubated with Cdc20C for 1 hr at room temperature in the presence of Mad2WT or Mad2 C. Associated Mad2 or Cdc20C was analyzed by immunoblotting. (F) GST-BubR1N or GST was affinity purified after incubation with Cdc20FL or Cdc20C that had been pre-incubated with Mad2 or buffer and analyzed for associated Cdc20 and Mad2 by immunoblotting. (G) Model for the production of the mitotic checkpoint through a cascade of two catalytic steps. Kinetochores generate a conformation change in Mad2, converting it from an open (O-Mad2) to a closed (C-Mad2) form. Binding of C-Mad2 to Cdc20 exposes Cdc20 for binding to BubR1 (1st catalytic step). BubR1, but not Mad2, functions as an inhibitor for APC/CCdc20. Following loading of BubR1 onto Cdc20, Mad2 dissociates in a partially active “closed” conformation (C-Mad2*), which then either binds another free Cdc20 molecule to produce the BubR1-Cdc20 inhibitor (2nd catalytic step) or converts back to O-Mad2 and is used again in kinetochore signaling (1st catalytic step). Thus one Mad2 molecule produces multiple BubR1-Cdc20 inhibitors through two interlocking catalytic steps.
Further, addition of wild type Mad2, but not the Cdc20-binding defective Mad2ΔC, to GST-Cdc20N and Cdc20C reduced association of the two Cdc20 domains (Fig. 7E). Most importantly, while BubR1N did not bind to Cdc20FL, Mad2 addition enabled this interaction, producing BubR1N-Cdc20FL-Mad2 complexes (Fig. 7F). By contrast, BubR1N bound Cdc20C directly and this interaction was unaffected by Mad2 addition. Thus, initial Mad2 binding to the N-terminus of Cdc20FL removed the inhibition of Cdc20 from binding to BubR1N, with only a substoichiometric amount of Mad2 remaining in the final Cdc20FL-BubR1N-Bub3 complex.
DISCUSSION
The initiating feature of the checkpoint signaling pathway is a first catalytic step at unattached kinetochores where immobilized heterodimers of Mad1-Mad2 act to catalyze a conformational change in inactive Mad2, producing an activated “closed” form (Kulukian et al., 2009). To this, our findings here have identified the mitotic checkpoint inhibitor to be produced by synergistic cooperation between Mad2 and BubR1. Mad2 binding reduces the affinity of the amino and carboxy-terminal Cdc20 domains for each other, exposing a previously poorly accessible BubR1 binding site for Cdc20. This Mad2-dependent functional priming in Cdc20 is essential for BubR1’s N-terminus to produce the BubR1-Cdc20 mitotic checkpoint inhibitor (Fig. 7C–F; modeled in Fig. 7G). BubR1-binding to Cdc20 has a key feature required in a bona fide anaphase inhibitor: the ability to inhibit activated APC/CCdc20. Added to this, an affinity of Mad2 in a closed conformation for BubR1 (Tipton et al., 2011) [or Mad3 (Chao et al., 2012)] could facilitate BubR1 recruitment to kinetochore-produced Mad2-Cdc20.
Our model for BubR1-bound to Cdc20 as the true mitotic checkpoint-derived inhibitor that blocks APC/C recognition of cyclin B is well supported by 1) the in vitro production of a Mad2-dependent inhibitory APC/CCdc20-BubR1-Bub3 complex containing little Mad2 (Kulukian et al., 2009), 2) long term production of a functional mitotic checkpoint inhibitor in cells depleted of most Mad2 (our evidence), 3) continued inhibition of APC/CCdc20 in mitotically arrested cells following in vivo targeted destruction of Mad2 (our evidence), and 4) the purification of a stable BubR1-Bub3-Cdc20 complex (free or APC/C-bound) containing little Mad2 from cells chronically mitotic checkpoint arrested (Kulukian et al., 2009; Nilsson et al., 2008). This model is also supported by prior works that have consistently shown Mad2 to have little ability to inhibit Cdc20 activation of APC/C at physiologically relevant concentrations (Fang, 2002; Kulukian et al., 2009; Sudakin et al., 2001).
Perhaps most importantly, we have shown that Mad2 released from the final inhibitory BubR1-Cdc20 complex can act catalytically to facilitate loading additional BubR1 molecules into inhibitory complexes with Cdc20 (Fig. 6). We propose this previously unappreciated catalytic step provides a means to cytosolically amplify initial kinetochore-derived Mad2-Cdc20 mitotic checkpoint complexes (Fig. 7G), thereby allowing a single unattached kinetochore to globally inhibit APC/CCdc20 activity. This model contrasts in a crucial way from a previous one that had proposed that cytosolic Cdc20-Mad2 complexes would amplify the number of Cdc20-Mad2 complexes, with each such complex in turn acting as an additional structural template (similar to unattached kinetochore-bound Mad1/Mad2) to promote the conversion of further open Mad2 molecule into Cdc20- bound closed Mad2 (De Antoni et al., 2005). This latter model had been challenged (Doncic et al., 2005; Mariani et al., 2012), including on the grounds that once initiated the proposed amplification loop (amplifying the number of activated Mad2 molecules – just as the kinetochore does) would obligatorily continue independently of the kinetochore-derived signal and could not be silenced following attachment of the kinetochores to spindle microtubules. Our model, on the other hand, remains responsive to kinetochore-derived signaling does not amplify the number of catalysts (i.e., activated, closed Mad2); instead, kinetochore-derived closed Mad2 is “reused” for two or more cycles, thereby providing an elevated number of APC/CCdc20 inhibited by bound BubR1.
Finally, rather than a stable component of the APC/CCdc20 inhibitory complex, Mad2’s continued association with APC/CCdc20-BubR1-Bub3 may instead contribute to the pathway of mitotic checkpoint silencing, where p31comet has been proposed to facilitate dissociation of Mad2 and/or BubR1 from Cdc20 (Chao et al., 2012; Jia et al., 2011; Reddy et al., 2007; Teichner et al., 2011; Westhorpe et al., 2011) or destabilize Cdc20 bound to BubR1 and/or Mad2 (Varetti et al., 2011). Since our results have established that Mad2 dissociation from Cdc20 leaves a stable BubR1-Cdc20 interaction that continues to inhibit Cdc20 even after Mad2 release (Fig. 3,4), our evidence makes it highly unlikely that Mad2 extraction from the inhibitory Mad2-BubR1-Bub3-Cdc20 complex by p31comet is a key step for checkpoint silencing. Instead, it is BubR1 dissociation from Cdc20 that is critical to reactivate APC/CCdc20 for cyclin B recognition. While many aspects of mitotic checkpoint inactivation and p31comet activity remain unresolved, we note that release from APC/CCdc20 inhibition could be achieved by stimulating deacetylation-derived BubR1 degradation (Choi et al., 2009) or action of some checkpoint protein(s) (including p31comet) stimulating Cdc20 degradation (Nilsson et al., 2008; Varetti et al., 2011), thereby facilitating generation of active APC/C upon binding of newly synthesized Cdc20.
EXPERIMENTAL PROCEDURES
Constructs
Full-length and fragments of the human BubR1 open reading frame were cloned into either a pcDNA5/FRT/TO-based vector (Invitrogen) modified to contain an amino-terminal Myc-LAP epitope tag for mammalian cell expression or a pFastBac1-based vector (Invitrogen) modified to contain an amino-terminal GST-HRV 3C site for insect cell expression. The LAP tag consists of GFP-HRV 3C-6xHis BubR1HRV and Mad2HRV mutants were generated using PCR-based site-directed mutagenesis to insert a HRV 3C site (LEVLFQGP) (QuikChange, Stratagene). For tetracycline-inducible expression of GFP-AID-BubR1 and Mad2-AID-YFP the corresponding genes were cloned into a pcDNA5/FRT/TO-based vector (Invitrogen). All other DNA constructs were previously described (Kulukian et al., 2009; Tang et al., 2001).
Generation of Stable Cell Lines and siRNA Treatment
Parental Flp-In™ TRex™-HeLa parental cells that stably express mRFP-tagged histone H2B (H2B-mRFP) were as previously described (Gassmann et al., 2010). Stable, isogenic cell lines expressing MycGFP-BubR1 were generated using FRT/Flp-mediated recombination (Tighe et al., 2004). Expression of MycGFP-BubR1 was induced with 1 μg/ml tetracycline. siRNA directed against 3′ untranslated region of BubR1 (5′-CUGUAUGUGCUGUAAUUUA-3′ [Figs. 1, 6]) or GAPDH (5′-UGGUUUACAUGAUCC-AAUA-3′) were purchased from Thermo Fisher Scientific. Cells were transfected with 50 nM of oligonucleotides using Lipofectamine RNAiMAX (Invitrogen). 14 h after transfection tetracycline was added to express MycGFP-BubR1 for 24 h before collecting cells for immunoblotting or analyzing by time-lapse microscopy. For IAA-inducible protein destruction TIR1-9Myc was introduced into Flp-In™ TRex™-DLD-1 parental cells (a kind gift of Stephen Taylor) using retroviral delivery (Shah et al., 2004). Stable integrates were selected in 2 μg/ml puromycin and single clones isolated using single cell sorting (FACS-Vantage; Becton Dickinson). Stable, isogenic cell lines expressing Mad2-AID-YFP or GFP-AID-BubR1 were generated using the FRT/Flp-mediated recombination and transgenes induced with tetracycline. To-induce protein destruction cells were treated with 500 μM of IAA. siRNA directed against 3′ untranslated region of BubR1 (5′-CUAAACAGACUCAUUACAA-3′ [Fig. 5]) or Mad2 (5′-GGAAGAGUCGGGACCACAG-3′ [Figs. 1,5,6]) or p31comet (5′-AGTGGTATGA GAAGTCCGAAG-3′ [Figs. 5,6]) was used to deplete endogenous protein.
Live-Cell Microscopy
To determine mitotic timing, cells were seeded onto poly-L-lysine coated coverglass chamber slides (Thermo Scientific) or poly-L-lysine coated 35 mm glass-bottomed tissue culture dishes (MatTek Inc) and 38–48 hr post transfection transferred to supplemented CO2 independent media (Invitrogen). Cells were maintained at 37°C in an environmental control station and images collected using a Deltavision RT system (Applied Precision) with a 40 × 1.35 NA oil lens at 5 min time intervals. For each time point, 6 × 3 μM z sections were acquired for RFP and maximum intensity projection created using softWoRx. Movies were assembled and analyzed using QuickTime (Apple) or Image J software.
Protein Purification
GST or His tagged human BubR1, Bub3 and Cdc20 were expressed in Sf9/High-Five insect cells using the Bac-to-Bac expression system (Invitrogen) and affinity purified over nickel-nitrilotriacetic acid beads (Qiagen) or glutathione sepharose beads (GE Healthcare Life Sciences). His-Mad21-102 and other GST-tagged proteins were expressed from Rosetta E.coli after induction with IPTG and purified (Kulukian et al., 2009). APC/C was immunoprecipitated from interphase Xenopus egg extracts as previously described (Kulukian et al., 2009).
APC/C Ubiquitination Activity Assay
The APC/C ubiquitination activity assay was performed as previously described (Tang and Yu, 2004) and activity assessed by ubiquitination-derived depletion of cyclin B1-102 substrate (Tugendreich et al., 1995). Quantitative analysis of cyclin B1-102 depletion was performed as previously described (Kulukian et al., 2009). Briefly, the level of cyclin B1-102 was determined against a series of dilution of the proteins.
APC/C Binding Assay
APC/C was immunoprecipitated from Xenopus interphase egg extracts for 2 h at 4°C using a peptide-derived anti-Cdc27 antibody crosslinked to Affiprep Protein A (Bio-Rad) beads. The APC/C beads were washed with TBS buffer supplemented with 0.4 M KCl and 0.1% Triton X-100 and incubated with Cdc20 and checkpoint proteins sequentially or simultaneously for the indicated time at room temperature. Unbound proteins were removed by washing the beads twice with 20 volumes of TBS buffer. APC/C complex was eluted from the beads by Cdc27 peptide competition as described (Herzog and Peters, 2005) and analyzed by immunoblotting.
In Vitro Binding Assay
In vitro binding assays were conducted in 50 mM Tris-HCl pH 7.7, 100 mM KCl, 0.1% Triton X-100, 10% glycerol, and 1 mM DTT. Glutathione Sepharose-bound GST-tagged protein and other recombinant proteins were combined and incubated at room temperature for 1 h. Bound protein complexes where washed four times with 20 volumes of the binding buffer, eluted from the beads in 15 mM glutathione buffer, and analyzed by immunoblotting.
Antibodies
The antibodies used in this study are as follows: BubR1 (SBR1.1, a gift from S. Taylor; A300-386, Bethyl Laboratories), Bub3 (SB3.2, a gift from S. Taylor), Mad1 (BB3-8, a gift from A. Musacchio), Mad2 (A300-300A, Bethyl Laboratories), Cdc20 (A301-180A, Bethyl Laboratories), p31comet (SC-134381, Santa Cruz Biotechnology), Myc (16–213, Millipore), Cdc27 (Herzog and Peters, 2005), α-Tubulin (DM1α, Sigma-Aldrich) His (A00186, GenScript), GST (SC-33613, Santa Cruz Biotechnology).
Supplementary Material
Research Highlights.
Mad2 binding induces a functional switch in Cdc20 that uncovers a BubR1 binding site
BubR1, not Mad2, is the mitotic checkpoint produced inhibitor of APC/CCdc20
Cytoplasmic amplification by Mad2 of the BubR1-Cdc20 mitotic checkpoint inhibitor
Dual catalytic steps by Mad2 produce the mitotic checkpoint inhibitor BubR1-Cdc20
Acknowledgments
We thank Stephen Taylor and Arshad Desai for providing reagents; Jennifer Meerloo of the University of California at San Diego Neuroscience Microscopy Shared Facility (NINDS P30 NS047101) for help with live-cell imaging; Weijie Lan for help with preparation for Xenopus egg extracts. This work was supported by grant GM20513 from the NIH to D.W.C. Salary for J.S.H and A.J.H was supported in part by a senior fellowship from the Leukemia and Lymphoma Society. Salary support for D.W.C is provided by the Ludwig Institute for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- Choi E, Choe H, Min J, Choi JY, Kim J, Lee H. BubR1 acetylation at prometaphase is required for modulating APC/C activity and timing of mitosis. EMBO J. 2009;28:2077–2089. doi: 10.1038/emboj.2009.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordingley MG, Callahan PL, Sardana VV, Garsky VM, Colonno RJ. Substrate requirements of human rhinovirus 3C protease for peptide cleavage in vitro. J Biol Chem. 1990;265:9062–9065. [PubMed] [Google Scholar]
- Davenport J, Harris LD, Goorha R. Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1. Exp Cell Res. 2006;312:1831–1842. doi: 10.1016/j.yexcr.2006.02.018. [DOI] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Doncic A, Ben-Jacob E, Barkai N. Evaluating putative mechanisms of the mitotic spindle checkpoint. Proc Natl Acad Sci USA. 2005;102:6332–6337. doi: 10.1073/pnas.0409142102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Holland AJ, Varma D, Wan X, Civril F, Cleveland DW, Oegema K, Salmon ED, Desai A. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010;24:957–971. doi: 10.1101/gad.1886810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F, Peters JM. Large-scale purification of the vertebrate anaphase-promoting complex/cyclosome. Methods Enzymol. 2005;398:175–195. doi: 10.1016/S0076-6879(05)98016-6. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Han JS, Cleveland DW. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci USA. 2012;109:e3350. doi: 10.1073/pnas.1216880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Izawa D, Pines J. Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation. J Cell Biol. 2012;199:27–37. doi: 10.1083/jcb.201205170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Li B, Warrington RT, Hao X, Wang S, Yu H. Defining pathways of spindle checkpoint silencing: functional redundancy between Cdc20 ubiquitination and p31(comet) Mol Biol Cell. 2011;22:4227–4235. doi: 10.1091/mbc.E11-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EM, van der Sar SJ, Hardwick KG. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS ONE. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Scott MI, Diez M, Sen O, Taylor SS. BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. J Cell Sci. 2011;124:4332–4345. doi: 10.1242/jcs.094763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22:R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Lau DT, Murray AW. Mad2 and Mad3 cooperate to arrest budding yeast in mitosis. Curr Biol. 2011;22:180–190. doi: 10.1016/j.cub.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Luo X, Fang G, Coldiron M, Lin Y, Yu H, Kirschner MW, Wagner G. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- Mapelli M, Filipp FV, Rancati G, Massimiliano L, Nezi L, Stier G, Hagan RS, Confalonieri S, Piatti S, Sattler M, et al. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 2006;25:1273–1284. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- Mariani L, Chiroli E, Nezi L, Muller H, Piatti S, Musacchio A, Ciliberto A. Role of the Mad2 dimerization interface in the spindle assembly checkpoint independent of kinetochores. Curr Biol. 2012;22:1900–1908. doi: 10.1016/j.cub.2012.08.028. [DOI] [PubMed] [Google Scholar]
- Nezi L, Rancati G, De Antoni A, Pasqualato S, Piatti S, Musacchio A. Accumulation of Mad2-Cdc20 complex during spindle checkpoint activation requires binding of open and closed conformers of Mad2 in Saccharomyces cerevisiae. J Cell Biol. 2006;174:39–51. doi: 10.1083/jcb.200602109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Pan J, Chen RH. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes Dev. 2004;18:1439–1451. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Sironi L, Mapelli M, Knapp S, De Antoni A, Jeang KT, Musacchio A. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 2001;20:6371–6382. doi: 10.1093/emboj/20.22.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- Tang Z, Yu H. Functional analysis of the spindle-checkpoint proteins using an in vitro ubiquitination assay. Methods Mol Biol. 2004;281:227–242. doi: 10.1385/1-59259-811-0:227. [DOI] [PubMed] [Google Scholar]
- Teichner A, Eytan E, Sitry-Shevah D, Miniowitz-Shemtov S, Dumin E, Gromis J, Hershko A. p31comet Promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc Natl Acad Sci USA. 2011;108:3187–3192. doi: 10.1073/pnas.1100023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe A, Johnson VL, Taylor SS. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J Cell Sci. 2004;117:6339–6353. doi: 10.1242/jcs.01556. [DOI] [PubMed] [Google Scholar]
- Tipton AR, Wang K, Link L, Bellizzi JJ, Huang H, Yen T, Liu ST. BUBR1 and Closed MAD2 (C-MAD2) Interact Directly to Assemble a Functional Mitotic Checkpoint Complex. J Biol Chem. 2011;286:21173–21179. doi: 10.1074/jbc.M111.238543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Mol Cell. 2011;44:710–720. doi: 10.1016/j.molcel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Vink M, Simonetta M, Transidico P, Ferrari K, Mapelli M, De Antoni A, Massimiliano L, Ciliberto A, Faretta M, Salmon ED, et al. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr Biol. 2006;16:755–766. doi: 10.1016/j.cub.2006.03.057. [DOI] [PubMed] [Google Scholar]
- Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J Cell Sci. 2011;124:3905–3916. doi: 10.1242/jcs.093286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Lan Z, Li W, Wu S, Weinstein J, Sakamoto KM, Dai W. p55CDC/hCDC20 is associated with BUBR1 and may be a downstream target of the spindle checkpoint kinase. Oncogene. 2000;19:4557–4562. doi: 10.1038/sj.onc.1203803. [DOI] [PubMed] [Google Scholar]
- Yang M, Li B, Liu CJ, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. Insights into mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric mad2 dimer. PLoS Biol. 2008;6:e50. doi: 10.1371/journal.pbio.0060050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li B, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131:744–755. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.