Zheng et al. report a comprehensive analysis of whole-genome sequencing and DNA copy number data sets on glioblastoma multiforme (GBM). A dense pattern of repeated focal amplifications involving the MDM2/CDK4 oncogenes was identified in 5% of GBMs and associated with poor survival. Recurrent breakpoints were noted, including in apoptosis regulator FAF1, resulting in protein depletion. Restoring FAF1 in glioma cells increased the FAS-mediated apoptosis response. These findings uncover a genomic mechanism of gene deregulation that can confer growth advantages.
Keywords: Glioblastoma multiforme, The Cancer Genome Atlas, intragenic breakpoint, gene fusion, genomic rearrangement, genomic instability
Abstract
With the advent of high-throughput sequencing technologies, much progress has been made in the identification of somatic structural rearrangements in cancer genomes. However, characterization of the complex alterations and their associated mechanisms remains inadequate. Here, we report a comprehensive analysis of whole-genome sequencing and DNA copy number data sets from The Cancer Genome Atlas to relate chromosomal alterations to imbalances in DNA dosage and describe the landscape of intragenic breakpoints in glioblastoma multiforme (GBM). Gene length, guanine–cytosine (GC) content, and local presence of a copy number alteration were closely associated with breakpoint susceptibility. A dense pattern of repeated focal amplifications involving the murine double minute 2 (MDM2)/cyclin-dependent kinase 4 (CDK4) oncogenes and associated with poor survival was identified in 5% of GBMs. Gene fusions and rearrangements were detected concomitant within the breakpoint-enriched region. At the gene level, we noted recurrent breakpoints in genes such as apoptosis regulator FAF1. Structural alterations of the FAF1 gene disrupted expression and led to protein depletion. Restoration of the FAF1 protein in glioma cell lines significantly increased the FAS-mediated apoptosis response. Our study uncovered a previously underappreciated genomic mechanism of gene deregulation that can confer growth advantages on tumor cells and may generate cancer-specific vulnerabilities in subsets of GBM.
Glioblastoma multiforme (GBM) is the most common and malignant brain tumor in adults (Furnari et al. 2007; Dunn et al. 2012). Despite multimodal therapy with radiation and the nitrosourea alkylating agent temozolomide, median survival is a dismal 15 mo (Stupp et al. 2005). Recent genomic studies have profiled large numbers of GBM samples, leading to multitiered characterization of core pathogenic pathways, identification of novel tumorigenic genes, and stratification into molecular subtypes (Beroukhim et al. 2007; The Cancer Genome Atlas Research Network 2008; Parsons et al. 2008; Noushmehr et al. 2010; Verhaak et al. 2010; Singh et al. 2012; Sturm et al. 2012; Zheng et al. 2012). These studies and the resources they provide have accelerated our understanding of gliomagenesis and laid the foundation for further research.
High-throughput sequencing technologies have fundamentally changed the nature of cancer research by enabling genome-wide analyses that were not previously feasible; for instance, by empowering the detection of structural alterations in cancer DNA (Chin et al. 2011). For example, paired-end DNA sequencing of colorectal carcinomas resulted in the identification of genomic lesions and fusion transcripts involving WNT pathway members TCF7L1 and TCF7L2 (Bass et al. 2011; The Cancer Genome Atlas Network 2012). Messenger RNA (mRNA) sequencing of GBMs identified a 70-kb inversion and associated FGFR3-TACC3 fusion transcript, which was then demonstrated to have transforming potential (Singh et al. 2012). More complex patterns identified have included chains of balanced, copy-neutral rearrangements in prostate cancer (Berger et al. 2011) and chromosome catastrophe events (“chromothripsis”) in which tens to hundreds of rearrangements occur in single chromosome arms (Stephens et al. 2011).
Investigations at base-pair resolution have revealed enrichment of sequence microhomologies at rearrangement breakpoints, suggesting that nonhomologous end-joining can generate functionally important genomic lesions in cancer (Lawson et al. 2011; Drier et al. 2012). Such defects in the homologous recombination repair mechanism have been related to the susceptibility of tumor cells to treatment with poly-ADP ribose polymerase (PARP) inhibitors (Patel et al. 2011). Whether increased frequency of genomic breakpoint lesions provides a biomarker for clinical efficacy of PARP inhibition remains a topic of active research (Ledermann et al. 2012).
The Cancer Genome Atlas Research Network (TCGA) is providing comprehensive genomic characterization of hundreds of tumor samples to generate a detailed catalog of genomic abnormalities in cancer. Multimodal genomic profiles of ∼600 glioblastomas, including whole-genome sequence (n = 42) and mRNA sequence (n = 164) for subsets of the samples, have been made available. Here, we leverage those data sets plus DNA copy number profiles to characterize the landscape of intragenic breakpoints in GBM.
Results
Genomic rearrangements are associated with changes in DNA copy number
We conducted genomic rearrangement analysis on 42 pairs of TCGA GBM samples and matching normal using paired-end whole-genome sequencing. BreakDancer (version 1.12) (Chen et al. 2009) was used to identify somatic genic rearrangements, and output candidates were filtered for a minimal 5-kb distance between breakpoints for intragenic events to obtain high confidence (the Materials and Methods). In total, 183 genic rearrangements were found in 29 samples, ranging from one to 27 per sample (median, 3) (Supplemental Table S1). The 183 events were divided into three groups: (1) interchromosomal (n = 48), (2) intrachromosomal/intergenic (n = 95), and (3) intragenic (n = 40). To investigate the relationship between rearrangements and DNA dosage, we compared these events with their matching DNA copy number profiles. Among the three groups, 41.7%, 66.3%, and 50.0%, respectively, co-occurred with an intragenic copy number difference in at least one of the two breakpoints, an overall percentage of 56.3%. The majority (161 of 183, or 88.0%) of rearrangements were within 1 Mb of a somatically altered region detected by SNP 6.0 arrays (Supplemental Table S1).
The high concordance between genomic rearrangements and intragenic copy number alterations motivated us to perform a systematic scan of intragenic breakpoints using DNA copy number profiles available through TCGA. By precisely mapping the boundaries of each copy number segment to their genomic locations and applying a sample-specific threshold for copy number differences between adjacent segments (Supplemental Fig. S1), we identified 57,464 breakpoints in 537 GBMs. Of those, 15,488 (27.0%) fell in genic regions (Fig. 1A). The number of intragenic breakpoints per sample ranged from one to 252, with median and mean values of 25 and 29, respectively. Four samples showed distinctly higher numbers of breakpoints (Supplemental Fig. S2) due to a large number of copy number changes on a single chromosome, perhaps representing the chromothripsis phenomenon. To examine further the possibility of chromothripsis, we analyzed mRNA sequencing data available for one of the four samples using our Pipeline for RNA-seq Data Analysis (PRADA) (http://sourceforge.net/p/prada). Fourteen gene fusions were detected related to the chromosome where high frequency of intragenic breakpoints was found. In contrast, only three other fusions were found in the sample (Supplemental Fig. S3; Supplemental Table S2).
Figure 1.
The landscape of intragenic breakpoints in GBM. (A) Circos plot of intragenic breakpoints identified using copy number profiles of 537 GBMs. The outermost circle represents chromosomes and cytogenetic bands. The next circle represents significant copy number alterations identified using genomic identification of significant targets in cancer (GISTIC) (Beroukhim et al. 2007). Green indicates deletion, and red indicates amplification. The purple circle represents the frequency of intragenic breakpoints. The black text circle lists selected genes with frequent breakpoints, and the innermost circle lists known GBM driver genes. (B) Genes with frequent breakpoints (n ≥ 10) show greater length and lower GC content. Each dot represents a gene. Green means the gene is close to or in a deletion peak, and red means the same for an amplification peak. The asterisk means the gene is in a chromosome-fragile site. Interquartile reference GC content and gene length are indicated by purple and blue bars. (C) Linear correlation of the number of intragenic breakpoints and chromosome arm lengths. Red and green indicate whether the arm is frequently targeted by amplification or deletion determined by GISTIC.
To explore the nature of genes affected by intragenic breakpoints, we evaluated gene length and the guanine–cytosine (GC) content of all genes that were found to harbor a breakpoint in at least 10 samples (n = 152) (Fig. 1B) and checked those genes for somatic alteration peaks and common genomic fragile sites (Lukusa and Fryns 2008). Genes with recurrent breakpoints in GBM were, overall, significantly longer (P ≤ 2.2 × 10−16, Kolmogorov-Smirnov test) and had a lower GC content (P = 2.2 × 10−16, Kolmogorov-Smirnov test). For example, 17 of 40 genes adjacent to a focal deletion were >500 kb (P = 0.0004, Fisher's exact test). That association is consistent with the earlier observation that larger genes are more frequently targeted by copy number loss (Beroukhim et al. 2010). Using different threshold values for breakpoint occurrence in genes did not change the patterns, suggesting that they are intrinsic and robust (data not shown). We found no association between common chromosome fragile sites and intragenic breakpoints.
In total, 3055 of 6139 intragenic breakage events (49.8%) were identified in more than one sample. When tallying the number of intragenic breakpoints across the genome, we found a pattern related to somatic copy number alterations (Fig. 1A): Genes that were frequently found to harbor intragenic copy number differences were frequently affected by whole-gene gains and losses. That relationship was most significant for EGFR (n = 154) and tumor suppressors PTEN (n = 59), NF1 (n = 29), RB1 (n = 20), and cyclin-dependent kinase N2A (CDKN2A) (n = 11). Genes such as PTPRD (n = 50), CAMTA1 (n = 37), and LRP1B (n = 40) that cover large territory and have been suggested to have tumor suppressor functions ranked among the genes most frequently targeted (Supplemental Table S3; Roversi et al. 2006; Veeriah et al. 2009; Henrich et al. 2011).
Intragenic deletions in EGFR are frequent in GBMs. Most well known are the vIII form, in which exons 2–7 are deleted, and the C terminus deletion variant (Fenstermaker and Ciesielski 2000; Cho et al. 2011). To evaluate those intragenic events in EGFR more fully, we used data from RNA sequencing (n = 164) to detect transcript patterns consistent with intragenic deletions. Transcriptome sequencing data were available for 48 of the 154 samples in which an intragenic EGFR breakpoint was predicted on the bases of their copy number profile. Evidence was found for expression of the EGFRvIII variant in 22 of 34 samples predicted to harbor an EGFR intron 1 breakpoint, and a transcription pattern consistent with a C-terminal deletion was shown in nine of 17 cases associated with a chromosomal C-terminal breakpoint (Supplemental Table S4). However, this percentage was likely underestimated, as the C-terminal deletion that removes the C terminus was not identifiable by the abnormal junctions that we examined in RNA sequencing data.
An association between chromosome arm length and copy number alteration has previously been reported (Beroukhim et al. 2010), and we found a similar correlation between frequency of intragenic breakpoints and size of chromosome arm (r = 0.54) (Fig. 1C). Several chromosome arms, including 7p, 9p, 10q, 12q, and 1p, showed an increased frequency of intragenic breakpoint events, suggesting selective pressure for copy number alteration. The enrichments for 7p, 9p, and 10q were associated with alterations targeting EGFR, CDKN2A/B, and PTEN, respectively.
A 12q14-15 dense pattern is indicative of local chromosome instability
To investigate the enrichment of intragenic breakpoints on chromosome 12q (Fig. 1C), we binned the arm into segments of 1 Mb and counted the number of breakpoints in each bin. A region of ∼18 Mb around 12q14-15, in which 185 protein-coding genes (Supplemental Table S5) are present, showed distinctly higher numbers of breakpoints (Fig. 2A,B). We refer to this region as a 12q14-15 “breakpoint-enriched region” (BER). Two oncogenes previously noted to be amplified frequently in GBMs are in the 12q14-15 region: CDK4 and murine double minute 2 (MDM2). We assessed the presence of the BER in individual samples by integrating the number of breakpoints and the amplitude difference between neighboring segments to compute a score and performed a permutation test to compare this score with a range of scores from randomly selected segments. A permutation-based empirical approach was used to calculate a false discovery rate (FDR) of the resulting set of P-values. By setting the FDR to 0.1, we classified the GBM cohort into 24 GBMs showing a 12q14-15 BER pattern (Supplemental Table S6; the Materials and Methods) and 469 samples lacking the BER. Seven cases could not be reliably classified. Evaluating the number of genomic rearrangements inferred from whole-genome sequencing data, including structural variants with <5 kb between breakpoints, revealed an increased number of genic rearrangements among the 185 genes in five of the six BER samples with available whole-genome sequences and thus verified the DNA segment-joining model manifested by the DNA copy number-shattering pattern (Supplemental Tables S7, S8). In the sixth case (TCGA-06-0152), no structural variants where detected across the entire genome, and we speculate that a data quality issue prevented us from identifying 12q14-specific rearrangements. Supervised analysis of nine samples with available RNA sequencing data detected 124 transcript fusions involving the 185 genes located around the 12q14-15 cytoband. Of these, 63% of the gene partners were within the set of 185 genes; the other 37% were with genes at other chromosomal locations (Supplemental Table S9). The number of fusions per sample ranged from four to 31, with a median value of 10. Interestingly, similar copy number levels (difference of segment copy number, <0.3) were found for 69% of the amplified fusion transcript partner genes (after excluding transcripts with partner genes originating from the same copy number segment), implying a balanced cojoining of DNA segments (Supplemental Table S9). Existence of the pattern was further validated using DNA copy number profiles from an independent data set obtained through the Repository for Molecular Brain Neoplasia Data (REMBRANDT) (National Cancer Institute REMBRANDT Database, http://rembrandt.nci.nih.gov), in which we found that five of 188 samples (2.7%) harbored elevated numbers of intragenic breakpoints around chromosome 12q14-15 (Supplemental Fig. S4). The lower frequency in the REMBRANDT data set may result from a less dense DNA copy number array platform (Affymetrix 100K arrays) and cohort differences due to sample accrual criteria.
Figure 2.

A shattered copy number pattern is observed on 12q14-15. (A) Breakpoint frequency on 12q per 1-Mb bin, across 537 GBMs. (B) Integrative genomics viewer (IGV) copy number screenshot of the 18-Mb region in GBMs. Red and blue represent copy number gain and loss, respectively.
The observation that the amplification segments dispersed in the 12q14-15 BER have similar copy number levels in each individual sample suggests a bridging mechanism that connects the segments and enables coamplification. This phenomenon resembles double minutes or homogeneously staining regions, which have identical amplicon composition and functional consequences but differ in structure and the way of presence; e.g., double minutes are extrachromosomal, whereas the homogenously staining regions insert into chromosomes (Storlazzi et al. 2010). As a means of amplifying oncogenes, double minutes have previously been reported in GBM and other cancer types (Lalic et al. 2004; Thomas et al. 2004; Vogt et al. 2004; Kuttler and Mai 2007). Recently, complex rearrangements labeled “chromoplexy” have been described in prostate cancer. However, since these are copy number-neutral, the events observed here do not match the chromoplexy characteristics, although similar mechanisms may contribute to both abnormalities (Baca et al. 2013).
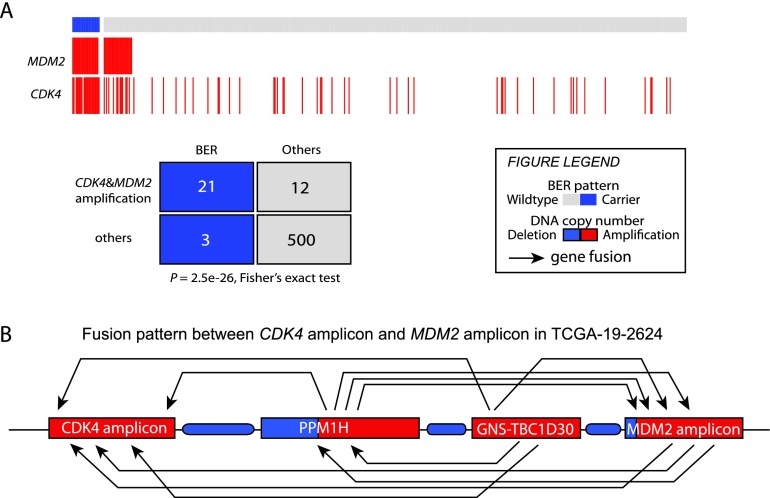
The 12q14-15 BER is associated with coamplification of CDK4 and MDM2
The two oncogenes in the 12q14-15 BER—CDK4 and MDM2—are among the most frequently amplified genes in GBMs (The Cancer Genome Atlas Research Network 2008). We found coamplification of CDK4 and MDM2 in 21 out of 24 samples that have the 12q14-15 BER pattern based on their copy number profiles (P = 2.5 × 10−26, Fisher's exact test) (Fig. 3A). That association was substantiated by transcript fusions between genes included in the CDK4 and MDM2 amplicon segments in four out of nine samples (Supplemental Table S9). For example, we identified 17 transcript fusions among the four amplicons in the 12q14-15 region between MDM2 and CDK4 in case TCGA-19-2624. Included were four transcript fusions involving genes of the MDM2 amplicon and CDK4 amplicon in both directions (Fig. 3B). Additionally, we found 12 cases in which both MDM2 and CDK4 were amplified but without clear evidence of the BER pattern. Inspection of RNA sequencing data showed that BER samples showed a significantly increased level of CDK4 and MDM2 transcription (Supplemental Fig. S7).
Figure 3.
The 12q14-15 BER samples show coamplification of CDK4 and MDM2. (A) Significant coamplification of CDK4 and MDM2 in BER samples. GISTIC thresholded copy number data in data freeze October 10, 2012, were used (n = 536). (B) A diagram shows transcript fusions in the 12q14-12q15 region in TCGA-19-2624. Red and blue represent amplification and deletion on DNA copy number.
To investigate the genomic context of samples harboring the 12q14-15 BER, we analyzed patterns of co-occurrence and mutual exclusivity using the TCGA catalog of genomic abnormalities. CDKN2A/B deletions, the most frequent focal deletions in GBMs, were found to be mutually exclusive with the region (P = 1.5 × 10−6, Fisher's exact test), perhaps a result of the interaction between those proteins in regulation of the RB1 cell cycle pathway. Furthermore, TP53 mutations were detected in statistically significantly fewer samples with the 12q14-15 BER pattern (P = 0.02, Fisher's exact test), potentially as a result of the inhibitory function of MDM2 on TP53. Of the 24 BER samples in our cohort, 11 showed focal EGFR amplification, a frequency that is similar to the frequency found in the general GBM population (45% vs 46%; P = 1.0, Fisher's exact test). We used the available RNA sequencing data to estimate expression levels and did not observe increased EGFR in samples harboring the 12q14-15 BER (Supplemental Fig. S6). Interestingly, fusion transcript analysis identified an MDM2-EGFR fusion transcript in one case (TCGA-19-2624) (Supplemental Table S9), and the matching DNA copy number profile showed an intragenic breakpoint in EGFR exon 1.
The 12q14-15 BER is associated with worse survival
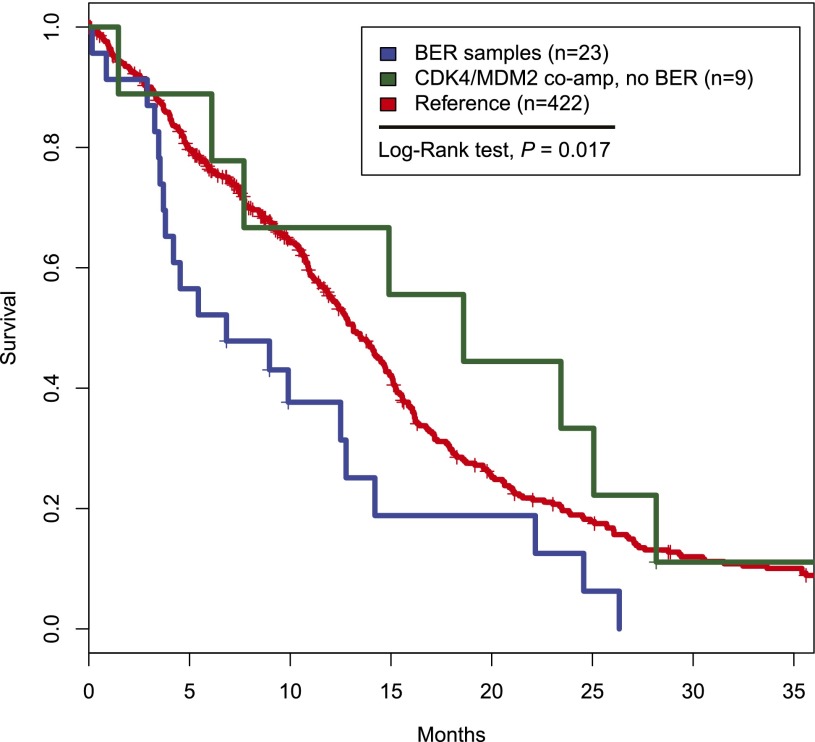
Next, we examined clinical parameters of the 12q14-15 BER-harboring samples. Median age at diagnosis was 60.7 yr, similar to that of the remainder of patients in the TCGA cohort (61.1 yr). Using available transcriptome profiles, we classified 23 of 24 samples into previously reported molecular subtypes (Verhaak et al. 2010) and found no significant association with any expression subtype (Proneural, n = 9; Neural, n = 7; Classical, n = 3; Mesenchymal, n = 4) (Supplemental Table S6), although there was a trend of more Proneurals (P = 0.08, Fisher's exact test). Clinical outcome annotation was available for 454 of the 493 classified cases and was used in survival analysis. Importantly, the 12q14-15 BER was associated with poor outcome (P = 0.007, log-rank test) (Fig. 4; Supplemental Fig. S5). The association with poor survival did not extend to samples that coamplified CDK4 and MDM2 but without further signs of 12q14-15 shattering (P = 0.29, log-rank test) (Fig. 4; Supplemental Fig. S5). G-CIMP cases were excluded from the outcome analysis because of their reported favorable outcome (Noushmehr et al. 2010).
Figure 4.
Kaplan-Meier survival curve of 12q14-15 BER samples and CDK4/MDM2 coamplified samples. GCIMP samples were excluded from the analysis because of their reported favorable outcomes. Of the 493 well-classified samples, 454 had clinical data that were used in the analysis.
FAF1 is a putative tumor suppressor gene in GBM
Chromosome arm 1p was a second outlier in terms of the number of intragenic breakpoints relative to arm length (Fig. 1C). There are two genes on chromosome 1p that showed frequent intragenic lesions: CAMTA1 and FAF1 (Fig. 1C; Supplemental Table S3). CAMTA1 has been reported to act as a tumor suppressor gene in gliomas (Henrich et al. 2011) and to fuse with WWTR1 in hemangioendotheliomas (Errani et al. 2012). FAF1 is adjacent to the tumor suppressor CDKN2C. It was first reported to potentiate FAS-induced apoptosis (Chu et al. 1995) and was found to play a role in pathogenesis of mesotheliomas (Altomare et al. 2009). Despite these reports, FAF1's function in GBMs is still elusive (Solomon et al. 2008), so we decided to analyze its function further.
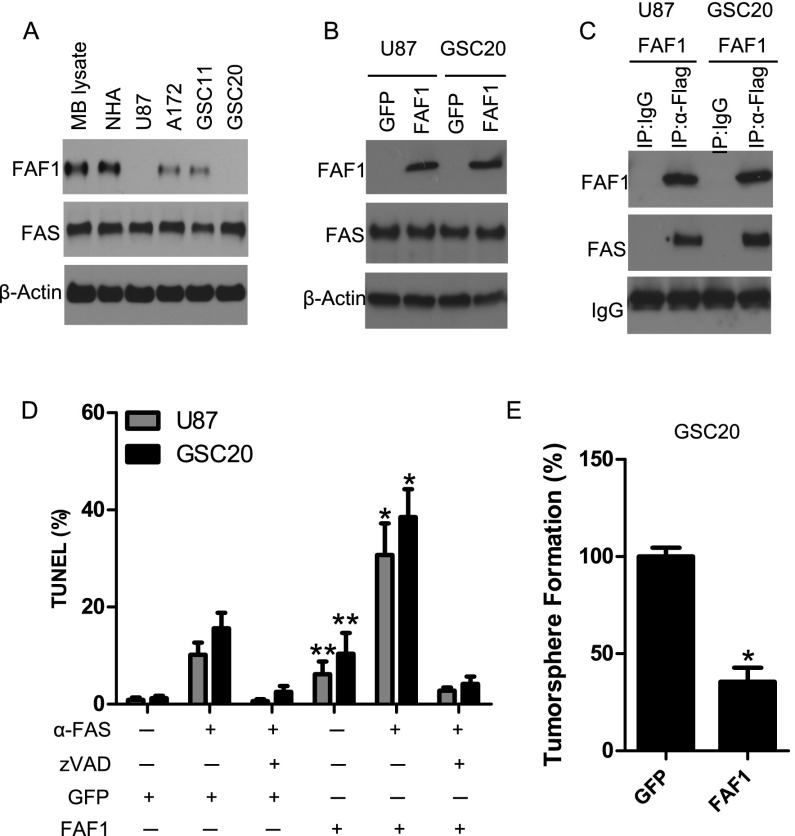
FAF1 intragenic breakpoints in our cohort (6.3%) were caused by deletion of the N terminus as a bystander effect of focal CDKN2C loss (Supplemental Figs. S8, S9A). Using RNA sequencing data, we found significantly decreased expression of FAF1 in the structurally disrupted cases (P = 4.53 × 10−10, t-test) (Supplemental Fig. S9B), possibly due to the loss of the promoter. To investigate the functional consequences of intragenic FAF1 breakpoints, we scanned copy number profiles of a panel of glioma cell lines, including two glioma stem cell lines (GSC11 and GSC20) (Supplemental Fig. S9C,D). Several cell lines, including U87 and GSC20, were found to have FAF1 N-terminal loss. The deletion status in U87 is further validated using recently published whole-genome sequencing data (Supplemental Fig. S9E; Clark et al. 2010). Western blotting using a C-terminal-specific antibody, which would be able to detect a trunctated FAF1, further validated the absence of FAF1 protein in these two cell lines (Fig. 5A).
Figure 5.
FAF1 intragenic break disrupts protein production and reduces FAS-mediated apoptosis. (A) Expression of endogenous FAF1 protein in the mouse brain (MB lysate), normal human astrocytes (NHA), established glioma cell lines, and glioma stem cell lines. A mouse monoclonal antibody was used to probe the C-terminal sequence of FAF1 protein. (B) Overexpression of the Flag-tagged FAF1 gene in glioma cells harboring N-terminal deletion of the FAF1 gene. Wild-type FAS levels are shown. Green fluorescent protein (GFP) was used as control. (C) FAF1 interacts with FAS in glioma cells. Cell lysate was immunoprecipitated by anti-Flag antibody. Isotype IgG was used as Immunoprecipitation control. (D) FAF1 enhances FAS-mediated induction of apoptosis in glioma cells. TUNEL assay was used to detect apoptotic cells following anti-FAS antibody or treatment with pan-caspase inhibitor zVAD. (*) P < 0.05 as compared with GFP control treated with anti-FAS antibody; (**) P < 0.05 as compared with GFP control. (E) FAF1 impairs the tumorsphere formation of glioma-initiating cells. GSC20 cells stably expressing GFP or FAF1 were dissociated into single cells and grown for 7 d. Tumorspheres in the GFP group were treated as 100%. (*) P < 0.05 as compared with GFP control.
We next transfected FAF1 into U87 and GSC20 and performed coimmunoprecipitation to show that FAF1 interacted with its natural ligand, FAS, in these cells (Fig. 5B,C). In both cell lines, we measured the levels of apoptotic cells through imaging of terminal deoxynucleotidyl transferase dUTP nick end-labeling (TUNEL) assays. The results show that cells with transfected FAF1 in the presence of the wild-type levels of FAS1 significantly enhanced the induction of apoptosis (Fig. 5D). Furthermore, transfecting FAF1 in the GSC20 impaired the tumorsphere formation of this glioma stem cell line (Fig. 5E).
Exome sequencing analysis of 291 TCGA cases found one somatic splicing site mutation in FAF1, suggesting that copy number deletion and intragenic disruption are the major mechanisms to abrogate its expression. No FAF1 rearrangement was found by our BreakDancer analysis. Given the adjacent position with CDKN2C, simultaneous loss of FAF1 may confer glioma cells with increased fitness in gliomagenesis and progression.
Discussion
In this study, we performed a multidimensional genomic analysis of intragenic breakpoints and associated gene fusions and rearrangements in GBM. The analyses were based on extensive data from TCGA's GBM project, supplemented with information from the REMBRANDT study, experiments with glioma cell lines, and the literature.
We found that occurrence of intragenic breakpoints is positively correlated with chromosome arm length but with several exceptions. Those exceptions appear to reflect defects that provide a growth advantage to the cancer cell or prevent its apoptotic death, as exemplified by chromosome arms 7p, 9p, and 10q (which harbor EGFR, CDKN2A/B, and PTEN, respectively). The dramatically increased rate of intragenic breakpoints associated with 12q led us to the identification of a shattered chromosome pattern (12q14-15 BER) in the DNA copy number profiles of ∼5% of non-GCIMP GBMs. Combining that analysis with data from RNA sequencing and whole-genome sequencing found elevated rearrangements and fusions in the region, suggesting local chromosome instability. We confirmed the 12q14-15 BER pattern in the independent REMBRANDT Database (National Cancer Institute, http://rembrandt.nci.nih.gov). Importantly, the presence of the 12q14-15 BER correlated with a relatively adverse outcome. It was also linked with simultaneous amplification of CDK4 and MDM2. Intriguingly, however, we found that coamplification of CDK4 and MDM2 without the context of local chromosome shattering do not correlate with clinical survival. Whether this local instability directly impacts tumorigenesis or is a feature of tumor cells with a high level of plasticity—and therefore an increased ability to adapt to chemotherapy- and radiotherapy-induced changes—remains a topic for further investigation.
Recent identification of high-frequency and complex genomic rearrangement events labeled as chromothripsis and chromoplexy revealed the enormous intricacy of the cancer genome (Stephens et al. 2011; Baca et al. 2013). Oscillating copy number patterns similar to the 12q14-15 BER were observed as concomitant with such rearrangements and have been suggested as a hallmark of chromothripsis (Korbel and Campbell 2013). TP53 mutation, a feature of genomic instability, was found to be associated with chromothripsis in Sonic Hedgehog medulloblastoma brain tumor and acute myeloid leukemia (Rausch et al. 2012). Our analysis, however, did not find any somatic TP53 mutations in 12q14-15 BER cases. We speculate that this mutual exclusivity may result from another feature of these cases; e.g., amplification of MDM2 and CDK4 due to the regulatory role of MDM2 in TP53 pathway. Those amplifications occur in a mutually exclusive fashion with deregulation of other members of the RB1 and TP53 pathways (Ciriello et al. 2011). Despite the involvement of MDM2 and CDK4, whether the 12q14-15 BER pattern is involved in gliomagenesis or progression or is a marker of small-scale chromosome shattering requires additional studies. In the first scenario, a dual inhibition of the MDM2 and CDK4 oncogenes may specifically benefit patients with the 12q14 BER pattern. Targeted inhibitors of CDK4 and MDM2 are in clinical development, and the presence of amplification in the context of double minutes should be considered when the clinical response is being evaluated (Lapenna and Giordano 2009; Liu et al. 2009). Otherwise, the driver of local instability, which is unlikely to be TP53 mutation due the absence of such mutations in BER samples, is to be identified.
Recent reports have suggested that chromothripsis occurs early in tumorigenesis and may thus be clonal; i.e., present in all cancer cells (Stephens et al. 2011; Rausch et al. 2012). Several groups have reported intratumoral heterogeneity in GBM, among others, showing that receptor tyrosine kinase amplifications such as of EGFR, PDGFRA, and MET are present only in subsets of glioma cells (Snuderl et al. 2011; Szerlip et al. 2012; Sottoriva et al. 2013). Our current data set does not allow discrimination between the clonal and subclonal presence of the 12q14-15 BER pattern, but this question could be answered through studies of multiple spatially separate samples of the same tumor (Gerlinger et al. 2012).
The results of our analysis suggest that intragenic disruption of genes adjacent to focal copy number alterations may have functional consequences and is able to provide tumor cells with a proliferative advantage. For example, we show here that focal deletion of CDKN2C converges with disruption of its neighbor gene, FAF1, which negatively impacts the apoptosis response. This finding provides further evidence for the hypothesis that focal DNA copy number gains and losses, such as those frequently found in GBMs, generate cancer-specific vulnerabilities beyond the target gene of the amplification or deletion.
In conclusion, the genome-wide analysis of intragenic breakpoints presented here described widespread heterogeneity, such as the presence of the chr12q14-15 local instability phenotype and breakpoints disrupting tumor suppressor candidate genes such as FAF1. Our findings complement and extend current literature on the genomic landscape of GBMs and provide a new angle from which to view the process of gliomagenesis as well as the possibilities for more effective therapy of subsets of this devastating disease.
Materials and methods
Tumor samples and GBM cell lines
Glioblastomas and matched normal samples were collected and processed by TCGA as described elsewhere. All TCGA data used in this study were included in the data freeze as of October 10, 2012, curated by the GBM working group of TCGA. RNA sequencing data and whole-genome sequencing data were deposited in the Cancer Genomics Hub database (https://cghub.ucsc.edu). GBM cell lines were obtained from the Brain Tumor Center, MD Anderson Cancer Center. Glioma cell line copy number data were downloaded from Cancer Cell Line Encyclopedia (Barretina et al. 2012).
Genomic rearrangement and gene fusion analysis
Somatic gene rearrangements were identified by BreakDancer (version 1.12) (Chen et al. 2009) using whole-genome sequencing data on 42 pairs of GBM samples and matched normal. Rearrangements that had 15 or more supporting reads and were not found in matched normal samples were obtained as somatic rearrangement events (referred to as the unsupervised rearrangement list). To correlate genic rearrangements with copy number data, we further filtered the list by removing intragenic events that had junction distance <5000 base pairs (bp), resulting in 183 events. The latter list was referred to as a supervised list and was considered a high-confidence event. Gene fusions were analyzed by PRADA (http://sourceforge.net/p/prada). A supervised module in PRADA named General User-Defined Supervise Search for Fusion Transcript (GUESS-ft) was used to perform supervised searches for fusion events.
Intragenic breakpoint scanning from copy number data
Affymetrix SNP 6.0 arrays for 537 GBMs were segmented using the circular binary segmentation algorithm (Venkatraman and Olshen 2007). Gene and exon boundaries were downloaded from the University of California at Santa Cruz Table Browser (http://genome.ucsc.edu) and were subsequently converted to range data structures using the R package IRanges. Each gene region was aligned to copy number segments. When one gene was found to overlap with more than one segment and the related copy number levels differed above a sample-specific threshold value, the gene was considered to harbor an intragenic breakpoint. Copy number threshold values were selected on a sample-by-sample basis. Specifically, two figures were generated for each sample to help determine the threshold: (1) a scatter plot of all intragenic segment-level differences and (2) a copy number-level histogram for all genes in the sample. The figures and the sample-specific cutoff values are available at http://odin.mdacc.tmc.edu/∼rverhaak/gbmbreaks. Tumor purity and ploidy were estimated using the ABSOLUTE algorithm (Carter et al. 2012) and are available through TCGA.
Identifying samples with the 12q14-15 BER
To stratify 12q14-15 BER samples, a sample × bin matrix was computed. Each bin represented a 1-Mb region on chromosome 12q. For one sample, the score of each bin was calculated as the sum of all segment differences in the bin that pass a sample-specific threshold. The sum of scores for the 18-Mb region was used as the aggregated score for the sample. To determine statistical significance, the score matrix was permutated by the bin index 10,000 times, and random scores were recomputed to approximate the background distribution. Sample-specific P-values were computed as the proportion of random scores larger than or equal to the real score in each row (sample). For multiple testing, an empirical FDR was computed for each frequency r as a/b, where a refers to the proportion of random scores that are larger than or equal to r using all permutation data, and b refers to the proportion of real scores from all samples that are larger than or equal to r.
Validation data set
Validation data from Affymetrix 100K arrays were downloaded from the REMBRANDT Database (National Cancer Institute, http://rembrandt.nci.nih.gov). Raw CEL files were processed using the Aroma package (Bengtsson et al. 2009), and probe-level copy numbers were then segmented using the circular binary segmentation algorithm (Venkatraman and Olshen 2007). In total, 188 GBMs were included in the data set.
Association analysis of 12q14-15 BER samples with copy number alterations and clinical outcome
Copy number levels were categorized into five levels (homozygous deletion, hemizygous deletion, copy number-neutral, low-level gain, and high-level gain) by the genomic identification of significant targets in cancer (GISTIC) algorithm (Beroukhim et al. 2007). Mutation data were obtained from TCGA and categorized as binary for each gene. The value was set to 1 if the gene had a nonsilent mutation. We tested associations of high-level gain, homozygous deletions, and gene mutations with 12q BER samples using Fisher's exact test. Clinical data were obtained from TCGA. The log-rank test and Kaplan-Meier analysis were used to test the survival relevance of the 12q14-15 BER samples.
Survival analysis
Survival analysis was done using the “Survival” package in R. Clinical information on 543 patients was included in the October 10, 2012, data freeze. Exclusion of GCIMP cases, unclassified cases, and cases that were not included in copy number analysis resulted into 454 cases that had clinical data, including 23 12q14-15 BER samples. Survival curves were estimated by Kaplan-Meier analysis and trimmed at 36 mo. The statistical significance of survival differences was calculated using the log-rank test in the “Survival” package.
Western blot analysis
Cells were harvested in a lysis solution as previously described (Fu et al. 2013) and subjected to Western blotting. Membranes were probed with the following primary antibodies: anti-FAS and anti-cleaved Caspase-3 (all from Cell Signaling). Mouse anti-FAF1 (recombinant fragment “EAIRLSLEQALPPEPKEENAEPVSKLRIRTPSGEFLERRFLASNKLQIVF-DFVASKGFPWDEYKLLSTFPRRDVTQLDPNKSLLEVKLFPQETLFLEAKE,” corresponding to amino acids 551–651 of human FAF1), was purchased from Abcam (ab56940). β-Actin and Flag antibodies were from Sigma-Aldrich.
Lentiviral vectors and virus production
Lentiviral vectors encoding nontargeting scramble control or shFAF1 was purchased from OpenBiosystem. Lentiviral vectors encoding green fluorescent protein (GFP) or FAF1 were purchased from Genecopoeia. The VSV-G pseudotyped lentiviral vectors were produced by transient cotransfection of three plasmids into 293FT cells. Viral supernatants were further enriched by Millipore centrifugal filter units (10K).
Immunoprecipitation analysis
Cells were harvested and lysed in immunoprecipitation buffer on ice for 20 min. The immunoprecipitation buffer consisted of 50 mM Tris (pH 7.5), 100 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1% NP-40, 5% glycerol, freshly added 1× Complete Mini protease inhibitor cocktail (Roche), 2 mM phenylmethylsulfonyl fluoride, and 2 mM NaF and 2 mM NaVO4 as phosphatase inhibitors. Cell extracts were clarified by centrifugation at 10,000 rpm for 5 min at 4°C. Protein concentrations were determined by the Bradford method with the Bio-Rad protein assay reagent. Immunoprecipitation assays were performed as described previously (Fu et al. 2013). Briefly, 300 μg of protein was incubated with 10 μL of anti-Flag M2 or control IgG agarose gel beads. The beads were washed three times with immunoprecipitation buffer and resuspended in sodium dodecyl sulfate sample buffer. Immunocomplexes were analyzed by immunoblotting.
TUNEL assay
Apoptosis was determined using the TUNEL method using in situ cell death detection reagent (Roche Applied Science). The percentage of TUNEL-labeled cells in each section was determined at a magnification of 400× by counting 500 cells in a randomly selected field.
Tumorsphere formation assay
For the tumorsphere-forming essay, equal numbers of cells infected with lentivirus containing GFP or FAF1 were seeded at low cell density (four cells per microliter) in wells of a 96-well plate. The total number of newly formed neurospheres was counted after 7 d in culture.
Acknowledgments
We thank all members of the TCGA GBM working group. This work is supported by award numbers 5 P50 CA127001 and 5 P50 CA083639-12 from the National Cancer Institute (NCI) to R.G.W., NCI grant number CA143883 (MD Anderson Genome Data Analysis Center) to J.N.W., and generous funding from the H.A. and Mary K. Chapman Foundation and the Michael and Susan Dell Foundation (honoring Lorraine Dell) to J.N.W. The results published here are in whole or part based on data generated by The Cancer Genome Atlas project established by the NCI and National Human Genome Research Institute (NHGRI). Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.213686.113.
References
- Altomare DA, Menges CW, Pei J, Zhang L, Skele-Stump KL, Carbone M, Kane AB, Testa JR 2009. Activated TNF-α/NF-κB signaling via down-regulation of Fas-associated factor 1 in asbestos-induced mesotheliomas from Arf knockout mice. Proc Natl Acad Sci 106: 3420–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, Macdonald TY, Ghandi M, et al. 2013. Punctuated evolution of prostate cancer genomes. Cell 153: 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, Sougnez C, Voet D, Saksena G, Sivachenko A, et al. 2011. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet 43: 964–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson H, Wirapati P, Speed TP 2009. A single-array preprocessing method for estimating full-resolution raw copy numbers from all Affymetrix genotyping arrays including GenomeWideSNP 5 & 6. Bioinformatics 25: 2149–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. 2011. The genomic complexity of primary human prostate cancer. Nature 470: 214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. 2007. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci 104: 20007–20012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. 2012. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 30: 413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. 2009. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat Methods 6: 677–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Hahn WC, Getz G, Meyerson M 2011. Making sense of cancer genomic data. Genes Dev 25: 534–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Pastorino S, Zeng Q, Xu X, Johnson W, Vandenberg S, Verhaak R, Cherniack AD, Watanabe H, Dutt A, et al. 2011. Glioblastoma-derived epidermal growth factor receptor carboxyl-terminal deletion mutants are transforming and are sensitive to EGFR-directed therapies. Cancer Res 71: 7587–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu K, Niu X, Williams LT 1995. A Fas-associated protein factor, FAF1, potentiates Fas-mediated apoptosis. Proc Natl Acad Sci 92: 11894–11898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Cerami EG, Sander C, Schultz N 2011. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res 22: 398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MJ, Homer N, O'Connor BD, Chen Z, Eskin A, Lee H, Merriman B, Nelson SF 2010. U87MG decoded: The genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet 6: e1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drier Y, Lawrence MS, Carter SL, Stewart C, Gabriel SB, Lander ES, Meyerson M, Beroukhim R, Getz G 2012. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res 23: 228–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, Agarwalla PK, Chheda MG, Campos B, Wang A, et al. 2012. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev 26: 756–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errani C, Sung YS, Zhang L, Healey JH, Antonescu CR 2012. Monoclonality of multifocal epithelioid hemangioendothelioma of the liver by analysis of WWTR1–CAMTA1 breakpoints. Cancer Genet 205: 12–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenstermaker RA, Ciesielski MJ 2000. Deletion and tandem duplication of exons 2-7 in the epidermal growth factor receptor gene of a human malignant glioma. Oncogene 19: 4542–4548 [DOI] [PubMed] [Google Scholar]
- Fu J, Koul D, Yao J, Wang S, Yuan Y, Colman H, Sulman EP, Lang FF, Alfred Yung WK 2013. Novel HSP90 inhibitor NVP-HSP990 targets cell-cycle regulators to ablate olig2-positive glioma tumor-initiating cells. Cancer Res 73: 3062–3074 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. 2007. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev 21: 2683–2710 [DOI] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. 2012. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich KO, Bauer T, Schulte J, Ehemann V, Deubzer H, Gogolin S, Muth D, Fischer M, Benner A, Konig R, et al. 2011. CAMTA1, a 1p36 tumor suppressor candidate, inhibits growth and activates differentiation programs in neuroblastoma cells. Cancer Res 71: 3142–3151 [DOI] [PubMed] [Google Scholar]
- Korbel JO, Campbell PJ 2013. Criteria for inference of chromothripsis in cancer genomes. Cell 152: 1226–1236 [DOI] [PubMed] [Google Scholar]
- Kuttler F, Mai S 2007. Formation of non-random extrachromosomal elements during development, differentiation and oncogenesis. Semin Cancer Biol 17: 56–64 [DOI] [PubMed] [Google Scholar]
- Lalic H, Volavsek C, Radosevic-Stasic B 2004. Chromosomal instability and double minute chromosomes in a breast cancer patient. Acta Med Okayama 58: 51–58 [DOI] [PubMed] [Google Scholar]
- Lapenna S, Giordano A 2009. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov 8: 547–566 [DOI] [PubMed] [Google Scholar]
- Lawson AR, Hindley GF, Forshew T, Tatevossian RG, Jamie GA, Kelly GP, Neale GA, Ma J, Jones TA, Ellison DW, et al. 2011. RAF gene fusion breakpoints in pediatric brain tumors are characterized by significant enrichment of sequence microhomology. Genome Res 21: 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott C, Meier W, Shapira-Frommer R, Safra T, et al. 2012. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med 366: 1382–1392 [DOI] [PubMed] [Google Scholar]
- Liu W, He L, Ramirez J, Ratain MJ 2009. Interactions between MDM2 and TP53 genetic alterations, and their impact on response to MDM2 inhibitors and other chemotherapeutic drugs in cancer cells. Clin Cancer Res 15: 7602–7607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukusa T, Fryns JP 2008. Human chromosome fragility. Biochim Biophys Acta 1779: 3–16 [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AG, Sarkaria JN, Kaufmann SH 2011. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci 108: 3406–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. 2012. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148: 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roversi G, Pfundt R, Moroni RF, Magnani I, van Reijmersdal S, Pollo B, Straatman H, Larizza L, Schoenmakers EF 2006. Identification of novel genomic markers related to progression to glioblastoma through genomic profiling of 25 primary glioma cell lines. Oncogene 25: 1571–1583 [DOI] [PubMed] [Google Scholar]
- Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S, et al. 2012. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337: 1231–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, Akhavanfard S, Cahill DP, Aldape KD, Betensky RA, et al. 2011. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 20: 810–817 [DOI] [PubMed] [Google Scholar]
- Solomon DA, Kim JS, Jenkins S, Ressom H, Huang M, Coppa N, Mabanta L, Bigner D, Yan H, Jean W, et al. 2008. Identification of p18 INK4c as a tumor suppressor gene in glioblastoma multiforme. Cancer Res 68: 2564–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S 2013. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci 110: 4009–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi CT, Lonoce A, Guastadisegni MC, Trombetta D, D'Addabbo P, Daniele G, L'Abbate A, Macchia G, Surace C, Kok K, et al. 2010. Gene amplification as double minutes or homogeneously staining regions in solid tumors: Origin and structure. Genome Res 20: 1198–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. 2005. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352: 987–996 [DOI] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, et al. 2012. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22: 425–437 [DOI] [PubMed] [Google Scholar]
- Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, et al. 2012. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci 109: 3041–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L, Stamberg J, Gojo I, Ning Y, Rapoport AP 2004. Double minute chromosomes in monoblastic (M5) and myeloblastic (M2) acute myeloid leukemia: Two case reports and a review of literature. Am J Hematol 77: 55–61 [DOI] [PubMed] [Google Scholar]
- Veeriah S, Brennan C, Meng S, Singh B, Fagin JA, Solit DB, Paty PB, Rohle D, Vivanco I, Chmielecki J, et al. 2009. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc Natl Acad Sci 106: 9435–9440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman ES, Olshen AB 2007. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 23: 657–663 [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt N, Lefevre SH, Apiou F, Dutrillaux AM, Cor A, Leuraud P, Poupon MF, Dutrillaux B, Debatisse M, Malfoy B 2004. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. Proc Natl Acad Sci 101: 11368–11373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Chheda MG, Verhaak RG 2012. Studying a complex tumor: Potential and pitfalls. Cancer J 18: 107–114 [DOI] [PMC free article] [PubMed] [Google Scholar]