Abstract
Myocardin related transcription factors A and B (MRTFs) activate serum response factor-driven transcription in response to Rho signaling and changes in actin dynamics. Myocardin and MRTFs have been implicated in anti-proliferative effects on a range of cell types. The precise mechanisms, however, remained elusive. We employed double knockdown of MRTF-A and MRTF-B in NIH 3T3 fibroblasts to evaluate its effects on cell cycle progression and proliferation. We show that transient depletion of MRTFs conveys a modest anti-proliferative effect and impinges on normal cell cycle progression, resulting in significantly shortened G1 phase and slightly extended S and G2 phase under normal growth conditions. Under serum-starved conditions we observed aberrant entry into the S and G2 phases without subsequent cell division. This was accompanied by downregulation of cyclin-CDK inhibitors p27Kip1, p18Ink4c and 19Ink4d as well as upregulation of p21Waf1 and cyclin D1. Extended knockdown led to increased formation of micronuclei, while cells stably depleted of MRTFs tend to become aneuploid and polyploid. Thus, MRTFs are required for accurate cell cycle progression and maintenance of genomic stability in fibroblast cells.
Keywords: Mrtf, actin, transcription, p27Kip1, aneuploidy, apoptosis
Introduction
Signaling to serum response factor (SRF) occurs mainly via the MAPK/Erk pathway and small GTPases of the Rho family.1,2 These pathways activate two families of transcriptional co-activators: ternary complex factors (TCFs: Elk-1, SAP-1 and Net) and myocardin-related transcription factors (MRTFs: MRTF-A/MAL/MKL1 and MRTF-B/MKL2).3 While TCFs regulate expression of a number of immediate early genes necessary for cell growth and proliferation,4,5 MRTFs couple SRF-dependent transcription to signals from Rho family GTPases and intracellular actin dynamics.2,6 MRTFs play an important role in a large number of developmental and physiological processes, including cardiovascular development,7,8 epithelial differentiation,9,10 neuronal plasticity11-13 and cell migration.14,15 In addition, the closely related SRF coactivator myocardin is a candidate tumor suppressor,16,17 while MRTFs have been implicated in experimental metastasis.15
There is increasing evidence for an involvement of the myocardin family in inhibiting proliferation and cell cycle progression. Both myocardin and MRTF-A exhibit anti-proliferative effects in various cell lines.18-20 MRTFs control the expression of anti-proliferative or pro-apoptotic genes, including Mig-6, Bok and Noxa.18,21 Whether downregulation of MRTFs leads to a proliferative advantage, however, remains poorly understood. This is at least in part due to the functional redundancy among the myocardin family of transcriptional coactivators.11,22
In this study we used siRNA to deplete both MRTF-A and MRTF-B in cells lacking myocardin expression. We show that depletion of MRTFs did not result in increased proliferation, but rather in proliferation impairment. This decreased proliferation was accompanied by changes in the duration of cell cycle phases, with a shorter G1 phase and slightly extended S and G2 phases. We identified key cell cycle regulators from the INK and CIP/KIP families of cyclin-CDK inhibitors, p18Ink4c, p19Ink4d and p27Kip1, to be downregulated upon MRTF double knockdown. In addition, we observed an increased number of cells containing micronuclei and nuclear buds during transient MRTF knockdown and enhanced aneuploidization of NIH 3T3 cells during stable MRTF depletion.
Results
MRTF-A/B knockdown leads to increase in S and G2/M populations in the absence of growth factors
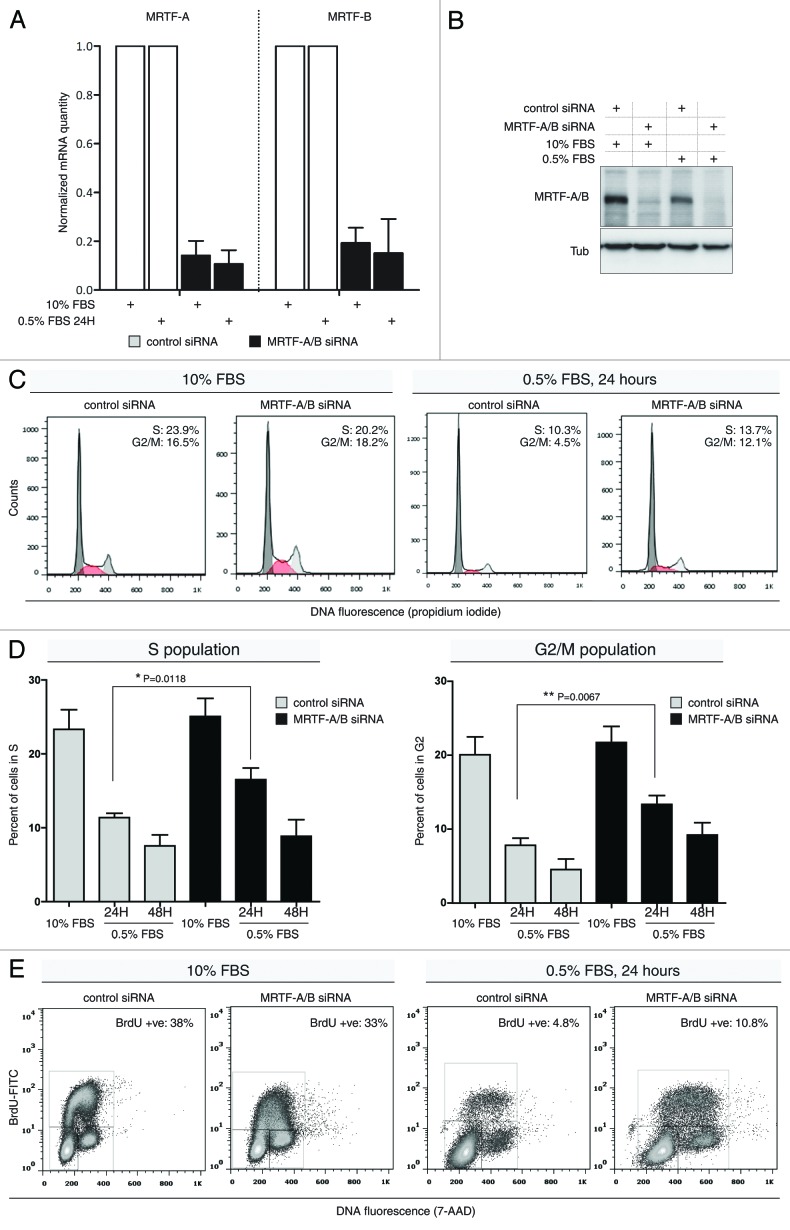
Myocardin and the myocardin-related transcription factors show antiproliferative effects when overexpressed in cells.18-20 To specifically analyze the effect of MRTFs on cell cycle progression, we employed transient siRNA-mediated knockdown of MRTF-A/B in NIH 3T3 mouse fibroblasts. The siRNA sequence used in this study targets both isoforms A and B, as previously described.15 Quantitative RT-PCR showed more than 84% decrease for MRTF-A mRNA and more than 70% decrease for MRTF-B mRNA at 24 h post-transfection (Fig. 1A). Western blotting revealed almost complete depletion of MRTF-A/B protein at the same time point (Fig. 1B).
Figure 1. Effects of transient MRTF-A/B knockdown on cell cycle profiles of NIH 3T3 cells. (A) mRNA quantitation upon siRNA-mediated MRTF-A/B double knockdown. MRTF-A and -B mRNA levels were normalized to Hprt (Error bars: +SEM, n = 3). (B) Representative western blot analysis of the MRTF knockdown efficiency 24 h post-transfection. (C) Cells transfected with control siRNA or siRNA against MRTF-A/B were either grown in 10% FBS-containing medium or serum-starved with 0.5% FBS for 24 h, followed by propidium iodide staining and FACS analysis. Cell cycle histograms generated from 10,000 events after gating out cell doublets/aggregates. Representative histograms from three independents experiments. (D) Quantification of S and G2/M phase distribution in NIH 3T3 cells transfected with siRNAs as in (C). Cell cycle histograms of propidium iodide-stained cells were de-convoluted using Dean-Jet-Fox model and FlowJo software (error bars: +SEM, n ≥ 3, p-value by unpaired Student’s t-test). (E) siRNA-transfected cells grown as in (A), pulse-labeled with BrdU for 20 min, stained with anti-BrdU, FITC-labeled antibody and 7-AAD and analyzed by FACS. Fifty thousand events were collected for each sample and dot plots generated after gating on single cell events. Gates on G1, S and G2/M populations are shown. Representative dot plots of three independent experiments.
The distribution of cell cycle phases was analyzed in either asynchronously growing cells in 10% FBS-containing medium or in cells incubated with 0.5% FBS for 24 h. MRTF-A/B knockdown did not lead to any significant differences in cell cycle phase distribution when cells proliferated in the presence of serum for a total of 42 h after the siRNA transfection (Fig. 1C). On the contrary, cells that were serum-starved with 0.5% FBS displayed elevated S and G2/M populations (Fig. 1C). Quantification of DNA histograms revealed a significant increase in cells in S phase and in cells with doubled DNA content upon MRTF knockdown after 24 h of serum starvation (Fig. 1D). At 48 h of serum starvation this increase was still obvious, but fell short of statistical significance (Fig. 1D).
To determine whether the observed increase in S phase was due to slippage through G1-S checkpoint, we pulse-labeled transfected cells with BrdU for 20 min and analyzed DNA content distribution vs. FITC-BrdU signal in cells grown with 10 or 0.5% FBS in culture medium. The results of three independent experiments confirmed that the increased S phase upon MRTF-A/B knockdown contained newly synthesized DNA (Fig. 1E). This indicates that the cells tend to override the G1 arrest induced by serum withdrawal upon MRTF depletion.
Consistent with propidium iodide staining, serum starved, BrdU-labeled cells displayed an increased fraction of cells in G2 phase. At 48 h of serum starvation, the BrdU-positive S population and G2 population were also increased upon MRTF knockdown, although to a lesser extent (data not shown). Cells grown in 10% FBS displayed similar DNA profiles to propidium iodide histograms, showing no discernible changes in cell cycle phases between control and MTRF siRNA transfections (Fig. 1E). Taken together, lack of MRTFs abolishes the ability of cells to arrest in G1 under starvation conditions.
MRTF-A/B knockdown impairs proliferation of NIH 3T3 fibroblasts
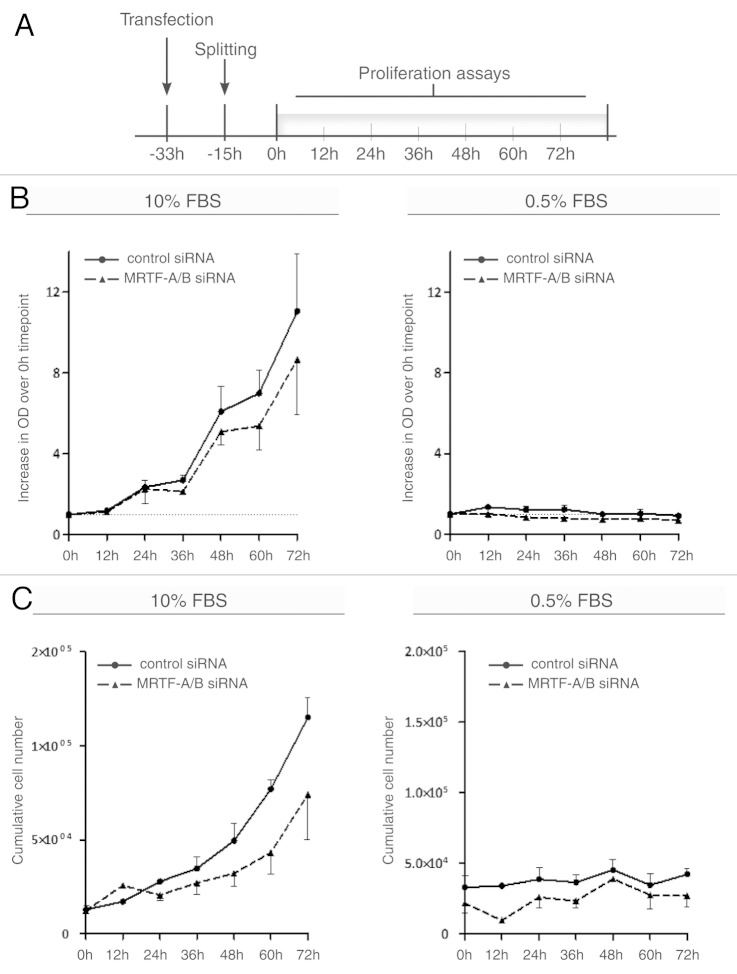
In order to determine whether increased S, G2 and M populations upon MRTF-A/B knockdown also correlate with the ability of these cells to proliferate in the presence of 0.5% FBS, we generated growth curves of NIH 3T3 cells transfected either with control or MRTF-A/B siRNA. MTT proliferation assays and direct cell counting were performed in parallel (Fig. 2B and C). Details of the experimental setup are depicted in Figure 2A: transfected cells were split 18 h post-transfection, and the first time point was measured at 33 h after the transfection, followed by measurements every 12 h. When maintained in 0.5% FBS, neither control cells, nor cells transfected with MRTF siRNA, showed any proliferation (Fig. 2B and C, right panel), demonstrating that the increase in S and G2/M populations upon MRTF knockdown is not followed by cell division and an increase in cell numbers. Thus, the cells that aberrantly entered S phase in the absence of serum are likely to be arrested and destined for apoptosis.
Figure 2. Proliferation of NIH 3T3 cells upon transient MRTF knockdown. (A) Scheme of experimental set up used for proliferation assays. (B) NIH 3T3 cells transfected with control or MRTF-A/B siRNAs were grown in 10 or 0.5% FBS-containing medium. MTT proliferation assay was performed at the indicated time points. Proliferation values are expressed as an increase in the optical density at 560 nm over the first time point (0 h). (C) In parallel, cells were grown as in (B) and counted using a Neubauer chamber. Total cell numbers plotted against time points. For all experiments, error bars indicate ± SEM (n = 3).
Moreover, when asynchronously grown in 10% FBS, MRTF knockdown caused a reproducible reduction in proliferation rate (Fig. 2B and C, left panel). These results indicate that lack of MRTFs impairs proper proliferation even in cycling cells, possibly by affecting mitotic cell division in both starved and non-starved cells.
MRTF-A/B knockdown changes the lengths of cell cycle phases
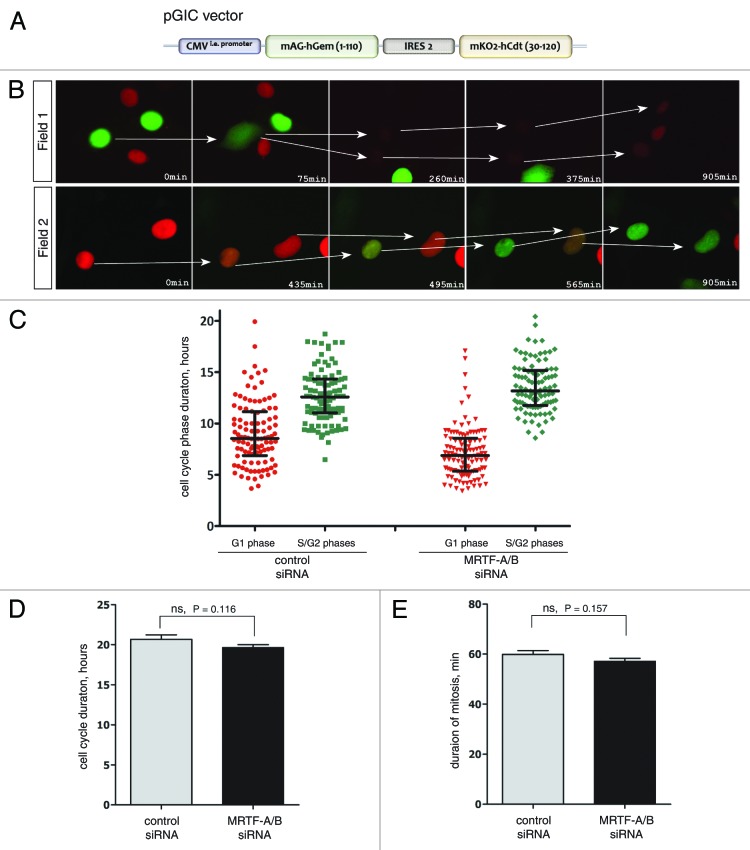
To gain a better understanding of the effects that MRTF-A/B knockdown has on cell cycle progression and to look for potential clues for delayed proliferation rate, we created two stable NIH 3T3 cell lines which allowed monitoring cell cycle phases in real time. The first contained a modified fluorescent indicator for cell cycle progression (FUCCI),23 where both markers were expressed from a single plasmid with the help of an IRES2 sequence (Fig. 3A). NIH 3T3-FUCCI cells display red fluorescence during G1 phase, when Cdt1 (30–120)-mKO2 fusion protein is accumulating. At the onset of S phase, it is quickly degraded by the SCFSkp2 E3 ligase complex, resulting in disappearance of red fluorescence. Simultaneous accumulation of Geminin (1–110)-mAG fusion protein during transition to S phase brings up green fluorescence, which persists through S, G2 and early M phases. During late mitosis, hGem (1–110)-mAG fusion is degraded by APCCdh1 E3 ligase complex, which remains active until the end of G1 phase. An example of cell cycle-dependent fluorescence changes in stable NIH 3T3 cell line is depicted in Figure 3B.
Figure 3. Analysis of the duration of cell cycle phases in NIH 3T3 cells using FUCCI markers. (A) The scheme of the FUCCI expression vector used to establish NIH 3T3 stable cell line. (B) The live-cell imaging of FUCCI-expressing NIH 3T3 cells showing cyclic changes of fluorescent markers during cell cycle. Field 1 follows two cells in G2 (green) undergoing nuclear envelope breakdown (75 and 375 min) followed by mitosis. Daughter cells in G1 become red. In field 2, two cells in G1 undergoing a change from red to green color, indicating progression from G1 to S phase. (C) Quantification of the live-cell imaging using FUCCI-expressing NIH 3T3 cells. Durations of the red and green phases were calculated in three independent experiments and the combined data are presented. n ≥ 100 cells for every plot. Median and interquartile distances are indicated. (D) Cell cycle duration was calculated as a total time between two divisions. n = 54 cells for control siRNA, n = 62 cells for MRTF siRNA (error bars: +SEM). (E) Quantification of the duration of mitosis in control and MRTF-A/B siRNA-transfected NIH 3T3-H2B-GFP cells during live-cell imaging (n ≥ 35 cells, error bars: +SEM). Statistical significance was determined by the unpaired Student’s t-test (ns: not significant).
Using live-cell imaging microscopy, the lengths of cell cycle phases were quantified in three independent experiments. MRTF double knockdown led to a significant shortening of the G1 phase when compared with the control. The average duration of the G1 phase was reduced from 9.028 h in the control siRNA transfection to 7.154 h in MRTF siRNA transfection (Fig. 3C). S/G2 populations have also been affected, but in the opposite direction—the mean duration had increased from mean 12.65 to 13.54 h in the knockdown (Fig. 3C). However, there was no significant difference in the total duration of the cell cycle between control siRNA and siRNA against MRTF-A/B, although we did notice that the cell cycle became slightly shorter when MRTFs were depleted (Fig. 3D).
The second stable NIH 3T3 cell line expressed a histone H2B-GFP fusion protein. This cell line allowed quantification of the duration of the mitosis. We scored the time that cells required from the nuclear envelope breakdown to the complete cytokinesis, and found that depletion of MRTFs did not have a significant effect on mitosis duration (Fig. 3E). Together this indicates that the S and G2 phases are extended on the expense of shorter G1 phases.
MRTF-A/B knockdown influences cell cycle protein levels
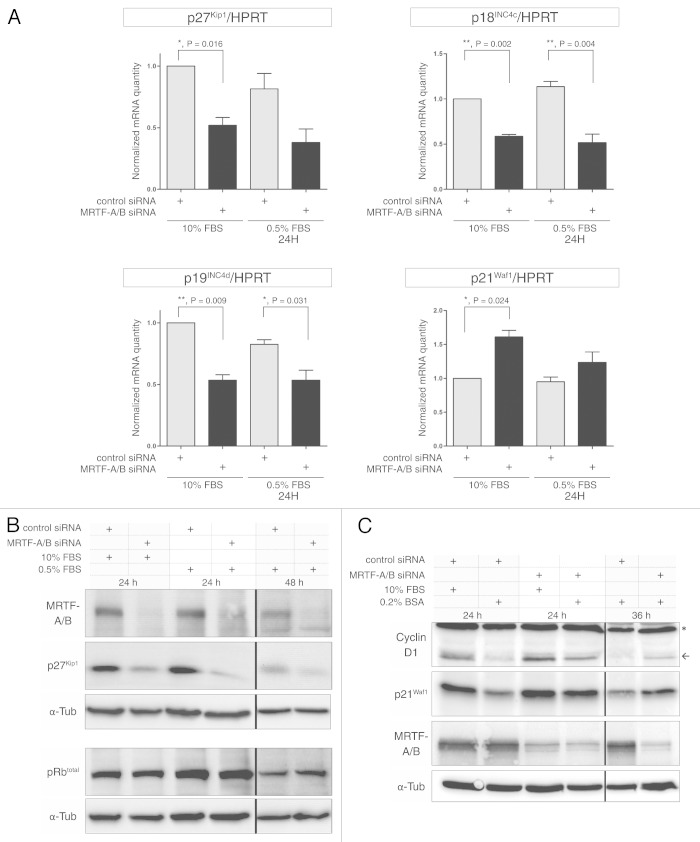
The observed changes in cell cycle dynamics raised the possibility that cyclin-CDK inhibitors might be affected by MRTF-A/B knockdown. Therefore, we examined the effects of the MRTF depletion on some key cell cycle-regulatory proteins. INK family members p18Ink4c and p19Ink4d were significantly downregulated on mRNA level after MRTF knockdown, both in the presence of 10% FBS and under starved conditions (0.5% FBS) (Fig. 4A). The CIP/KIP family members p21Waf1 and p27Kip1 appeared to be differentially affected: while p21Waf1 was slightly upregulated, p27Kip1 was significantly downregulated (Fig. 4A). Three other INK family members, p15INK4b, p16INK4a and p19ARF, could not be detected using quantitative RT-PCR in NIH 3T3 cells (data not shown).
Figure 4. Effect of MRTF-A/B knockdown on cell cycle regulators in NIH 3T3 cells. (A) Real-time PCR analysis of selected INK4 and Cip/Kip family of cyclin-CDK inhibitors upon transient MRTF-A/B knockdown. Error bars: +SEM, n = 3, p value by unpaired Student’s t-test. (B) Western blot analysis of p27Kip1 and total Rb proteins after MRTF knockdown. Representative pictures from three independent experiments are shown. (C) Western blot analysis of cyclin D1 and p21Waf1 proteins following MRTF knockdown. Representative pictures from three independent experiments.
Western blotting analysis confirmed a strong downregulation of p27Kip1 on the protein level upon MRTF depletion. Under starvation conditions, this downregulation was even more pronounced,due to the lack of serum-induced p27Kip1 degradation in control-transfected cells (Fig. 4B). p21Waf1 protein levels have also been slightly upregulated when the cells were grown in 10% FBS-containing medium after MRTF knockdown, however the protein seemed to be stabilized in the absence of serum (0.2% BSA), while in the control transfection, levels of p21Waf1 were significantly reduced (Fig. 4C). Total Rb protein levels were not affected by the knockdown (Fig. 4B). Cyclin D1 protein level displayed a similar pattern to the p21Waf1 protein, with a weak upregulation in full medium (10% FBS) and impaired degradation in the absence of serum growth factors (0.2% BSA) (Fig. 4C). Together, the results show that MRTF depletion critically affects several cell cycle regulators, which may underly the observed alterations in cell cycle progression.
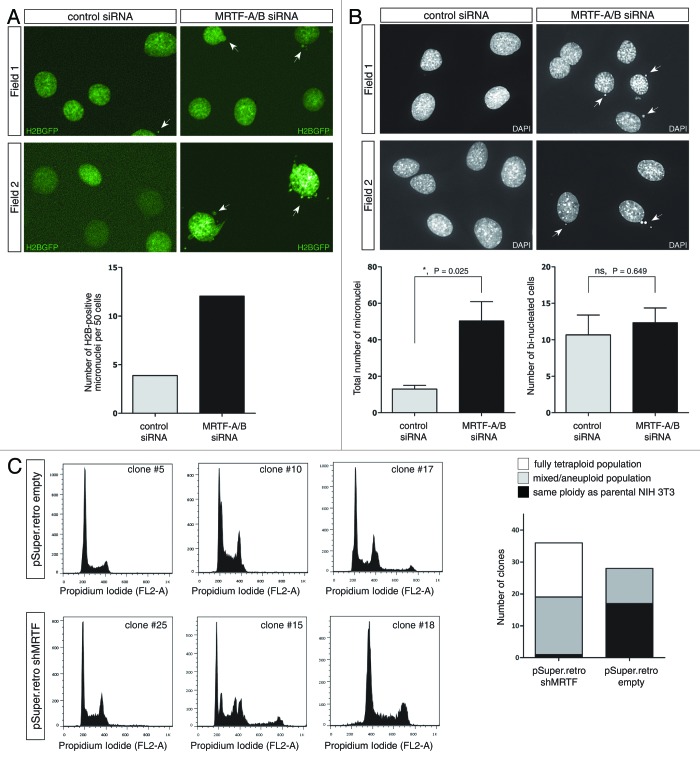
MTRF-A/B knockdown leads to defects in chromosomal stability
While performing live-cell imaging experiments with H2B-GFP-expressing NIH 3T3 cells, we noticed a significant increase in micronuclei and nuclear bud formation upon MRTF double knockdown (Fig. 5A). This increase first became apparent after more than 48 h post-transfection, and at 60 h, there was approximately 3-fold increase in cells, displaying nuclear defects in the form of either micronuclei or nuclear buds (Fig. 5A, lower panel).
Figure 5. Effects of MRTF-A/B knockdown on genomic stability of NIH 3T3 cells. (A) Stable NIH 3T3-H2BGFP cells were transfected either with control or MRTF-A/B siRNA and 24 h post-transfection live imaging was performed for 48 h. Number of micronuclei-containing cells was counted at the last time point and normalized to 50 cells. Representative micrographs and the quantified results are shown. Significance was calculated using the unpaired Student’s t-test (ns, not significant). (B) NIH 3T3 cells transfected with control or MRTF siRNA for 72 h were fixed and stained with DAPI. Bi-nucleated cells and micronuclei-containing cells were counted in three independent experiments. At least 200 cells were counted per experiment. Representative micrographs and quantified results are shown (error bars: +SEM, p values indicate unpaired Student’s t-test). (C) Clonal selection of the NIH 3T3 cells stably transfected with empty vector or expressing shRNA against MRTF-A/B. After 3 wk of selection, clonal lines were analyzed for DNA content using propidium iodide staining and flow cytometry. DNA content was scored as tetraploid compared with parental NIH 3T3 cells, aneuploid/mixed, or the same ploidy as parental cells. Representative histograms and the quantified distribution of the DNA content in the clonal lines are shown.
To quantify these defects, we transfected parental NIH 3T3 cells with either control siRNA or MRTF siRNA for 72 h, fixed the cells on coverslips and stained with DAPI (Fig. 5B, upper panel). Three independent experiments confirmed the findings from the H2B-GFP cell line and demonstrated a significant increase in nuclear defects-containing cells in the presence of siRNA against MRTFs (Fig. 5B, lower panel). We note, however, that the number of bi-nucleated cells remained unchanged in comparison to the control transfection.
When MRTFs were stably knocked down in NIH 3T3 cells using retrovirally delivered shRNA, the resulting stable pool of cells became aneuploid after 3 weeks of selection (data not shown). We further sub-cloned this pool and analyzed DNA content in 37 individual clones. In parallel, DNA profiles of 28 clones derived from pSuper.retro.puro empty vector stable pool were analyzed. In empty vector control clones, 11 out of 28 clones displayed varying degrees of aneuploidization (Fig. 5C, upper panel), which points toward some extent of inherent genomic instability of the NIH 3T3 cell line. In shRNA-expressing cells, however, all except one clone became aneuploid, with 17 out of 37 clones fully doubling their DNA content (Fig. 5C, lower panel). It is worth noting that there was no apparent growth deceleration in stable MRTF shRNA expressing clones, and some of them were proliferating faster than parental NIH 3T3 cells (data not shown).
Discussion
In the present study, we have analyzed the effects of MRTF depletion in NIH 3T3 fibroblasts and established novel links between the myocardin-related transcription factors and cell cycle progression. We demonstrate that under normal growth conditions (10% FBS), transient double knockdown of MRTF-A and -B leads to the shortening of G1 phase and slight lengthening of the S and G2 phases. In serum-starved conditions, MRTF-A/B-depleted cells show a tendency to slip from G1 into S and G2 phases, but this does not result in proliferation. Rather, the knockdown decreases proliferation with concomitant accumulation of cells carrying chromosomal segregation defects (micronuclei and nuclear buds). Persistent depletion of MRTFs by stable knockdown gives rise to aneuploid and polyploid cell lines. Finally, we link MRTF-A/B knockdown to the changes in the expression levels of several important cell cycle regulators, which could at least in part explain the observed phenotype.
The results suggest an important and complex role for MRTFs in maintaining proper cell cycle progression and genomic stability. Indeed, myocardin family members have previously been implicated in proliferation in a range of cell lines: overexpression of MRTF-A confers strong cytotoxicity in NIH 3T3 cells that requires the transcriptional activation domain of MRTF-A, and is in part due to the induction of apoptosis.18 Myocardin was shown to markedly impair proliferation of vascular smooth muscle (VSMC) and CHO cells20 as well as leiomyosarcoma cells.19 In HT1080 fibrosarcoma cells, myocardin inhibits proliferation at low cell density and abrogates colony formation in soft agar.16 These observations lead to the hypothesis that the depletion of MRTFs, which presumably control a similar set of target genes, might give cells a proliferative advantage. However, our data presented here do not support this hypothesis.
The suggested molecular mechanisms underlying myocardin-induced growth impairment, however, remain conflicting. Kimura and colleagues showed that myocardin in conjunction with SRF directly binds to the p21Waf1 promoter and induces its expression, which leads to a G1 arrest. In contrast, Tang et al. showed that p21Waf1 and p27Kip1 levels are not affected by the overexpression of myocardin, but they observed downregulation of c-myc, CDK1, CDK2 and S6K, which, in concert, results in a G2-M arrest and accumulation of polyploid cells.
Our data presented here do not formally exclude that MRTFs can directly activate the p21Waf1 promoter. However, the results at least suggest additional regulatory mechanisms ensuring that p21Waf1 levels do not decrease upon MRTF knockdown. Moreover, a gain-of-function experiment using inducible MRTF-A expression failed to detect increased levels of p21Waf1 mRNA and protein in our hands (A. Descot and G. Posern, unpublished).
It has been shown that in fibroblasts, p21Waf1 protein is upregulated by serum, PDGF or TGF-β.24,25 Consistently, we observe higher levels of p21Waf1 in asynchronously cycling NIH 3T3 cells in comparison to serum-starved cells, while MRTF knockdown leads to the increase in p21Waf1 protein in both conditions. In the absence of serum, upregulation of p21Waf1 mRNA was minimal, but we also observed upregulation of cyclin D1, which is known to sequester p21Waf1 and prevent its degradation.26 Therefore, the elevated levels of p21Waf1 possibly represent a combined effect of the increased mRNA production during normal culture conditions in the presence of serum and the delayed degradation in serum-starved conditions. Alternatively, the upregulation of the p21Waf1 gene could reflect stabilized p53 which may accumulate as a consequence of elevated genomic instability and the micronuclei formation observed.
We showed that MRTF-depleted cells slip into S and G2 phases despite serum starvation. This could in part be explained by the downregulation of p27Kip1 upon MRTF double knockdown. The observed decrease in mRNA levels suggests an involvement of transcriptional regulation or mRNA stability. However, under serum-starved conditions, the protein appears to be more affected than the mRNA. It is known that in serum-deprived cells, p27Kip1 accumulates in the nucleus and is stabilized there via phosphorylation by the Mirk/Dyrk1b kinase on Serine-10 (ref. 27 and references therein). Similar to the effects observed by MRTF knockdown, depletion of Mirk/Dyrk1b leads to increased degradation of p27Kip1 and progression into the S phase.28 Intriguingly, Mirk/Dyrk1b is induced by RhoA, which is also a potent inducer of the actin-MRTF-SRF pathway.29,30 Thus, it would be interesting to see if Mirk/Dyrk1b is affected by MRTF knockdown.
It remains possible that the downregulation of p27Kip1 alone is sufficient to allow NIH 3T3 cells to progress into S and G2 phase in the absence of serum; however, additional downregulation of p18Ink4c and p19Ink4d on mRNA level might also contribute to this phenomenon. Despite slippage into S-G2 phases, MRTF-depleted cells fail to proliferate, suggesting that they become arrested before or during mitosis. Elevated p21Waf1, which is known to target cdc2 kinase for degradation31 might contribute to this arrest.
p21Waf1 expression is controlled by Ras, Raf and the TCF Elk-1.32-34 In serum-starved Swiss 3T3 cells (but not in NIH 3T3), activated Ras leads to accumulation of p21Waf1 and G1 arrest, while the presence of serum inhibits Ras-dependent p21Waf1 accumulation through RhoA activity.33,35,36 One could therefore speculate that p21Waf1 is subject to the described competition between Ras- and Rho-dependent SRF coactivators, and that MRTF depletion somehow elevates a Ras-Elk1 dependent p21Waf1 expression.3,37-39
Slight lengthening of S-G2 phases in NIH 3T3 cells grown in full medium upon MRTF knockdown also resembles previously observed 1 h delay in entering mitosis upon blocking RhoA with C3 exoenzyme in HeLa cells.40 It is possible that RhoA function during G2-M transition, via its effects on the actin cytoskeleton, could involve MRTFs. Moreover, it is clear that the role of RhoA in cell cycle extends beyond MRTF-dependent regulation of transcription, because RhoA-null phenotype confers more severe defects in proliferation when compared with MRTF knockdown.41
While MRTF-A/B-depleted cells do not display obvious defects in mitosis short-term (up to 48 h), at longer time points increased micronucleation points to the errors in chromosomal segregation, which is reminiscent of the phenotype seen in H-Ras overexpression and activation of MAPK pathway studies, once more hinting at the potential imbalance of two pathways activating SRF.42 Similarly, RhoA-null MEFs displayed micronuleation and increased number of bi-nucleated cells.41 While we could not detect an increase in bi-nucleated cells up to 72 h of MRTF-A/B knockdown, clonal selection of stable MRTF-depleted NIH 3T3 cells revealed that 36 out of 37 clones became aneuploid or completely doubled their DNA, suggesting that MRTFs are indeed required for maintenance of genomic stability.
Material and Methods
Plasmids, antibodies and reagents
siRNA and shRNA sequences against murine MRTF-A/B were described previously.15 Control siRNA was Silencer® Negative Control siRNA (Applied Biosystems, AM4635). MRTF-A/B shRNA was cloned into pSuper.retro.puro vector (Oligoengine) as previously reported.21 H2B-GFP fusion was expressed from pBOS-H2BGFP (BD PharMingen). FUCCI markers were expressed from pGIC plasmid. Complete sequences are available upon request. Antibodies used were rabbit anti-MRTF-A/B serum #79 (home-made), monoclonal mouse anti-α-tubulin, clone DM1A (Sigma, T9026), monoclonal mouse anti-p21Waf1, Ab-4 (Calbiochem, OP76), mouse monoclonal anti-cyclin D1 (Millipore, 05-815), monoclonal mouse anti-p27Kip1 (BD Biosciences, 610242) and rabbit anti-Rb (C-15; Santa Cruz Biotechnology, sc-50).
Cell lines, transfections and infections
NIH 3T3 mouse fibroblasts were maintained at 37°C, 10% CO2, in high-glucose (4.5 g/L) DMEM (Invitrogen), supplemented with 10% FBS and 100 units Pen-Strep (Invitrogen). For generation of stable cell lines, NIH 3T3 cells were transfected using Lipofectamine 2000 (Invitrogen) in 10 cm dishes, according to manufacturer’s protocol. Twenty-four hours post-transfection, cells were split into 15-cm dishes and incubated in medium containing appropriate antibiotics. NIH-pGIC stable pool was generated by FACS sorting the cells six times, using BD Aria II. The first and the second sorts included mAG-hGem (1–110)-only positive cells; the third sort included mKO2-hCdt (1–130)-only positive cells, and sorts 4–6 were for double-positive cells. NIH-H2BGFP cells were FACS-sorted for GFP-positive cells two times. NIH-pGIC pool was maintained in presence of 800 µg/mL G418 and NIH-H2BGFP pool in 1 µg/mL puromycin. For generation of a stable MRTF-A/B knockdown cell line, retroviral infections were employed. Detailed procedure is described elsewhere.18 Briefly, 6 × 106 cells of the Phoenix E packaging cell line were transfected with 20 µg of pSuper.retro.puro-shMRTF vector using the calcium phosphate method. Viruses were produced for 24 h, subsequently concentrated from supernatant with Vivaspin 20 columns (30 kDa cut-off, Sartorius, VS2032) and used to infect 1.5 × 105 NIH 3T3 cells. Infection was repeated once more 8 h later. Twenty-four hours post-infection, cells were split 1:20 into medium containing 1 µg/mL puromycin and astable pool was selected for 3 wk. For clonal selection, the stable pools were split at 1:25 into 15 cm dishes, and 2 wk later single clones were picked using cloning cylinders (Sigma, C1059). Transient transfections with MRTF-A/siRNA were done using Lipofectamine RNAiMAX (Invitrogen) according to manufacturer’s instructions. 100 pmol of siRNA was used for 6 cm dishes.
Cell cycle analysis
Propidium iodide staining was done as described.43 Briefly, 18 h post-transfection, cells were split into 6-cm plates and grown for indicated periods of time. Cells were collected by trypsinization and fixed with 70% ethanol. After washing with PBS, cells were stained for 30 min with 50 µg/mL propidium iodide (Sigma, 81845) in PBS, supplemented with 200 µg/mL RNase A (Sigma, R6513). BD FACSCalibur (BD Biosciences) was employed to measure fluorescence from 10,000 events per sample. Data was analyzed using FlowJo software v7 (TreeStar). DNA histograms were generated by gating on single cell events and cell cycle phases de-convoluted using built-in Dean-Jet-Fox algorithm. BrdU staining was performed using FITC BrdU Flow kit (BD Pharmigen, 559619) according to manufacturer’s protocol. Briefly, cells were pulse-labeled with 10 µM BrdU for 20 min in medium, trypsinized and fixed. After DNA digestion, cells were stained with FITC-labeled anti-BrdU antibody for 20 min, co-stained with 7-AAD and analyzed with FACSCalibur flow cytometer. Fifty thousand events were collected per sample, and dot plots of single events were generated with the FlowJo software.
Live-cell imaging
Cells were seeded into non-coated, four-chambered Cellview™ glass-bottom dishes (Greiner Bio-One, 627871) 18 h post-transfection and incubated for 6 h before live imaging was started. During live imaging, cells were maintained at 37°C, 10% CO2 in a Carl Zeiss Observer.Z1 microscope equipped with incubator XL S1 (Pecon), TempModule S, CO2 module S and heating unit XL S. Images were captured every 5 min for 48 to 65 h with AxioCam MRm camera and Plan-Apochromat 10×/0.45 objective (FUCCI markers) or Plan-Apochromat 20×/0.8 objective (H2BGFP). GFP and mAG fluorescence was imaged using Zeiss Filter Set 38HE (BP470/40, FT 495, BP 525/50), mKO2 fluorescence was imaged using Zeiss Filter Set 43HE (BP 545/25, FT570, BP 605/70). Exposures were 200 ms for mAG, 400 ms for mKO2 and 90 ms for GFP. Images were saved with AxioVision 4.7 software. For analysis, TIFFs were exported to ImageJ software (US National Institute of Health) and manually analyzed for individual cell phase duration, total cell cycle length and duration of mitosis.
Proliferation assays
For MTT assay, cells were seeded into 96-well plates in quadruplicates 18 h post-transfection and incubated for further 15 h before the first time point. One hour before the time point, 40 µl of MTT solution (5 mg/mL; Sigma, M5655) was added to the well, and cells were incubated for 1 h. Medium was removed, and MTT Formazan crystals were dissolved in 150 µl DMSO. Absorbance was measured at 560 nm using the Labsystems Multiscan RC (Thermo Scientific). For cell counting, cells were seeded into 12-well plates in duplicates (10,000 for 10% FBS-containing medium, 35,000 for 0.5% FBS-containing medium). At the indicated time points, cells were trypsinized and counted using an improved Neubauer chamber. Nine quadrants were counted per sample.
Quantitative RT-PCR
Total RNA was prepared using RNeasy kit (Qiagen, 74104). First-strand cDNA was synthesized from 1 µg of RNA using oligo-dT primer and Verso cDNA synthesis kit (Thermo Scientific, AB-1453) according to manufacturer’s instructions. qPCR was performed using 2.5 µl cDNA (1:5 dilution), 0.5 mM primers and Fast SYBR Green master mix (Applied Biosystems, 4385612) in 15 μl reaction volume using the StepOne Plus instrument (Applied Biosystems). Primer sequences used were as follows: Hprt, forward: TCA GTC AAC GGG GGA CAT AAA, reverse: GGG GCT GTA CTG CTT AAC CAG; MRTF-A, forward: CCA GGA CCG AGG ACT ATT TG, reverse: CGA AGG AGG AAC TGT CTG CTA; MRTF-B, forward: CCC ACC CCA GCA GTT TGT TG, reverse: TGC TGG CTG TCA CTG GTT TCA TC; p27Kip1, forward: TAA TTG GGT CTC AGG CAA ACT C; reverse: AGA ATC TTC TGC AGC AGG TC; p21Waf1, forward: ACA AGA GGC CCA GTA CTT CC; reverse: TGG AGT GAT AGA AAT CTG TCA GG; p18Ink4c, forward: GCT GCA GGT TAT GAA ACT TGG; reverse: GTT AAC ATC AGC CTG GAA CTC; p19Ink4d, forward: CTT GCA GGT CAT GAT GTT TGG; reverse: GTC CAG GGC ATT GAC ATC AG.
Immunofluorescent staining
Cells were grown on coverslips, fixed with 4% PFA for 15 min and permeabilized with 0.2% Triton X-100 for 5 min. Coverslips were subsequently incubated with 1:10,000 dilution of DAPI (Invitrogen, D1306) in PBS for 20 min and mounted onto glass slides with Mowiol reagent (Sigma, 324590), supplemented with 2.5% DABCO (Sigma, D2522). Pictures were acquired with Plan-Apochromat 20×/0.8 objective on Observer.Z1 microscope (Carl Zeiss) using Zeiss Filter Set 49 (G365, FT395, BP 445/50) and AxioVision 4.7 software.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft, grant PO1032/2-2, and by the European Regional Development Fund of the European Commission.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24839
References
- 1.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 2.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–96. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Zaromytidou A-I, Miralles F, Treisman R. MAL and ternary complex factor use different mechanisms to contact a common surface on the serum response factor DNA-binding domain. Mol Cell Biol. 2006;26:4134–48. doi: 10.1128/MCB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gille H, Kortenjann M, Strahl T, Shaw PE. Phosphorylation-dependent formation of a quaternary complex at the c-fos SRE. Mol Cell Biol. 1996;16:1094–102. doi: 10.1128/mcb.16.3.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–15. doi: 10.1016/S0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 6.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–65. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Zhu X, Chen M, Cheng L, Zhou D, Lu MM, et al. Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc Natl Acad Sci U S A. 2005;102:8916–21. doi: 10.1073/pnas.0503741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh J, Richardson JA, Olson EN. Requirement of myocardin-related transcription factor-B for remodeling of branchial arch arteries and smooth muscle differentiation. Proc Natl Acad Sci U S A. 2005;102:15122–7. doi: 10.1073/pnas.0507346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connelly JT, Gautrot JE, Trappmann B, Tan DW, Donati G, Huck WT, et al. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat Cell Biol. 2010;12:711–8. doi: 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- 10.Luxenburg C, Pasolli HA, Williams SE, Fuchs E. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat Cell Biol. 2011;13:203–14. doi: 10.1038/ncb2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mokalled MH, Johnson A, Kim Y, Oh J, Olson EN. Myocardin-related transcription factors regulate the Cdk5/Pctaire1 kinase cascade to control neurite outgrowth, neuronal migration and brain development. Development. 2010;137:2365–74. doi: 10.1242/dev.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith-Hicks C, Xiao B, Deng R, Ji Y, Zhao X, Shepherd JD, et al. SRF binding to SRE 6.9 in the Arc promoter is essential for LTD in cultured Purkinje cells. Nat Neurosci. 2010;13:1082–9. doi: 10.1038/nn.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stern S, Debre E, Stritt C, Berger J, Posern G, Knöll B. A nuclear actin function regulates neuronal motility by serum response factor-dependent gene transcription. J Neurosci. 2009;29:4512–8. doi: 10.1523/JNEUROSCI.0333-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leitner L, Shaposhnikov D, Mengel A, Descot A, Julien S, Hoffmann R, et al. MAL/MRTF-A controls migration of non-invasive cells by upregulation of cytoskeleton-associated proteins. J Cell Sci. 2011;124:4318–31. doi: 10.1242/jcs.092791. [DOI] [PubMed] [Google Scholar]
- 15.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol. 2009;11:257–68. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milyavsky M, Shats I, Cholostoy A, Brosh R, Buganim Y, Weisz L, et al. Inactivation of myocardin and p16 during malignant transformation contributes to a differentiation defect. Cancer Cell. 2007;11:133–46. doi: 10.1016/j.ccr.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 17.Shats I, Milyavsky M, Cholostoy A, Brosh R, Rotter V. Myocardin in tumor suppression and myofibroblast differentiation. Cell Cycle. 2007;6:1141–6. doi: 10.4161/cc.6.10.4251. [DOI] [PubMed] [Google Scholar]
- 18.Descot A, Hoffmann R, Shaposhnikov D, Reschke M, Ullrich A, Posern G. Negative regulation of the EGFR-MAPK cascade by actin-MAL-mediated Mig6/Errfi-1 induction. Mol Cell. 2009;35:291–304. doi: 10.1016/j.molcel.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Kimura Y, Morita T, Hayashi K, Miki T, Sobue K. Myocardin functions as an effective inducer of growth arrest and differentiation in human uterine leiomyosarcoma cells. Cancer Res. 2010;70:501–11. doi: 10.1158/0008-5472.CAN-09-1469. [DOI] [PubMed] [Google Scholar]
- 20.Tang R-H, Zheng X-L, Callis TE, Stansfield WE, He J, Baldwin AS, et al. Myocardin inhibits cellular proliferation by inhibiting NF-kappaB(p65)-dependent cell cycle progression. Proc Natl Acad Sci U S A. 2008;105:3362–7. doi: 10.1073/pnas.0705842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaposhnikov D, Descot A, Schilling J, Posern G. Myocardin-related transcription factor A regulates expression of Bok and Noxa and is involved in apoptotic signalling. Cell Cycle. 2012;11:141–50. doi: 10.4161/cc.11.1.18499. [DOI] [PubMed] [Google Scholar]
- 22.Pipes GCT, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–56. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 23.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, et al. Visualizing spatiotemporal dynamics of multicellular cell cycle progression. Cell. 2008;132:487–98. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 24.Macleod KF, Sherry N, Hannon G, Beach D, Tokino T, Kinzler K, et al. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 1995;9:935–44. doi: 10.1101/gad.9.8.935. [DOI] [PubMed] [Google Scholar]
- 25.Kivinen L, Laiho M. Ras- and mitogen-activated protein kinase kinase-dependent and -independent pathways in p21Cip1/Waf1 induction by fibroblast growth factor-2, platelet-derived growth factor, and transforming growth factor-beta1. Cell Growth Differ. 1999;10:621–8. [PubMed] [Google Scholar]
- 26.Coleman ML, Marshall CJ, Olson MF. Ras promotes p21(Waf1/Cip1) protein stability via a cyclin D1-imposed block in proteasome-mediated degradation. EMBO J. 2003;22:2036–46. doi: 10.1093/emboj/cdg189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Susaki E, Nakayama KI. Multiple mechanisms for p27(Kip1) translocation and degradation. Cell Cycle. 2007;6:3015–20. doi: 10.4161/cc.6.24.5087. [DOI] [PubMed] [Google Scholar]
- 28.Deng X, Mercer SE, Shah S, Ewton DZ, Friedman E. The cyclin-dependent kinase inhibitor p27Kip1 is stabilized in G(0) by Mirk/dyrk1B kinase. J Biol Chem. 2004;279:22498–504. doi: 10.1074/jbc.M400479200. [DOI] [PubMed] [Google Scholar]
- 29.Deng X, Ewton DZ, Pawlikowski B, Maimone M, Friedman E. Mirk/dyrk1B is a Rho-induced kinase active in skeletal muscle differentiation. J Biol Chem. 2003;278:41347–54. doi: 10.1074/jbc.M306780200. [DOI] [PubMed] [Google Scholar]
- 30.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–42. doi: 10.1016/S0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 31.Dash BC, El-deiry WS. Phosphorylation of p21 in G 2 / M Promotes Cyclin B-Cdc2 Kinase Activity. Mol Biol Cell. 2005;25:3364–87. doi: 10.1128/MCB.25.8.3364-3387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravi RK, McMahon M, Yangang Z, Williams JR, Dillehay LE, Nelkin BD, et al. Raf-1-induced cell cycle arrest in LNCaP human prostate cancer cells. J Cell Biochem. 1999;72:458–69. doi: 10.1002/(SICI)1097-4644(19990315)72:4<458::AID-JCB2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 33.Olson MF, Paterson HF, Marshall CJ. Signals from Ras and Rho GTPases interact to regulate expression of p21Waf1/Cip1. Nature. 1998;394:295–9. doi: 10.1038/28425. [DOI] [PubMed] [Google Scholar]
- 34.Shin SY, Kim CG, Lim Y, Lee YH. The ETS family transcription factor ELK-1 regulates induction of the cell cycle-regulatory gene p21(Waf1/Cip1) and the BAX gene in sodium arsenite-exposed human keratinocyte HaCaT cells. J Biol Chem. 2011;286:26860–72. doi: 10.1074/jbc.M110.216721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahai E, Olson MF, Marshall CJ. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 2001;20:755–66. doi: 10.1093/emboj/20.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adnane J, Bizouarn FA, Qian Y, Hamilton AD, Sebti SM. p21(WAF1/CIP1) is upregulated by the geranylgeranyltransferase I inhibitor GGTI-298 through a transforming growth factor beta- and Sp1-responsive element: involvement of the small GTPase rhoA. Mol Cell Biol. 1998;18:6962–70. doi: 10.1128/mcb.18.12.6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murai K, Treisman R. Interaction of serum response factor (SRF) with the Elk-1 B box inhibits RhoA-actin signaling to SRF and potentiates transcriptional activation by Elk-1. Mol Cell Biol. 2002;22:7083–92. doi: 10.1128/MCB.22.20.7083-7092.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gineitis D, Treisman R. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J Biol Chem. 2001;276:24531–9. doi: 10.1074/jbc.M102678200. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–9. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 40.Ando Y, Yasuda S, Oceguera-Yanez F, Narumiya S. Inactivation of Rho GTPases with Clostridium difficile toxin B impairs centrosomal activation of Aurora-A in G2/M transition of HeLa cells. Mol Biol Cell. 2007;18:3752–63. doi: 10.1091/mbc.E07-03-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melendez J, Stengel K, Zhou X, Chauhan BK, Debidda M, Andreassen P, et al. RhoA GTPase is dispensable for actomyosin regulation but is essential for mitosis in primary mouse embryonic fibroblasts. J Biol Chem. 2011;286:15132–7. doi: 10.1074/jbc.C111.229336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saavedra HI, Fukasawa K, Conn CW, Stambrook PJ. MAPK mediates RAS-induced chromosome instability. J Biol Chem. 1999;274:38083–90. doi: 10.1074/jbc.274.53.38083. [DOI] [PubMed] [Google Scholar]
- 43.Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Protoc. 2006;1:1458–61. doi: 10.1038/nprot.2006.238. [DOI] [PubMed] [Google Scholar]