Abstract
Schimke Immunoosseous Dysplasia (SIOD) is a rare, autosomal recessive disorder of childhood with classical features of spondyloepiphyseal dysplasia, renal failure, and T cell immunodeficiency. SIOD has been associated with several malignancies, including non-Hodgkin lymphoma and osteosarcoma. About half of SIOD patients have biallelic mutations in SMARCAL1 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin, subfamily a-like 1). This gene encodes an annealing helicase and replication stress response protein that localizes to damage-stalled DNA replication forks. We report a child with SIOD and a novel S859P missense mutation in SMARCAL1 who developed undifferentiated carcinoma of the sinus.
Keywords: Schimke Immunoosseous Dysplasia, SMARCAL1, Cancer Predisposition Syndrome, DNA Damage Response, Replication Stress Response
Introduction
Schimke Immunoosseous Dysplasia is a rare, autosomal recessive, and pleiotropic disorder of childhood with classical features of spondyloepiphyseal dysplasia and primordial dwarfism, proteinuria preceding glomerulosclerosis and renal failure, and T cell immunodeficiency1. Less common features include premature atherosclerosis, malformed teeth and other ectodermal abnormalities, neurologic complications such as transient ischemic attacks and chronic headaches, autoimmune disease, and cancer, including non-Hodgkin lymphoma and osteosarcoma2-5. SIOD is normally fatal by the adolescent years in the absence of stem cell and/or renal transplantation, though patients may live into adulthood6.
About half of SIOD patients have biallelic mutations in SMARCAL17. The SMARCAL1 protein, also referred to as HARP (HepA Related Protein) and DNA-dependent ATPase A, belongs to the SNF2 family of chromatin remodeling ATPases8,9. Members of this family use energy from ATP hydrolysis to move along DNA, and participate in a number of cellular processes, including DNA transcription, replication, and repair9. SMARCAL1 in particular is a replication stress response protein with annealing helicase activity that localizes to stalled DNA replication forks where it is thought to help stabilize these forks8,10-12. We report a case of undifferentiated carcinoma of the sinus in a child with SIOD and a novel S859P missense mutation in SMARCAL1.
Case Report
A 3 year old Caucasian male came to medical attention due to failure to thrive (weight 3rd percentile), short stature (height < 3rd percentile). At 4 years of age he developed lymphopenia (ALC of 899/μl) and neutropenia (ANC of 756/μl). Bone marrow evaluation revealed a hypocellular marrow with partial myeloid arrest and a prominent population of hematogones identified by flow cytometry. Testing for Fanconi anemia, autoimmune neutropenia, autoimmune lymphoproliferative syndrome (ALPS), Schwachman-Diamond syndrome, and deficiencies in the complement cascade were negative. A karyotype and chromosomal microarray were normal.
At 6 years of age he developed abdominal ascites with nephrosis. A renal biopsy revealed focal segmental glomerulosclerosis. He progressed over 8 months to renal failure requiring peritoneal dialysis. A radiograph of the femur was obtained at this time and revealed a small femoral capital epiphysis and mild irregularity of the acetabulum and distal femoral metaphysis. Immunologic evaluation revealed decreased CD4 and CD8 T cell numbers (< 100 cells/μl). Mitogen stimulation testing showed his lymphocytes to be unresponsive to CD3 and PHA stimulation. This, combined with his renal failure and skeletal abnormalities, supported a diagnosis of SIOD, which was confirmed with SMARCAL1 mutational analysis that revealed a c.1190delT mutation in one allele, a known SIOD-associated frameshift mutation (p.L397fsX40), and a c.2575C>T mutation in the other allele, a novel missense mutation (p.S859P). This missense mutation was not found in 130 normal chromosomes and S859 appears to be conserved in vertebrates. These heterozygous mutations were defined by direct sequencing of both sense and antisense strands of the SMARCAL1 coding exons as previously described13. Exons and intronic splice junctions were PCR amplified from genomic DNA. The amplification products were purified and sequenced by automated dideoxy sequencing using fluorescent dye primers. Sequencing analyses from the parents of the child revealed segregation of p.L397fsX40 from the father and segregation of p.S859P from the mother.
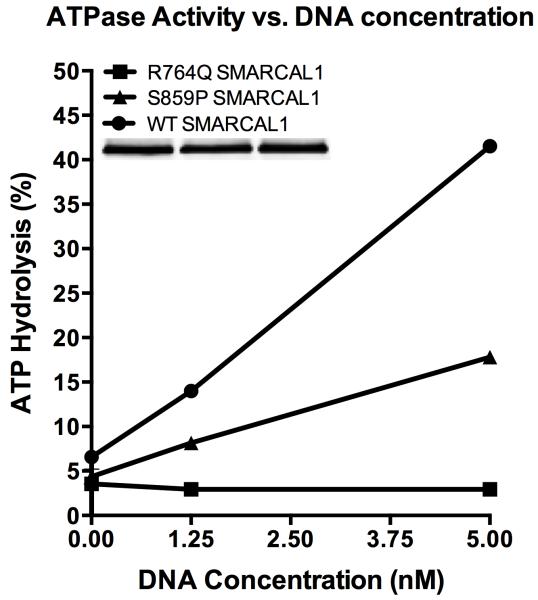
SMARCAL1 has several critical domains (Figure 1). The novel S859P mutation described in this report is predicted to lie just within the C terminal boundary of the ATPase domain. The S859P mutant does retain some enzymatic activity, as seen in a side-by-side comparison to SMARCAL1 with a R764Q missense mutation that abolishes enzymatic activity (Figure 2).
Figure 1.

Wild-type SMARCAL1. SMARCAL1, which is 954 amino acids in length, has several critical domains, including an N-terminal DNA binding domain (RPA) comprised of the first 34 amino acids of the protein; two HARP domains, each 55 amino acids in length and connected by a 40 amino acid linker, that are critical for annealing helicase activity; and an ATPase enzymatic domain, separated from the second HARP domain by 47 amino acids and divided into two regions by a 115 amino acid chain, necessary for ATP hydrolysis. Our patient’s S859P missense mutation is predicted to lie just within the C terminal boundary of the ATPase domain (*).
Figure 2.
To purify SMARCAL1 from human cells, HEK-293T cells were transfected with pLPCX-Flag-SMARCAL1, pLPCX-Flag-S859P SMARCAL1, or pLPCX-Flag-R764Q SMARCAL1 plasmids using Lipofectamine 2000 (Invitrogen). The latter two destination vectors were made by site directed mutagenesis of pENTR-SMARCAL1 with an 11x wobble mutation and then cloned into the pLPCX vector using the Clonase system (Invitrogen). Seventy-two hours after transfection, the cells were lysed in NETN buffer (150 mM NaCl, 20 mM Tris pH 8, 1 mM EDTA, 0.5% IGEPAL CA-630) for 30 min on ice. After high-speed centrifugation, the cleared lysates were incubated with Flag-M2 beads (Sigma) for 3 h at 4°C. The beads were washed three times in NETN lysis buffer and twice in SMARCAL1 buffer (20 mM HEPES at pH 7.6, 20% glycerol, 0.1 M KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.2 mM PMSF, 0.01% IGEPAL CA-630). The bound proteins were eluted in SMARCAL1 buffer containing 0.3 mg/mL Flag peptide on ice, flash-frozen, and stored at −80°C. For the ATPase assay increasing concentrations of oligonucleotides (0, 1.25, or 5 nM final concentration) were incubated with purified SMARCAL1 (5 nM final concentration) in a final volume of 10 μL, and the reactions were incubated for 30 minutes at 30°C. The results are presented as the percent ATP hydrolyzed during the reaction. The assay was performed at least three times with the graph depicting means and standard deviation error bars. The splayed arm DNA substrate used in this assay was prepared by annealing the following two oligonucleotides: CCAGTGAATTGTTGCTCGGTACCTGCTAAC and GACATTTGATACCGAGCAACAATTCACTGG. The differences between the S859P SMARCAL1 and WT SMARCAL1 curves were significant at all oligonucleotide concentrations by two-way ANOVA. The differences between the R764Q SMARCAL1 and S859P SMARCAL1 curves were significant at 1.25 nM and 5 nM oligonucleotide concentrations by two-way ANOVA.
At 8 years of age the patient developed persistent purulent nasal discharge. A CT scan revealed an expansile and hyperdense mass filling the left nasal and paranasal sinuses with destruction of the left ethmoid air cells and cribriform plate. Biopsy revealed a high-grade, poorly differentiated carcinoma with squamous and glandular differentiation, and large areas of necrosis. Human Papilloma virus (HPV) testing was negative. A metastatic work-up was negative. Due to his immunodeficient state chemotherapy was not utilized. He instead received local radiation therapy (5,940 cGy in 33 fractions) and is now 3 months out from radiation with no evidence of recurrent disease.
Discussion
Biallelic mutations in SMARCAL1 are associated with Schimke immunoosseous dysplasia. SMARCAL1 is a replication stress response protein that localizes to damage-stalled DNA replication forks through an interaction with RPA, a single strand DNA binding protein. SMARCAL1 is thought to stabilize stalled replication forks, preventing fork collapse and double strand DNA break formation8,10-12. It may also play a role in transcription and recombination14. SMARCAL1 is an annealing helicase capable of resolving RPA-coated bubbles in plasmid DNA15. Other in vitro activities include an ability to hydrolyze ATP in a DNA-dependent manner, to bind DNA with single and double strand components, including gaps and overhangs, and to resolve complex DNA intermediate structures that mimic stalled or damaged replication forks16. This ability to process DNA intermediates is shared by several helicases implicated in well-characterized cancer predisposition syndromes, including the Bloom syndrome protein, Werner syndrome protein, and RECQL4 helicase associated with the Rothmund-Thomson syndrome17-19.
A number of SIOD-associated SMARCAL1 mutations have been described. SMARCAL1 missense mutations are typically associated with later onset disease, while nonsense, frameshift, and splicing mutations are more often seen in severe disease20. Patient-derived SMARCAL1 mutants fail to rescue the genome maintenance defects seen in cells in which native SMARCAL1 is silenced, suggesting that SIOD is at least in part a genome maintenance disorder21. However, there is no clear genotype-phenotype correlation in SIOD, with reports of the same SMARCAL1 mutations in both milder and severe disease, and evidence of environmental factors shaping SIOD phenotypes in fruit flies, mice, and humans22,23.
In a review of 71 patients with SIOD, four developed malignancy, two with Epstein-Barr virus (EBV) positive non-Hodgkin lymphoma, one with EBV negative non-Hodgkin lymphoma, and one with osteosarcoma14. The literature on SMARCAL1 and its relation to human malignancy is extremely limited; however, members of the SWI/SNF2 family of ATPases have been implicated in several cancers, including BRM and BRG1 in non-small cell lung cancer and SMARCA4 in WNT-driven medulloblastomas24,25. Our case represents the first of undifferentiated carcinoma described in a patient with SIOD. Whether malignancies in this population are coincidental, stem from underlying immunodeficiency, or are related to defects in the genome maintenance function of SMARCAL1, is not entirely clear. EBV-driven lymphomas may reasonably be attributed to immunodeficiency. EBV negative lymphoma, osteosarcoma, and now our case of HPV negative carcinoma are less clearly linked to immune dysfunction. High-grade undifferentiated carcinomas, such as that seen in our patient, have been described in patients with Bloom syndrome, Werner syndrome, and the Rothmund-Thomson syndrome, all well-established cancer predisposition syndromes26-28. Presently, SIOD is not considered a cancer predisposition syndrome, and the incidence of cancer in this population may never accurately be known due to the limited number of patients and the short life expectancy of SIOD patients. Additional studies are certainly needed to better evaluate the risk of cancer in this population, but with several disparate malignancies now associated with this very rare disorder, it is reasonable to consider SIOD a cancer predisposition syndrome.
Acknowledgment
We would like to thank Alisha McCord for her administrative support and Dr. Cornelius Boerkoel for his advice and critical review of the manuscript.
Support
C.C. is funded by the St. Baldrick’s Foundation as a St. Baldrick’s Foundation Fellow. D.C. is funded by R01GM160342.
Footnotes
Conflict of Interest Statement:
None of the authors have conflicts of interest to report.
References
- 1.Boerkoel CF, O’Neill S, Andre JL, Benke PJ, Bogdanovic R, Bulla M, et al. Manifestations and treatment of Schimke immuno-osseous dysplasia: 14 new cases and a review of the literature. Eur J Pediatr. 2000;159(1-2):1–7. doi: 10.1007/s004310050001. [DOI] [PubMed] [Google Scholar]
- 2.Lev A, Amariglio N, Levy Y, Spirer Z, Anikster Y, Rechavi G, et al. Molecular assessment of thymic capacities in patients with Schimke immuno-osseous dysplasia. Clin Immunol. 2009;133(3):375–81. doi: 10.1016/j.clim.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 3.Lucke T, Clewing JM, Boerkoel CF, Hartmann H, Das AM, Knauth M, et al. Cerebellar atrophy in Schimke-immuno-osseous dysplasia. Am J Med Genet A. 2007;143A(17):2040–5. doi: 10.1002/ajmg.a.31878. [DOI] [PubMed] [Google Scholar]
- 4.Morimoto M, Kerouredan O, Gendronneau M, Shuen C, Baradaran-Heravi A, Asakura Y, et al. Dental abnormalities in Schimke immuno-osseous dysplasia. J Dent Res. 2012;91(7 Suppl):29S–37S. doi: 10.1177/0022034512450299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zieg J, Krepelova A, Baradaran-Heravi A, Levtchenko E, Guillen-Navarro E, Balascakova M, et al. Rituximab resistant evans syndrome and autoimmunity in Schimke immuno-osseous dysplasia. Pediatr Rheumatol Online J. 2011;9(1):27. doi: 10.1186/1546-0096-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lou S, Lamfers P, McGuire N, Boerkoel CF. Longevity in Schimke immuno-osseous dysplasia. J Med Genet. 2002;39(12):922–5. doi: 10.1136/jmg.39.12.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clewing J, Fryssira H, Goodman D, Smithson S, Sloan E, Lou S, et al. Schimke immunoosseous dysplasia: suggestions of genetic diversity. Hum Mutat. 2007;28(3):273–83. doi: 10.1002/humu.20432. [DOI] [PubMed] [Google Scholar]
- 8.Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23(20):2405–14. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34(10):2887–905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, et al. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009;23(20):2415–25. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009;23(20):2394–9. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yusufzai T, Kong X, Yokomori K, Kadonaga JT. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 2009;23(20):2400–4. doi: 10.1101/gad.1831509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boerkoel C, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet. 2002;30(2):215–20. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- 14.Baradaran-Heravi A, Raams A, Lubieniecka J, Cho K, DeHaai K, Basiratnia M, et al. SMARCAL1 deficiency predisposes to non-Hodgkin lymphoma and hypersensitivity to genotoxic agents in vivo. Am J Med Genet A. 2012;158A(9):2204–13. doi: 10.1002/ajmg.a.35532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science. 2008;322(5902):748–50. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, et al. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26(2):151–62. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croteau DL, Rossi ML, Ross J, Dawut L, Dunn C, Kulikowicz T, et al. RAPADILINO RECQL4 mutant protein lacks helicase and ATPase activity. Biochim Biophys Acta. 2012;1822(11):1727–34. doi: 10.1016/j.bbadis.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machwe A, Karale R, Xu X, Liu Y, Orren DK. The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) holliday junctions to functional replication forks. Biochemistry. 2011;50(32):6774–88. doi: 10.1021/bi2001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281(32):22839–46. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 20.Bokenkamp A, deJong M, van Wijk JA, Block D, van Hagen JM, Ludwig M. R561C missense mutation in the SMARCAL1 gene associated with mild Schimke immuno-osseous dysplasia. Pediatr Nephrol. 2005;20(12):1724–8. doi: 10.1007/s00467-005-2047-x. [DOI] [PubMed] [Google Scholar]
- 21.Bansbach CE, Boerkoel CF, Cortez D. SMARCAL1 and replication stress: an explanation for SIOD? Nucleus. 2010;1(3):245–8. doi: 10.4161/nucl.1.3.11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baradaran-Heravi A, Cho KS, Tolhuis B, Sanyal M, Morozova O, Morimoto M, et al. Penetrance of biallelic SMARCAL1 mutations is associated with environmental and genetic disturbances of gene expression. Hum Mol Genet. 2012;21(11):2572–87. doi: 10.1093/hmg/dds083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucke T, Billing H, Sloan EA, Boerkoel CF, Franke D, Zimmering M, et al. Schimke-immuno-osseous dysplasia: new mutation with weak genotype-phenotype correlation in siblings. Am J Med Genet A. 2005;135(2):202–5. doi: 10.1002/ajmg.a.30691. [DOI] [PubMed] [Google Scholar]
- 24.Reisman D, Sciarrotta J, Wang W, Funkhouser W, Weissman B. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–66. [PubMed] [Google Scholar]
- 25.Robinson G, Parker M, Kranenburg T, Lu C, Chen X, Ding L, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488(7409):43–8. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma B, Corry J, Rischin D, Leong T, Peters LJ. Combined modality treatment for locally advanced squamous-cell carcinoma of the oropharynx in a woman with Bloom’s syndrome: a case report and review of the literature. Ann Oncol. 2001;12(7):1015–7. doi: 10.1023/a:1011106202939. [DOI] [PubMed] [Google Scholar]
- 27.Marin-Bertolin S, Amorrortu-Velayos J, Aliaga Boniche A. Squamous cell carcinoma of the tongue in a patient with Rothmund-Thomson syndrome. Br J Plast Surg. 1998;51(8):646–8. doi: 10.1054/bjps.1998.0050. [DOI] [PubMed] [Google Scholar]
- 28.Saeki H, Kondo S, Morita T, Yamaguchi O, Ishizuka G, Koizumi Y. Bladder carcinoma with Werner syndrome. Urology. 1987;30(5):494–5. doi: 10.1016/0090-4295(87)90392-x. [DOI] [PubMed] [Google Scholar]