Abstract
Breast cancer is a heterogeneous tumor type characterized by a complex spectrum of molecular aberrations, resulting in a diverse array of malignant features and clinical outcomes. Deciphering the molecular mechanisms that fuel breast cancer development and act as determinants of aggressiveness is a primary need to improve patient management. Among other alterations, aberrant expression of microRNAs has been found in breast cancer and other human tumors, where they act as either oncogenes or tumor suppressors by virtue of their ability to finely modulate gene expression at the post-transcriptional level. In this study, we describe a new role for miR-181a/b as negative regulators of the DNA damage response in breast cancer, impacting on the expression and activity of the stress-sensor kinase ataxia telangiectasia mutated (ATM). We report that miR-181a and miR-181b were overexpressed in more aggressive breast cancers, and their expression correlates inversely with ATM levels. Moreover we demonstrate that deregulated expression of miR-181a/b determines the sensitivity of triple-negative breast cancer cells to the poly-ADP-ribose-polymerase1 (PARP1) inhibition. These evidences suggest that monitoring the expression of miR-181a/b could be helpful in tailoring more effective treatments based on inhibition of PARP1 in breast and other tumor types.
Keywords: breast cancer, microRNA, DNA damage response, ATM, BRCA1, BRCAness, PARP inhibitors
Introduction
The relevance of the DNA damage response (DDR) pathway in providing a cell-intrinsic barrier against cancer progression has clearly emerged in the last years. Experimental and clinical data indicated that DDR activation occurs at early stages of transformation as a consequence of oncogene deregulation, and bypassing its growth-suppressive outcomes (apoptosis or senescence)1 is required for cancer progression.2 Consequently, cancer cells are under positive selective pressure for DDR inactivation, as frequently observed in breast cancer, where inherited inactivating mutations of critical DDR components including ATM and the breast cancer susceptibility gene 1 and 2 (BRCA1/2) predispose to the development of hereditary breast carcinomas.3,4 In contrast, in sporadic breast cancers, which account for nearly 90% of all mammary tumors, ATM and BRCA1 mutations are detected in only 2% of cases (www.sanger.ac.uk/genetics/CGP/cosmic). Nonetheless, reduced expression and activity of BRCA1 and ATM are frequent events in sporadic breast tumors.5,6 This has been reported to occur as a consequence of either promoter methylation,7 deregulated transcriptional control8 or aberrant regulation by microRNAs (miRNAs).9-12 In particular, downregulation of ATM and/or BRCA1 has been frequently observed in more aggressive breast cancers, such as the Basal-like and triple-negative (TNBC, i.e., ER-/PR-/HER-2 tumors) breast cancers subtypes. These two groups of tumors show a high degree of overlap and frequently display a phenotype defined “BRCAness”13 that is characterized by traits similar to BRCA-mutated breast tumors, including lack of estrogen receptor, high grade, aggressiveness and frequent TP53 mutations.14 Despite this role in malignancies, the molecular basis of BRCAness is still largely unclear.
Filling this gap in knowledge would be of particular relevance from a therapeutic perspective, since deficiency in proteins involved in the DDR and in DNA double-strand break repair by homologous recombination (HR) is considered a major determinant of response to chemotherapy.15 For instance, ATM or BRCA1-deficient tumors display an extreme sensitivity to radiotherapy and chemotherapeutic agents (i.e., platinum-derivates),16 and a selective “synthetic lethal” effect can be achieved with the pharmacological inhibition of the DNA repair protein poly (ADP-Ribose) polymerase 1 (PARP1).17
In addition to genetic and epigenetic changes, aberrant post-transcriptional modulation of gene expression by miRNAs is emerging among major factors contributing to the unbalance of oncogenes and tumor suppressors in human cancers.18 miRNAs are small RNAs that finely regulate gene expression at the post-transcriptional level by interacting with the 3′UTR of their target transcripts through partial sequence complementarity,19 dampening mRNA translation or triggering its degradation.20 Several reports indicate that altered expression of specific microRNAs strongly contributes to tumorigenic hallmarks of breast cancer, including stemness,21 deregulated proliferation,22 genomic instability11 and metastatic potential,23 and recently it has been suggested that miRNAs directly targeting BRCA1 (e.g., miR-182 and miR-146) might be involved in establishing BRCAness traits.11,12
In this study, we highlight a role for miR-181a/b in determining the BRCAness phenotype in aggressive breast cancers. We demonstrate that miR-181a/b negatively impact on ATM levels and activity, dampen DDR thereby conferring to breast cancer cells highly expressing miR-181a/b a sensitivity to treatment with the PARP1 inhibitor Olaparib.
Results
Increased expression of miR-181a/b correlates with breast cancer aggressiveness
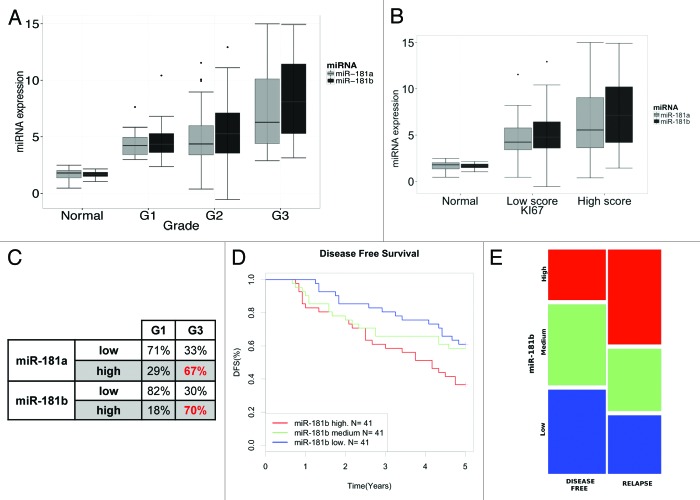
To identify miRNAs that may be deregulated in breast cancer, we performed a survey of public breast cancer data sets for miRNA expression. This search highlighted that miR-181b is frequently overexpressed in tumor samples included in three different breast cancer collections (Fig. S1).24-26 To validate this data, we investigated the expression of miR-181b and of its sibling miR-181a (originated from the same polycristronic transcripts) in a panel of 104 snap-frozen primary breast cancers and eight normal tissue counterparts (104 BC data set, Table S1). As shown in Figure 1A, higher levels of miR-181a/b were detected in tumor samples as compared with normal tissue, and, importantly, their expression levels correlated positively with tumor aggressiveness, with grade 3 (G3) tumors showing highest expression of the two miRNAs with respect to grade 1 (G1) and 2 (G2) tumors (Fig. 1A) as well as with proliferation index, as judged by KI67 protein expression (Fig. 1B). In particular, in G3 tumors, high levels of miR-181a/b were detected in 67% (miR-181a) and 70% (miR-181b) of cases, while in G1 samples, only 18% of them showed high levels of expression for miR-181a and 29% of them for miR-181b (Fig. 1C).
Figure 1. miR-181a/b expression levels correlate with breast cancer aggressiveness. (A) Plot of miR-181a and miR-181b expression (ΔΔCt) according to tumor grade (grade 1 tumors n = 17, grade 2 n = 60, grade 3 n = 27). miR-181a and miR-181b levels were evaluated by RT-qPCR and normalized to the expression of U6B RNA (see “Materials and Methods” for details). p < 10−7 (miR-181a) and p < 10−8 (miR-181b), Anova test on linear regression models. (B) Plot of miR-181a and miR-181b expression (ΔΔCt) according to Ki67 staining evaluated by IHC. p < 10−09 for both miR-181a and miR-181b, Anova test on linear regression models. (C) miR-181a/b expression was classified as high or low relative to the average value of expression in all samples. Percentage of miR-181a/b high or low expression within grade 1 or grade 3 tumors is reported. p = 0.0014 for miR-181a and p = 4.2 × 10−6 for miR-181b, Pearson’s Chi-square test. (D) Kaplan-Meier survival curves of disease free survival (DFS) of breast cancer patients classified according to the expression miR-181b, LogRank test, p = 0.0066, n = 123) (E) Mosaic plot showing the distribution of tumors with high (blue bars), medium (green bars) or low (red bars) miR-181b levels, assessed by comparing miRNA abundance in breast tumor samples that developed (relapse) or not (disease-free) a metastasis within 5 y after diagnosis. p-value (p < 0.05, Pearson’s Chi-square test) was calculated comparing “low” vs “high” miR-181b expression (see “Materials and Methods” for details).
We next investigated the association of miR-181 expression (stratified by tertiles) with the clinical outcome in a cohort of 123 primary breast cancers with annotated clinical history (123 BC B, Table S2). Of note, high expression levels of miR-181b were associated with shorter disease-free survival (Fig. 1D), with a nearly 3-fold increased risk of developing recurrence (HR = 2,89, Cox univariate regression analysis, 95% CI 1.36, 6.16, p < 0.006). Accordingly, primary tumors that gave rise to early metastatic events within 5 y since diagnosis (“relapse”) displayed more frequent overexpression of miR-181b (“high” expression) as compared with non-metastatic primary tumors (“disease-free,” Fig. 1E).
Altogether, these data obtained in the clinical setting suggest that miR-181a/b play a role in promoting breast cancer aggressiveness.
miR-181a/b regulate ATM levels in breast cancer cell lines
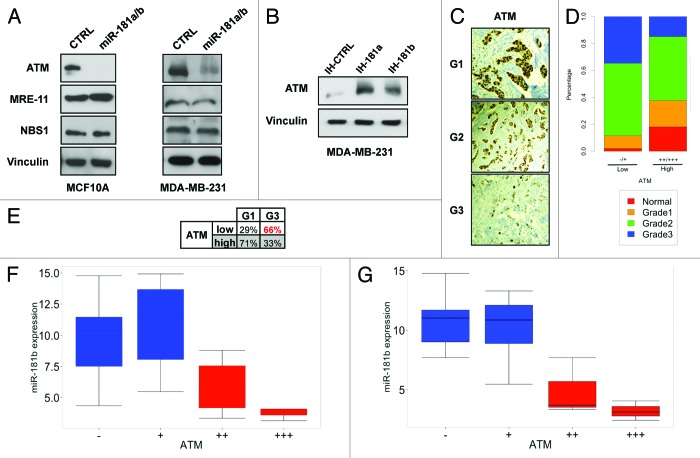
To investigate which functions of miR-181a/b may be determinant for breast tumorigenesis, we analyzed their predicted targets by TargetScan and Miranda algorithms. Interestingly, several candidate targets belonged to the DDR pathway, including Mre11, NBS1 and the stress sensor kinase ATM. We investigated whether the expression of these genes is regulated by miR-181a/b in both normal breast epithelial MCF10A cells and MDA-MB-231 breast cancer cells. As reported in Figure 2A, ectopic overexpression of miR-181a/b was associated with decreased expression of ATM protein in both cell lines. In contrast, we could not detect significant changes in Mre11 and NBS1 protein levels upon miR-181a/b overexpression. Moreover, we verified that miR-181a/b overexpression caused a decrease of ATM protein levels similar to that obtained by RNAi-mediated silencing (Fig. S2A and B). Conversely, when we transfected MDA-MB-231 with specific inhibitors of miR-181a and miR-181b (IH-181a and IH-181b), a consistent accumulation of ATM was obtained (Fig. 2B). These effects were also confirmed in other cell lines derived from breast (MDA-MB-468, SUM159PT), ovarian (OVCAR3), pancreatic (PANC1) and lung cancers (H1299) (see ahead Fig. 5C and D; Figs. S2 and 3). We then verified that the ATM 3′UTR is indeed a direct target of miR-181a/b, since miR-181a/b overexpression caused a robust reduction of luciferase-ATM 3′UTR reporter activity, and this was not observed upon introducing mutations in the two predicted miR-181a/b binding sites (Fig. S4).
Figure 2. miR-181a/b regulate ATM levels in breast cancer. (A) MCF10A and MDA-MB-231 cells were transfected with a combination of miR-181a/b or control siRNA and western blot analysis was performed after 72 h to detect ATM, Mre11, NBS1 and vinculin (loading control) protein levels. (B) MDA-MB-231 cells were transfected with miR-181a (anti-miR-181a), miR-181b (anti-miR-181b) or control inhibitors (anti-CTRL). Western blot analysis was performed after 96h to detect ATM and vinculin (loading control) protein levels. (C) Representative images of ATM staining in a tumor of grade 1, 2 and 3 (see “Materials and Methods” for details). (D) Mosaic plot showing the distribution of tumor grade according to ATM low (−/+) or high (++/+++) expression detected by IHC in each breast cancer sample p < 10−06, Pearson’s Chi-square test. (E) Percentage of ATM high or low expression within grade 1 or grade 3 tumors. p = 0.0014, Pearson’s Chi-square test. (F) Plot of miR-181b expression (ΔΔCt) according to ATM staining intensity in grade 3 breast cancer samples. p = 0.004665, Anova test on linear regression models. (G) Plot of miR-181b expression (ΔΔCt) according to ATM staining intensity in triple-negative breast cancer samples (n = 15). p = 0.005395, Anova test on linear regression models.
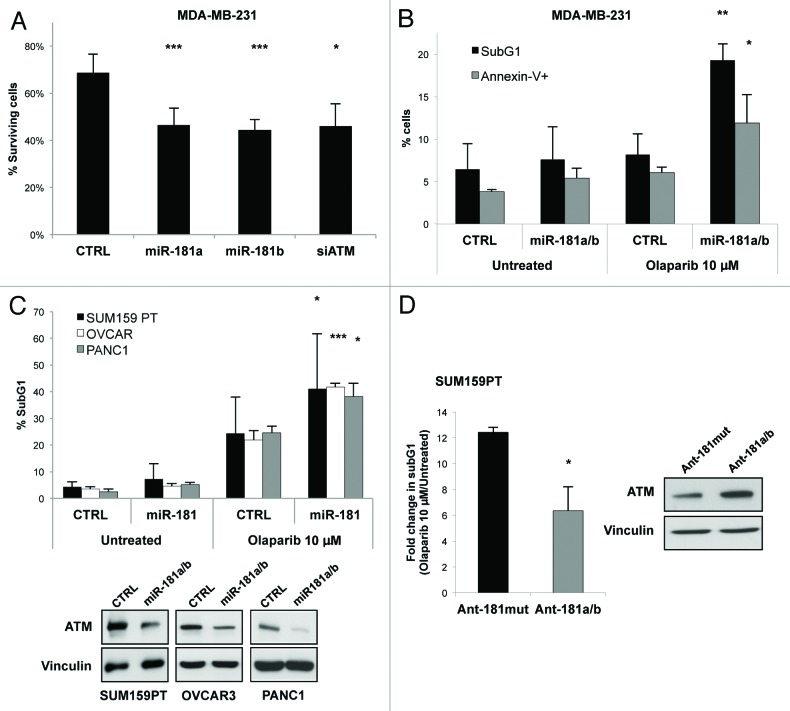
Figure 5. miR-181a/b sensitize cancer cells to Olaparib treatment. (A) Colony formation of MDA-MB-231 cells transfected with miR-181a, miR-181b, ATM (siATM) or control siRNA (CTRL) and treated with 1 µM Olaparib for 10 d. Percentage of surviving fraction are reported. Surviving fractions were calculated as ratio between plating efficiency in Olaparib-treated cells and plating efficiency in untreated cells. Plating efficiency was obtained according the following formula: # of colonies/# of plated cells. (B) MDA-MB-231 cells were transfected with a combination of miR-181a and miR-181b or control siRNA and 72 h hour later splitted and treated with 10 µM Olaparib. After 7 d, cells were harvested and half of them permeabilized and stained with propidium iodide, and the other half stained with PI/Annexin V, and analyzed by flow cytometry. The percentage of sub G1 population (black columns) and Annexin V positive cells (gray columns) are reported. (C) SUM159PT, OVCAR-3 and PANC1 cells transfected with a combination of miR-181a and miR-181b or control siRNA, were treated with 10 µM Olaparib for 4 (SUM159PT and PANC1) or 7 (OVCAR) d. Cells were then harvested, stained with propidium iodide and subjected to FACS analysis. The percentage of SubG1 population is reported. A representative western blot showing the levels of ATM and Vinculin as loading control is reported. (D) SUM159PT cells transfected with a combination of antagomiR against miR-181a and miR-181b (ant-181a/b) or a control antagomir mutated in five residues (Ant-181mut), and treated as in (C). A representative western blot showing the levels of ATM and Vinculin as loading control is reported. Graphs show means and s.d. for at least three independent experiments. p values were calculated with two tailed t-test: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
miR-181a/b inversely correlate with ATM levels in breast cancer
Analysis of ATM expression in the 104 BC data set by immunohistochemistry revealed low expression of ATM in G3 as compared with G1 tumor or normal samples (Fig. 2C and D). Low levels of ATM were detected in the 67% of G3 tumors while in G1 samples only 29% of cases showed low levels of ATM (Fig. 2E). Importantly, supporting the hypothesis that miR-181a/b regulate ATM levels in breast cancer tumors, we demonstrated an inverse correlation between the expression levels of ATM and miR-181a/b in this data set (Fig. S5A and B), and in particular in G3 tumor samples (Fig. 2F; Fig. S5C) and in the triple-negative cases present in this data set (Fig. 2G; Fig. S5D).
Altogether these data strongly support a role of miR-181a/b as crucial regulators of ATM activity in more aggressive breast cancers.
miR-181a/b dampen the DDR
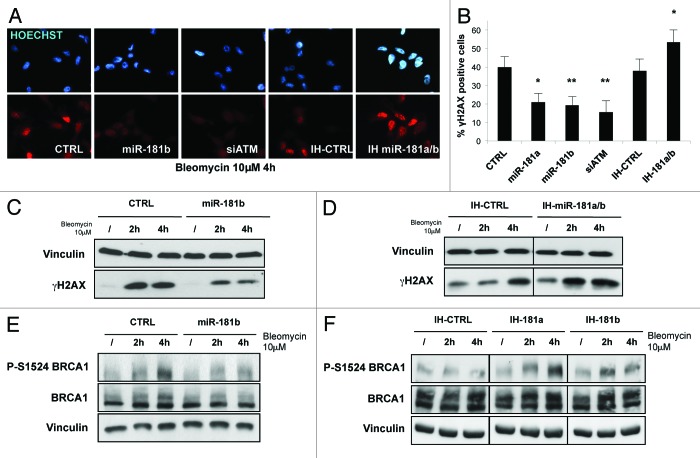
Activation of ATM triggered by DNA double-strand-breaks is crucial to initiate an efficient DDR and to induce DNA repair by HR.27,28 To investigate the extent to which miR-181a/b may impact on these processes, we first analyzed its effect on histone H2AX phosphorylation as a marker of ATM kinase activity induced by genotoxic stress. Overexpression of miR-181a/b in MDA-MB-231 cells caused a strong decrease in the number of γ-H2AX foci (Figs. 3A and B) and in the extent of H2AX phosphorylation (Fig. 3C) upon DNA damage. Importantly, this effect was comparable to that observed upon RNAi-mediated knockdown of ATM (Fig. 3A and B). On the contrary, miR-181a/b inhibitors significantly increased both H2AX phosphorylation and the number of foci (Fig. 3A, B and D). These data were also confirmed in normal breast epithelial MCF10A cells transformed with RASV12 upon stable overexpression of miR-181a/b (Fig. S6A and B). Following DNA damage, ATM promotes the phosphorylation and activation of several factors, among them the BRCA1 tumor suppressor.27 Phosphorylation of BRCA1 on Ser1524 after DNA damage29 was reduced upon overexpression of miR-181a/b (Fig. 3E), while it was increased upon inhibition of endogenous miR-181a/b (Fig. 3F).
Figure 3. miR-181a/b impairs ATM signaling. (A) MDA-MB-231 cells were transfected with miR-181a, miR-181b, siATM, control siRNA (CTRL) and with a combination of miR-181a and miR-181b inhibitors (IH-miR-181a/b) or control inhibitor (IH-CTRL). After 72 h cells, were split, and after 24 h treated with bleomycin (10 µM) for additional 4 h. Cells were then fixed, and immunofluorescence assay was performed to detect cells positive for γH2AX foci. A representative picture is shown for each condition. (B) Graph represents percentage of cells positive for γH2AX foci and shows means and s.d. for three independent experiments. (C) MDA-MB-231 cells transfected with miR-181b or control siRNA (CTRL) and (D) with a combination of miR-181a and miR-181b inhibitors (IH-miR-181a/b) or control inhibitor (IH-CTRL) were treated with bleomycin (10 µM) for 2 or 4 h. Western blot analysis was performed to detect phosphorylated H2AX and vinculin levels (loading control). (E) The same lysates analyzed in (C) were subjected to western blot analysis to detect phosphorylation on serine 1524 of BRCA1, total BRCA1 and vinculin levels (loading control). (F) MDA-MB-231 cells transfected with miR-181a (IH-miR-181a), miR-181b (IH-miR-181b) or control inhibitors (IH-CTRL) were treated with bleomycin (10 µM) for 2 or 4 h and subjected to western blot analysis to detect phosphorylation on serine 1524 of BRCA1, total BRCA1 and vinculin levels (loading control). p values were calculated with two tailed t-test, *, p < 0.05; **, p < 0.01; ***, p < 0.001.
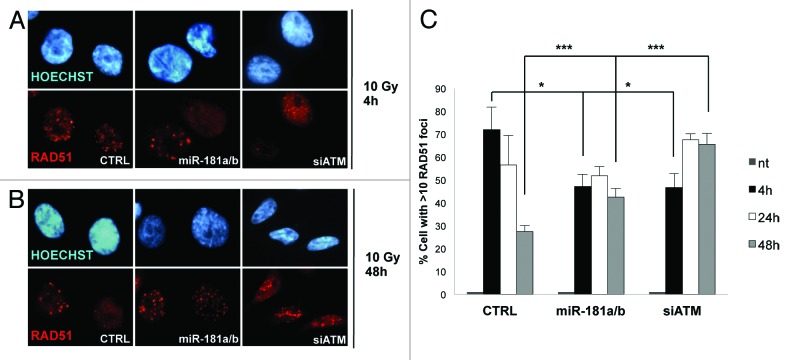
The above results suggest that aberrant overexpression of miR-181a/b may have a major impact on the cellular response to DNA damage. In particular, we evaluated the impact of miR-181a/b overexpression on the efficiency of DNA repair by HR by monitoring the appearance of RAD51 foci as a consequence of γ-radiation. Notably, the number of foci-containing cells 4 h after irradiation was lower upon overexpression of miR-181a/b as compared with controls in two different cell lines, and this effect is similar to that observed upon knocking down ATM expression by RNAi (p < 0.05, Fig. 4A and C; Fig. S7, black columns). Conversely, 48 h after DNA damage, cells overexpressing miR-181a/b were still highly positive for RAD51 foci, while controls showed only a residual signal, thus suggesting that the unscheduled expression of miR-181a/b leads to persistence of unrepaired DNA lesions (p < 0.001, Fig. 4B and C; Fig. S7, gray columns).
Figure 4. miR-181a/b alters the assembly of RAD51 foci. (A and B) MDA-MB-231 cells were transfected with a combination of miR-181a/b, ATM (siATM) or control siRNA and treated with 10 Gy of IR. Cells were fixed after 4, 24 and 48 h and immunofluorescence assay was performed to detect cells positive for RAD51 foci. A representative picture is shown for cells fixed after 4 (A) and 48 (B) h. (C) Graph represents percentage of cells positive for RAD51 foci and shows means and s.d. for three independent experiments. p values were calculated with two-tailed t-test: *, p < 0.05; **, p < 0.01; ***, p < 0.001-
In summary, the above results indicate that miR-181a/b are able to dampen both DDR and the DNA repair process by HR in breast cancer cells, causing the accumulation of unfixed lesions in DNA.
miR-181a/b act as determinants of sensitivity to PARP inhibition
Defects in proteins involved in DNA double-strand breaks repair and HR (e.g., ATM and BRCA1/2) display synthetic lethality with inhibition of base excision repair factors, such as poly (ADP-ribose) polymerase (PARP) 1.17 Based on our evidence, we reasoned that overexpression of miR-181a/b could sensitize breast cancer cells to pharmacological inhibition of PARP by means of their ability to reduce ATM expression and curb the activation of BRCA1. To evaluate this hypothesis, we treated MDA-MB-231 cells overexpressing miR-181a/b with the PARP inhibitor Olaparib (AstraZeneca) and performed colony formation assays to evaluate cell survival. As shown in Figure 5A (and Fig. S8A), expression of miR-181a/b significantly reduced the number of colonies counted after 10 d as compared with control, and this effect was comparable to that observed upon ATM silencing. Analysis of DNA content by propidium iodide staining followed by FACS analysis of cells treated for 72 h with Olaparib highlighted a cell cycle block at the G2/M transition upon either miR-181a/b overexpression or ATM knockdown (Figs. S8B and 9). Interestingly, a similar effect of PARP inhibitors has been previously reported for cells lacking functional BRCA1/2.30 Moreover, cells overexpressing miR-181a/b showed an increase of apoptotic markers (Annexin V staining and cleaved Caspase-3) upon prolonged administration of Olaparib (Fig. 5B; Figs. S8B and 10A). Similar effects were observed in a panel of different cancer cell lines of breast (SUM159PT), ovarian (OVCAR3), pancreas (PANC1) and colon (HT29) origin (Fig. 5C; Figs. S10 and 11).
Finally, we sought to demonstrate whether endogenous miR-181a/b expression could mediate the sensitivity of these cells to PARP inhibition. To this aim, SUM159PT cells were transfected with antagomiRs against miR-181a and miR-181b and then treated with Olaparib. As reported in Figure 5D, inhibition of endogenous miR-181a/b significantly decreased cell death caused by Olaparib treatment as compared with control, and this effect was concomitant with the rescue of ATM expression.
Altogether, these results indicate that deregulated expression of miR-181a/b sensitizes cancer cells to PARP inhibition.
Discussion
Breast cancer is a heterogeneous disease that comprises a range of distinct tumor types differing in their biological and clinical features. The most aggressive breast cancer subtypes, such as the Basal-like and triple-negative breast cancer, are characterized by high rates of relapse and of chemoresistant metastasis and greatly suffer from lack of therapeutic options.31 Several evidences indicate that the DNA damage response process is frequently impaired in more aggressive breast cancers, as a consequence of either mutation or altered expression of critical components, such as BRCA1, ATM and p53. While p53 mutations are reported with high frequency,32 mutations in genes coding for ATM or BRCA1 represent rare events in sporadic breast cancers. Nevertheless, functional impairment of BRCA (“BRCAness” phenotype) has been frequently observed in sporadic breast cancers.5,13 Likely, the existence of alternative mechanisms curbing the expression and functions of either BRCA1 or its regulators, such as ATM, may underlie this phenomenon. Among these mechanisms, aberrant activity of miRNAs could play a relevant role, as reported for miR-146 and miR-182, which directly target BRCA1.11,12
Our study now demonstrates that deregulation of miR-181a/b, by dampening the DDR, represents a key mechanism to establish BRCAness traits in aggressive breast cancer. High levels of miR-181a/b may dampen ATM and BRCA1 functions in tumors bearing functional alleles of these genes. We demonstrated that ATM levels are strongly downregulated by miR-181a/b in breast cancer cells, in agreement with previous findings by Wang et al.,33 who identified the miR-181 family as TGF-β-regulated miRNAs that contribute to the expansion of breast cancer stem-like cells by acting on ATM levels. We provide evidence that miR-181a/b are overexpressed in breast cancer, where they correlate with aggressive features, that they are associated with the likelihood to develop distant metastases (as also recently reported by Taylor and colleagues)34 and, most notably, we found an inverse correlation between miR-181a/b and ATM expression in human breast tumors. Accordingly, we found that miR-181a/b overexpression impairs the proper induction of both the DDR and the repair of DNA double-strand breaks in breast cancer cells.
miR-181a/b overexpression has been ascribed to aberrant activation of major pathways involved in breast tumorigenesis, including IL6/Stat3,35 TGF-β,34,36 HIF-1,37 WNT/β-catenin38 and HMGA1,39 suggesting that it may represent a crucial mechanism fuelling mammary gland transformation. Remarkably, the miR-181 family has been shown to be deregulated also in several other solid tumors (e.g., pancreas, prostate, gastric and colon) and to be able to target other tumor suppressors, including TIMP3, CYLD, PTEN and p27.35,36,40,41 Therefore, unscheduled expression of miR-181a/b may represent a common step of cellular transformation, contributing to the acquisition of different hallmarks of cancer.
Our findings have relevant clinical implications for the treatment of basal-like and TNBCs, the most aggressive breast cancer subtype characterized by high rates of relapse, visceral metastases and early death,31 and for which no effective therapies are available. A subset of TNBCs has shown sensitivity to PARP inhibitors, either alone or in combination with chemotherapy.16 However, lack of reliable biomarkers other than BRCA1 and BRCA2 to predict sensitivity to PARP inhibitors has curbed the success of phase III clinical trials in TNBC patients.42 Having shown compelling evidence that the expression of miR-181a/b determines the sensitivity of TNBC cells to the PARP inhibitor Olaparib, we may expect that quantifying the expression of miR-181a/b in basal-like breast cancers or TNBCs could help in predicting the patients’ response to PARP inhibitors. Notably, tumors showing the “BRCAness” traits are also sensitive to platinum-derived anticancer drugs.13 Since miR-181a expression has been shown to sensitize cancer cells to cisplatin,43 we may expect that miR-181a/b expression could also influence the efficacy of platinum-derived compounds.44,45 Besides breast cancer, our data suggest that the miR-181a/b-ATM axis may sensitize to the effects of PARP inhibition also in ovarian, pancreatic and colon tumors, where ATM is rarely found mutated (5, 3 and 14%, respectively, according to COSMIC database), while miR-181a/b have been found frequently overexpressed,24,46,47 thus providing an effective mechanism to functionally downregulate ATM, the DDR and the repair of DNA damages also in these tumors.
Therapeutic strategies based on ectopic expression or inhibition of cancer-related miRNAs have been proposed.48 Although the control of aberrant expression of miR-181a/b and other oncomiRNAs may represent an appealing therapeutic strategy, several challenges need to be addressed before these approaches could meet clinical application, including tumor delivery, safety and constant target inhibition.49 An alternative strategy exploiting miRNA deregulation in tumors would be to take advantage of cancer-related miRNAs to predict the clinical response to available treatments. Our data suggest that monitoring miR-181a/b expression alone or in combination with ATM could be a valuable strategy to select patients that would benefit from treatment with PARP inhibitors or platinum-based chemotherapy (both exploiting defects of HR-mediated DNA damage repair), thereby improving the therapeutic options for more aggressive breast cancers.
Materials and Methods
Cell culture, transfections and retroviral transduction
MDA-MB-231, HEK 293GP, MDA-MB-468, SUM159PT, OVCAR, HT29, PANC1 and Sk-Br-3 cells were cultured in DMEM medium supplemented with 10% FCS. H1299 cells were cultured in RPMI medium with 10% FCS. MCF10A cells were maintained in DMEM:F12 Ham’s medium 1:1, supplemented with 5% horse serum, insulin (10 μg/ml), hydrocortisone (0.5 μg/ml) and epidermal growth factor (20 ng/ml). All media were added with penicillin and streptomycin antibiotics (100IU/ml for each). For drug treatments, Bleomycin (Calbiochem 203401) and Olaparib (Selleck AZD2281) were used as indicated.
Transfections of H1299 were performed with Lipofectamine 2000 (Invitrogen), following manufacturer’s instructions. For miRNA/siRNA transfections, cells were transfected with 40 nM siRNA oligonucleotides (MWG biotech), 3 nM miRNAs (Ambion, PM10421 and PM12442) or 20nM miRNA inhibitors [Dharmacon IH-300553-07, IH-300553-08, using Lipofectamine RNAiMax (Invitrogen), following manufacturer’s instructions]. Sequence of siRNA against ATM is CCUGUUUGUUAGUUUAUUA. Control siRNA was from Qiagen (1027281) and control-miRNA inhibitor was from Dharmacon (IN-001005-01-05). For retroviruses production, low confluent HEK 293GP packaging cells were transfected by calcium phosphate precipitation. After 48–72 h the virus-containing medium was filtered and added to target cells. Cells were selected with blasticidin (2 μg/ml) and/or puromycin (0.5 μg/ml) and kept under selection for the entire experiment.
Cell cycle analysis and Annexin V staining
For cell cycle analysis by flow cytometry, both adherent and floating cells were collected and fixed in ethanol. After rehydration, cells were treated with 200 μg/mL RNaseA and 50 μg/mL propidium iodide (PI), and at least 5 × 104 cells were analyzed on a flow cytometer (FacsCalibur, BD). FACS data were processed using FlowJo software, and cell cycle profiles were determined using the Watson pragmatic model (Tree Star). For Annexin V staining and analysis by flow cytometry both adherent and floating cells were collected and stained with PI and Annexin V (Trevigen #4830-01-K).
Immunofluorescence assay
For immunofluorescence assay cells were seeded on coverslips, fixed with 3% paraformaldehyde for 20 min and then permeabilized with 0.1% Triton X-100. For detection of γ-H2AX foci cells were treated with 10 µM bleomycin for 4 h. Primary anti-phospho-Ser139-H2AX was used together with anti-mouse tetramethylrhodamine B isothiocyanate-conjugated secondary antibody (Sigma T5393). For detection of RAD51foci, cells were treated with 10 Gy IR and fixed after 4, 24 and 48 h. Primary anti-RAD51 was used together with orange-red-fluorescent Alexa Fluor 568 anti-mouse IgG (Life Technologies A-11004). Cellular nuclei were then stained with Hoechst.
Statistical analysis
104 BC data set: Differences between groups were analyzed using a linear regression analysis. Specific linear models were used to explore the relationships between miR-181a and -181b, and ATM and also to assay any relationship between miR-181a and -b expression, grade and tumor size. The expression levels of ATM and Ki67 were obtained by immunohistochemistry, and the expressions level of miR-181a and miR-181b were obtained by quantitative RT-PCR (see “Supplementary Methods”). Anova test was used to explore any significant difference in miR-181a, -b expression among the evaluated clinical categories. p values < 0.05 were considered statistically significant. (Figs. 1A and C, 2D, F and G; Fig. S5 A–D). To build and perform categorical data analysis (Figs. 1B and 2E) the expressions level of miR-181a, miR-181b were categorized on the quartile distribution. Pearson’s chi-square test with Yates’s correction for continuity was then performed.
123 BC data set: the expressions level of miR-181a, miR-181b were obtain by humanMI_V2 chip array analysis (see “Supplementary Methods”) and were categorized on the tertile distribution. Pearson’s chi-square test with Yates’s correction for continuity was performed for Figure 2E. Kaplan-Meier survival curves of disease free survival (DFS) time of Breast Cancer patients classified according to the categorized expression of miR-181b (Fig. 2D).
Statistical analysis and graphical representation were performed in R/Bioconductor, using the following packages: gplots, ggplot2, survival, ca and reshape.
Experimental data on cell models: p-values were obtained by performing type-two t-test (assuming equal variances) using Microsoft Excel.
Supplementary Material
Acknowledgments
We thank A. Testa for reading and editing the manuscript and F. Mantovani and L. Collavin for reading and discussion. We thank G. Pacini and F. Galardi for their technical assistance for IHC and data collection; M. Giacca and M. Bestagno for FACS analysis.
This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) Special Program Molecular Clinical Oncology “5 per mille” to G.D.S., Italian University and Research Ministerium (MIUR-PRIN), FIRB and Friuli-Venezia-Giulia (regional grant AITT) to G.D.S., AIRC and Ministero della Salute (Tumori Femminili) to M.G.D., and AIRC Grant-6251 to L.S.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/24757
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24757
References
- 1.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 2.Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed M, Rahman N. ATM and breast cancer susceptibility. Oncogene. 2006;25:5906–11. doi: 10.1038/sj.onc.1209873. [DOI] [PubMed] [Google Scholar]
- 4.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller CR, Roskelley CD. Regulation of BRCA1 expression and its relationship to sporadic breast cancer. Breast Cancer Res. 2003;5:45–52. doi: 10.1186/bcr557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tommiska J, Bartkova J, Heinonen M, Hautala L, Kilpivaara O, Eerola H, et al. The DNA damage signalling kinase ATM is aberrantly reduced or lost in BRCA1/BRCA2-deficient and ER/PR/ERBB2-triple-negative breast cancer. Oncogene. 2008;27:2501–6. doi: 10.1038/sj.onc.1210885. [DOI] [PubMed] [Google Scholar]
- 7.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–9. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 8.Baldassarre G, Battista S, Belletti B, Thakur S, Pentimalli F, Trapasso F, et al. Negative regulation of BRCA1 gene expression by HMGA1 proteins accounts for the reduced BRCA1 protein levels in sporadic breast carcinoma. Mol Cell Biol. 2003;23:2225–38. doi: 10.1128/MCB.23.7.2225-2238.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song L, Lin C, Wu Z, Gong H, Zeng Y, Wu J, et al. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS ONE. 2011;6:e25454. doi: 10.1371/journal.pone.0025454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci USA. 2010;107:1506–11. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med. 2011;3:279–90. doi: 10.1002/emmm.201100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011;41:210–20. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–9. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 14.Walerych D, Napoli M, Collavin L, Del Sal G. The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis. 2012;33:2007–17. doi: 10.1093/carcin/bgs232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer. 2012;12:587–98. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- 16.Narod SA. BRCA mutations in the management of breast cancer: the state of the art. Nature reviews. Clin Oncol. 2010;7:702–7. doi: 10.1038/nrclinonc.2010.166. [DOI] [PubMed] [Google Scholar]
- 17.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–94. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 18.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brodersen P, Voinnet O. Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol. 2009;10:141–8. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 21.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 22.Biagioni F, Bossel Ben-Moshe N, Fontemaggi G, Canu V, Mori F, Antoniani B, et al. miR-10b*, a master inhibitor of the cell cycle, is down-regulated in human breast tumours. EMBO Mol Med. 2012;4:1214–29. doi: 10.1002/emmm.201201483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothé F, Ignatiadis M, Chaboteaux C, Haibe-Kains B, Kheddoumi N, Majjaj S, et al. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS ONE. 2011;6:e20980. doi: 10.1371/journal.pone.0020980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–69. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 28.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–8. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 29.Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–6. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 30.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 31.Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–81. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 32.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Yu Y, Tsuyada A, Ren X, Wu X, Stubblefield K, et al. Transforming growth factor-beta regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene. 2011;30:1470–80. doi: 10.1038/onc.2010.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123:150–63. doi: 10.1172/JCI64946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang B, Hsu SH, Majumder S, Kutay H, Huang W, Jacob ST, et al. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29:1787–97. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–67. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji J, Yamashita T, Wang XW. Wnt/beta-catenin signaling activates microRNA-181 expression in hepatocellular carcinoma. Cell Biosci. 2011;1:4. doi: 10.1186/2045-3701-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mansueto G, Forzati F, Ferraro A, Pallante P, Bianco M, Esposito F, et al. Identification of a New Pathway for Tumor Progression: MicroRNA-181b Up-Regulation and CBX7 Down-Regulation by HMGA1 Protein. Genes Cancer. 2010;1:210–24. doi: 10.1177/1947601910366860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cuesta R, Martínez-Sánchez A, Gebauer F. miR-181a regulates cap-dependent translation of p27(kip1) mRNA in myeloid cells. Mol Cell Biol. 2009;29:2841–51. doi: 10.1128/MCB.01971-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–95. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guha M. PARP inhibitors stumble in breast cancer. Nat Biotechnol. 2011;29:373–4. doi: 10.1038/nbt0511-373. [DOI] [PubMed] [Google Scholar]
- 43.Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70:1793–803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- 44.Rajan A, Carter CA, Kelly RJ, Gutierrez M, Kummar S, Szabo E, et al. A phase I combination study of olaparib with cisplatin and gemcitabine in adults with solid tumors. Clin Cancer Res. 2012;18:2344–51. doi: 10.1158/1078-0432.CCR-11-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28:1145–53. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang J, Zheng X, Xu X, Zhou Q, Yan H, Zhang X, et al. Prognostic significance of miR-181b and miR-21 in gastric cancer patients treated with S-1/Oxaliplatin or Doxifluridine/Oxaliplatin. PLoS ONE. 2011;6:e23271. doi: 10.1371/journal.pone.0023271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakajima G, Hayashi K, Xi Y, Kudo K, Uchida K, Takasaki K, et al. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are Associated with Chemoresponse to S-1 in Colon Cancer. Cancer Genomics Proteomics. 2006;3:317–24. [PMC free article] [PubMed] [Google Scholar]
- 48.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–64. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.