Here, Elbarbary et al. define a role for the dsRNA-binding protein Staufen 1 (STAU1) in regulating mRNAs containing inverted repeat Alu elements (IRAlus) within their 3′ untranslated regions (UTRs). The investigators delineate a mechanism whereby STAU1 augments the nuclear export—and enhances the translation—of 3′ UTR IRAlus mRNAs. This study addresses a new function for IRAlus and STAU1 in mRNA export and global translation and also demonstrates a role for STAU1 during myogenesis.
Keywords: STAU1, inverted repeat Alu elements, p54nrb, nuclear export, PKR, translation, myogenesis
Abstract
For a number of human genes that encode transcripts containing inverted repeat Alu elements (IRAlus) within their 3′ untranslated region (UTR), product mRNA is efficiently exported to the cytoplasm when the IRAlus, which mediate nuclear retention, are removed by alternative polyadenylation. Here we report a new mechanism that promotes gene expression by targeting mRNAs that maintain their 3′ UTR IRAlus: Binding of the dsRNA-binding protein Staufen1 (STAU1) to 3′ UTR IRAlus inhibits nuclear retention so as to augment the nuclear export of 3′ UTR IRAlus-containing mRNAs (IRAlus mRNAs). Moreover, we found that 3′ UTR IRAlus-bound STAU1 enhances 3′ UTR IRAlus mRNA translation by precluding protein kinase R (PKR) binding, which obviates PKR activation, eukaryotic translation initiation factor 2α (eIF2α) phosphorylation, and repression of global cell translation. Thus, STAU1 binding to 3′ UTR IRAlus functions along with 3′ UTR IRAlus-mediated nuclear retention to suppress the shutdown of cellular translation triggered by PKR binding to endogenous cytoplasmic dsRNAs. We also show that a changing STAU1/PKR ratio contributes to myogenesis via effects on the 3′ UTR IRAlus of mRNA encoding the microRNA-binding protein LIN28.
Alu elements, which are a type of short interspersed element (SINE), are the most abundant repetitive sequences in human DNA. At ∼1.4 million copies per cell, they constitute >10% of the human genome (Lander et al. 2001). While initially thought of as “junk” DNA, Alu elements are often transcribed and can function within RNAs to regulate primate gene expression by diverse mechanisms (Walters et al. 2009; Berger and Strub 2011). In one role, certain intronic Alu elements create a splice donor or splice acceptor site within pre-mRNAs (for review, see Walters et al. 2009). Additionally, single Alu elements within the 3′ untranslated region (UTR) of translationally active mRNAs can trigger STAU1-mediated mRNA decay (SMD) when they base-pair with a partially complementary Alu element of a long noncoding RNA (lncRNA) (Gong and Maquat 2011).
Apart from the narrowly defined intermolecular Alu element-formed STAU1-binding sites (SBSs) and the 19-base-pair (bp) intramolecular SBS formed within the 3′ UTR of ADP ribosylation factor 1 (ARF1) mRNA (Kim et al. 2005, 2007), little is known about what defines a mammalian cell SBS. While existing studies of intermolecular SBSs have been limited to sequences containing two or more consecutive nucleotide mismatches, SBSs are likely to be more tolerant of mismatches depending on the lengths of their perfectly duplexed regions. Even though STAU1 binds dsRNA with low sequence specificity in vitro (Wickham et al. 1999), it may manifest a preference for certain sequences given that adenosine deaminase, RNA-specific, B1 (ADAR2; which, like STAU1, contains dsRNA-binding domains) recognizes certain nucleotides in the minor groove of dsRNA (Stefl et al. 2010). The ability of STAU1 to dimerize if not multimerize (Martel et al. 2010; Gleghorn et al. 2013) may also influence binding specificity, especially to longer SBSs.
Multiple Alu elements can also reside within RNA polymerase II (Pol II) transcripts. Inverted repeat Alu elements (IRAlus) that base-pair with one another within RNAs undergo adenosine-to-inosine (A-to-I) editing at multiple sites by ADAR proteins (e.g., see Athanasiadis et al. 2004; Levanon et al. 2004). In fact, >90% of ADAR-mediated editing in human cells occurs at IRAlus. Given the length of Alu elements (∼300 bp) and their high (>70%) similarity (Sen et al. 2006), the coexistence of inverted Alu elements within a 3′ UTR often results in the formation of imperfect duplexes of >100 bp (e.g., see Bass 2002; Deininger 2011). In human (h) cells, mRNAs with edited 3′ UTR IRAlus are retained within nuclei to varying degrees by their association with paraspeckles, the formation of which depends on the nuclear lncRNA hNEAT1 and its binding partner, p54nrb (Prasanth and Spector 2007; Chen and Carmichael 2008). Considering that STAU1 exists in both the cytoplasm and nucleus (Martel et al. 2006) and SBSs can consist of imperfectly base-paired Alu elements residing in trans (Gong and Maquat 2011), we hypothesized that nuclear 3′ UTR IRAlus might also bind STAU1. Additionally, we aimed to address the long-standing issue of how cytoplasmic IRAlus-containing mRNAs (IRAlus mRNAs) evade the actions of protein kinase R (PKR), another dsRNA-binding protein that senses cytoplasmic viral dsRNA in infected cells and provides a means to inhibit cellular and, by so doing, viral translation (Chen and Carmichael 2008; Hundley and Bass 2010).
Here we demonstrate that 3′ UTR IRAlus indeed constitute SBSs. We show that STAU1 binding to nuclear 3′ UTR IRAlus reduces nuclear retention by competing with p54nrb binding and, to some extent, ADAR1 binding without significantly decreasing A-to-I editing. As a consequence, 3′ UTR IRAlus mRNA is exported to the cytoplasm, where bound STAU1 activates its translation by precluding the binding of PKR, thereby inhibiting (1) PKR activation, (2) the resulting PKR-mediated phosphorylation of eukaryotic translation initiation factor 2α (eIF2α), and (3) global cell translational repression. Our findings indicate that STAU1 regulates the export and translation of 3′ UTR IRAlus mRNAs and also obviates a PKR-mediated innate immune response to cytoplasmic 3′ UTR IRAlus. We demonstrate the importance of STAU1 and PKR to the differentiation of human primary myoblasts (MBs) via effects on the 3′ UTR IRAlus of mRNA for the microRNA-binding protein LIN28.
Results
STAU1 increases the amount of protein produced by 3′ UTR IRAlus mRNAs
Several human mRNAs that contain 3′ UTR IRAlus are sequestered within nuclei by complexes containing the 54-kDa nuclear RNA-binding protein p54nrb, PSF, paraspeckle component 1 (PSPC1), and hNEAT1 RNA; thus, these mRNAs are inefficiently exported to the cytoplasm (Chen and Carmichael 2008, 2009; Bond and Fox 2009). Supporting the possibility that 3′ UTR IRAlus might constitute SBSs, of 333 3′ UTR IRAlus mRNAs computationally identified by Chen et al. (2008), 23% (77 out of 333) were reported to coimmunoprecipitate with hemagglutinin-tagged STAU155 (STAU155-HA3) in four independently performed microarray analyses of human embryonic kidney 293T (HEK293T) cell transcripts (Supplemental Table S1; Furic et al. 2008). In other words, of the 1384 HEK293T cell mRNAs found to coimmunoprecipitate with STAU155-HA3, 5.6% (77 out of 1384) contain at least one 3′ UTR IRAlus.
Consistent with STAU1 binding to SBSs within nuclei, STAU155-HA3 coimmunoprecipitated with not only HEK293T cell ARF1 mRNA, but also its intron-containing pre-mRNA (Supplemental Fig. S1A,B). Thus, it is possible that STAU1 begins to bind 3′ UTR IRAlus mRNAs within nuclei so as to inhibit their nuclear retention and allow their export to the cytoplasm.
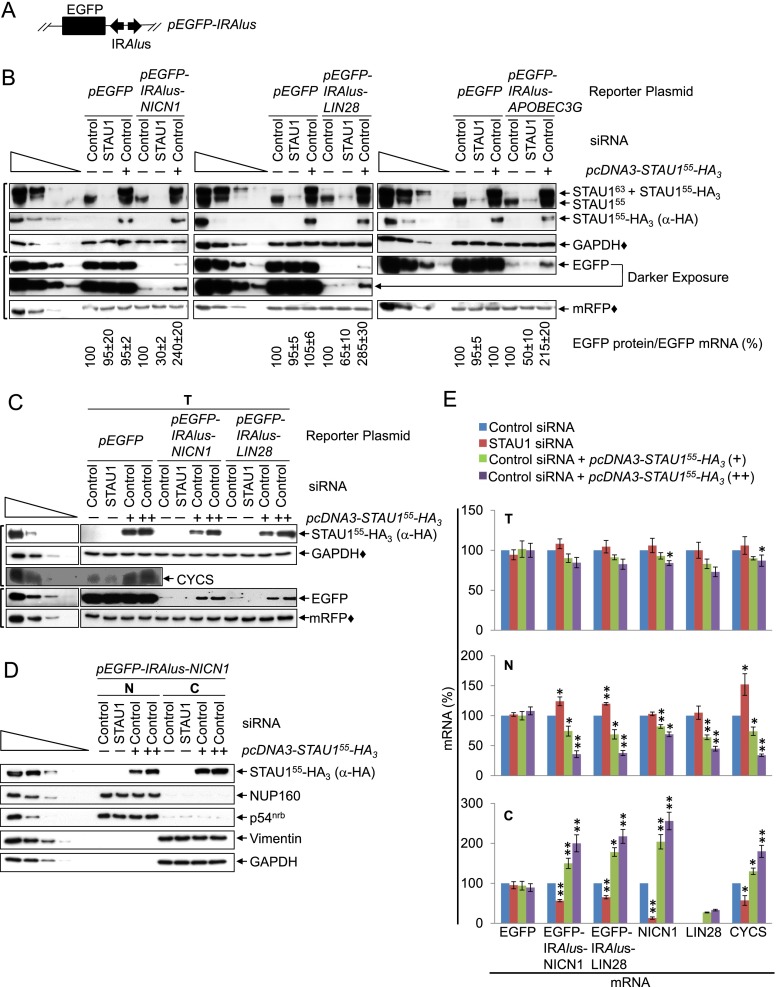
To test this hypothesis, cultured HEK293T cells were transiently transfected with (1) a pmRFP reference plasmid (Campbell et al. 2002) that produces monomeric red fluorescent protein (mRFP) and (2) a pEGFP-IRAlus-NICN1, pEGFP-IRAlus-LIN28, pEGFP-IRAlus-APOBEC3G, or, as a negative control, pEGFP reporter plasmid (Chen et al. 2008), each of which produces monomeric enhanced green fluorescent protein (EGFP) (Shcherbo et al. 2007) from a translational reading frame that resides upstream of, respectively, the 3′ UTR IRAlus of NICN1 (Nicolin1) mRNA or LIN28 mRNA, the intronic IRAlus of APOBEC3G (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G) pre-mRNA (Fig. 1A), or no IRAlus. We used a completely heterologous ORF (EGFP) upstream of each IRAlus in this and subsequent experiments to confirm that the results obtained are due to the IRAlus and not other mRNA sequences. Furthermore, we extended our analyses of reporter mRNAs to endogenous IRAlus mRNAs. The IRAlus in the three EGFP-IRAlus reporter mRNAs are extensively edited and retain their host mRNAs within nuclei, repressing EGFP expression in HEK293T cells (Chen et al. 2008). To assess the effects of variations in STAU1 abundance on the levels of IRAlus mRNAs, we subsequently titrated STAU1 dosage by transfecting cells with control siRNA or STAU1 siRNA (Kim et al. 2005) together with either the effector plasmid pcDNA3-STAU155-HA3 (Kim et al. 2005), which produces human STAU155-HA3, or pcDNA3-HA, which produces only HA.
Figure 1.
STAU1 increases the production of EGFP from EGFP-IRAlus mRNAs by augmenting nuclear mRNA export to the cytoplasm. (A) Diagram of the pEGFP-IRAlus reporter plasmids. (Black box) EGFP translational reading frame; (diverging arrows) 3′ UTR IRAlus. (B) Western blotting using lysates of HEK293T cells (8 × 106 per 100-mm dish) that had been transiently transfected with 2.5 μg of pmRFP and 2.5 μg of the designated pEGFP reporter, 1 d later with 50 nM control siRNA or STAU1 siRNA, and 2 d later with 10 μg of pcDNA3-HA (−) or pcDNA3-STAU155-HA3 (+). EGFP levels were normalized to the level of mRNA from which they derived (Supplemental Fig. S1C), and the normalized level in the presence of control siRNA + pcDNA3-HA is defined as 100. STAU155 and STAU163 are, respectively, 55-kDa and 63-kDa isoforms of STAU1 that are the products of alternative splicing (Wickham et al. 1999). Here and elsewhere, each bracket to the left denotes blots loaded so the amounts of reference protein, marked with a solid diamond (♦), were comparable. (C) Western blotting of lysates of HEK293T cells (1.8 × 107 per 150-mm dish) that had been transiently transfected with 5 μg of pmRFP and 5 μg of the specified pEGFP reporter, 1 d later with 50 nM control siRNA or STAU1 siRNA, and 2 d later with pcDNA3-HA (−) or 15 μg (+) or 25 μg (++) of pcDNA3-STAU155-HA3 (where pcDNA3-HA was added to keep the total micrograms of introduced plasmid constant). See Supplemental Table S3 for quantitations. (D) Western blotting of nuclear (N) and cytoplasmic (C) fractions prepared from cells analyzed in C. (E) Histograms of RT-qPCR quantitations (Supplemental Fig. S1D,I; Supplemental Table S2) of samples shown in C and D, where the level of each EGFP reporter mRNA was normalized to the level of mRFP mRNA, and the levels of cellular mRNAs were normalized to the level of GAPDH mRNA in total cell (T) and cytoplasmic (C) extracts or the level of U6 snRNA in nuclear (N) extracts. The normalized level in the presence of control siRNA + pcDNA3-HA is defined as 100. Note that the level of cytoplasmic LIN28 mRNA was normalized to the level of total cell LIN28 mRNA, since its cytoplasmic level in the absence of STAU155-HA3 was undetectable. The left lanes below wedges analyzed serial threefold dilution of total cell protein. All results are representative of three independently performed experiments. Additionally, they recapitulate quantitations of blots that analyzed larger amounts of pEGFP-IRAlus-derived proteins. (*) P < 0.05; (**) P < 0.01; n ≥ 3.
Western blotting of cell lysates using anti-STAU1 or anti-HA demonstrated that the level of STAU155 was ∼30% of normal (STAU1 siRNA), normal (control siRNA), or ∼200% of normal (pcDNA3-STAU155-HA3) (Fig. 1B). After normalizing the level of EGFP expression (Fig. 1B, where a comparable amount of mRFP protein [♦] was loaded per lane) to the level of mRNA from which it derived (Supplemental Fig. S1C; Supplemental Table S2), EGFP protein produced by each EGFP-IRAlus mRNA in the presence of control siRNA was ∼10% the level produced by EGFP mRNA (Fig. 1B). This is consistent with the previously reported nuclear retention of these EGFP-IRAlus mRNAs (Chen et al. 2008). Furthermore, decreasing the amount of STAU1 using STAU1 siRNA decreased the level of EGFP protein produced by each EGFP-IRAlus mRNA approximately twofold to threefold, whereas increasing the amount of STAU1 by expressing STAU155-HA3 increased this level approximately twofold to threefold (Fig. 1B).
These findings are consistent with the hypothesis that STAU1 binding to 3′ UTR IRAlus may compete with the association of ADAR1, which mediates A-to-I editing, and/or p54nrb, which colocalizes with nuclear-retained EGFP-IRAlus mRNAs (Chen et al. 2008), thereby allowing the export of EGFP-IRAlus mRNAs to the cytoplasm, where they could be translated.
STAU1 augments the nuclear export of 3′ UTR IRAlus mRNAs
To test whether STAU1 inhibits the nuclear retention of 3′ UTR IRAlus mRNAs, the reporter plasmid transfections were repeated (excluding pEGFP-IRAlus-APOBEC3G), and our STAU1 titrations were extended using two different concentrations (+ and ++) of pcDNA3-STAU155-HA3 that produced STAU1 at approximately twofold and approximately threefold its normal cellular abundance in total cell (Fig. 1C) as well as nuclear and cytoplasmic (Fig. 1D) lysates. Nuclear lysates were free of detectable cytoplasmic contamination, as measured using vimentin and GAPDH, and cytoplasmic fractions were free of detectable nuclear contamination, as measured using the nucleoporin (NUP)160 and p54nrb (Fig. 1D). The cellular distribution of tRNA and U6 snRNA was unchanged in lysates from different transfections (data not shown). Thus, any differences observed in the nuclear to cytoplasmic (N/C) ratio among 3′ UTR IRAlus mRNAs are not due to variations in cellular fractionation.
After normalizing the level of EGFP protein (Fig. 1C) to the level of the mRNA from which it derived in total cell lysates (Fig. 1E; Supplemental Fig. S1D), we observed that STAU1 siRNA decreased EGFP production from EGFP-IRAlus-NICN1 and EGFP-IRAlus-LIN28 mRNAs to nondetectable levels, whereas increasing levels of STAU155-HA3 increased EGFP production approximately twofold (+) and approximately threefold (++) (Fig. 1C; Supplemental Table S3). In the presence of control siRNA, the N/C ratios of EGFP-IRAlus-NICN1 and EGFP-IRAlus-LIN28 mRNAs were approximately sixfold to sevenfold higher than for EGFP mRNA, consistent with the nuclear enrichment of these 3′ UTR IRAlus mRNAs (Supplemental Fig. S1E). Varying the cellular concentration of STAU1 did not significantly affect the cellular distribution of EGFP mRNA lacking an IRAlus (Fig. 1E; Supplemental Fig. S1D). In stark contrast, the nuclear levels of EGFP-IRAlus mRNAs increased by ∼20%–25% with STAU1 siRNA and decreased by ∼25%–30% and ∼60%–65% with increasing levels of STAU155-HA3, whereas the cytoplasmic levels of these mRNAs decreased by ∼35%–40% with STAU1 siRNA and increased by ∼50%–80% and ∼100%–120% with increasing levels of STAU155-HA3 (Fig. 1E; Supplemental Fig. S1D). From the results of these combined STAU1 titration studies, we conclude that STAU1 promotes the export of nuclear-retained 3′ UTR IRAlus mRNAs. Furthermore, using fluorescent in situ hybridization (Supplemental Fig. S1F,G), we reproduced our findings that EGFP mRNA is largely cytoplasmic (while EGFP-IRAlus-NICN1 mRNA is retained in the nucleus) and that STAU155-HA3 expression promotes the export of nuclear-retained EGFP-IRAlus-NICN1 mRNA.
We also demonstrated that results obtained with reporter EGFP-IRAlus mRNAs are recapitulated by cellular mRNAs by deep sequencing poly(A)+ HEK293T cell transcripts. Sequences from 1,883,737 poly(A) site-containing reads revealed that 1069 out of 9311 (11.5%) genes analyzed and 2134 out of 32,020 (6.7%) mRNA 3′ UTR isoforms analyzed contain one or more 3′ UTR IRAlus (Supplemental Table S4). Several 3′ UTR IRAlus mRNAs, including those for NICN1, LIN28, apopotic factor cytochrome c, somatic (CYCS), and phosphoribosylaminoimidazole carboxylase (PAICS), were enriched in the nuclear fraction (Supplemental Fig. S1H). Changes in the cellular distributions of these four mRNAs with varying STAU1 levels recapitulated those of the reporter EGFP-IRAlus mRNAs (Fig. 1E; Supplemental Fig. S1I; data not shown for PAICS mRNA). While we could not analyze cellular NICN1 or LIN28 proteins (due to the unavailability of suitable antibody or undetectable expression levels, respectively), changes in the level of CYCS protein with varying STAU1 levels recapitulated changes obtained with EGFP-IRAlus mRNA-encoded proteins (Fig. 1C). Thus, STAU1 augments protein production from those reporter and cellular 3′ UTR IRAlus mRNAs that were tested.
Interestingly, and consistent with previous reports (Hundley and Bass 2010; Capshew et al. 2012; Fitzpatrick and Huang 2012), some 3′ UTR IRAlus mRNAs that we studied—e.g., eukaryotic elongation factor 2-kinase (eEF2K) and Caspase8 (Casp8) mRNAs (Supplemental Table S4)—manifested an N/C distribution like mRNAs that lack 3′ UTR IRAlus; e.g., GAPDH and SMG7 mRNAs (Supplemental Fig. S1H). It is worth noting that eEF2K and Casp8 mRNAs responded to changes in STAU1 abundance differently from one another and from the category of 3′ UTR IRAlus mRNAs that is the focus of this study; i.e., those that are enriched in nuclei in untransfected HEK293T cells (data not shown). These additional two categories of 3′ UTR IRAlus mRNAs will not be further discussed.
Notably, for those 3′ UTR IRAlus mRNAs analyzed, we found no evidence that STAU1 binding triggers SMD. For one, the half-life of cytoplasmic FLUC-IRAlus NICN1 mRNA was not prolonged by STAU1 siRNA (Supplemental Fig. S1J,K). Second, relative to control siRNA, UPF1 siRNA failed to increase the levels of cytoplasmic isoforms of NICN1, CYCS, or PAICS mRNAs that contain a 3′ UTR IRAlus (cytoplasmic LIN28 mRNA was undetectable) (data not shown). Thus, we conclude that, for those 3′ UTR IRAlus mRNAs studied here, STAU1 does not function in a degradative capacity but rather promotes the cytoplasmic disposition of these mRNAs.
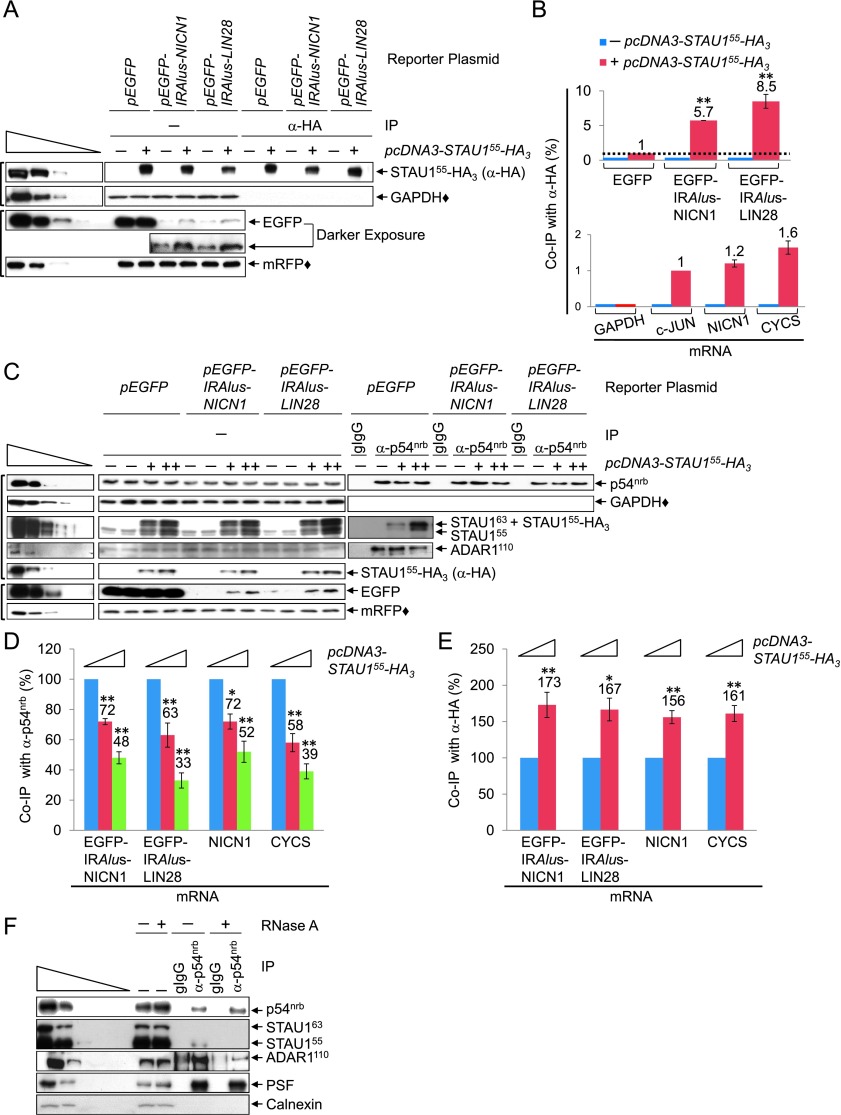
STAU1 coimmunoprecipitates with 3′ UTR IRAlus
To begin to test the mechanism behind STAU1-mediated nuclear export of 3′ UTR IRAlus mRNAs, we examined whether STAU1 physically associates with the 3′ UTR IRAlus element. HEK293T cells were transiently transfected with (1) pcDNA-STAU155-HA3 or pcDNA3-HA, (2) one of the above-described pEGFP reporters that did or did not harbor a 3′ UTR IRAlus, and (3) pmRFP. Transfected cells were exposed to formaldehyde cross-linking prior to lysis to ensure that any protein–protein and protein–RNA interactions observed were formed within cells and not as an experimental artifact after lysis. Lysates were analyzed before and after immunoprecipitation using anti-HA (Supplemental Table S5).
STAU155-HA3, which was expressed at approximately twofold the level of cellular STAU1 (data not shown), increased the production of EGFP protein by EGFP-IRAlus mRNAs but not EGFP mRNA (Fig. 2A), as it did before (Fig. 1B,C). Compared with EGFP mRNA, EGFP-IRAlus-NICN1 mRNA and EGFP-IRAlus-LIN28 mRNA were enriched, respectively, approximately sixfold and approximately ninefold after anti-HA immunoprecipitation in the presence of STAU155-HA3 (Fig. 2B; Supplemental Fig. S2A), indicating that STAU155-HA3 binds primarily to the 3′ UTRs of each IRAlus mRNA. Cellular NICN1 and CYCS mRNAs were immunoprecipitated, respectively, ∼1.2-fold and 1.6-fold more efficiently than was the bona fide SMD target c-JUN mRNA (Fig. 2B; Supplemental Fig. S2B). These data support the idea that STAU1 binding to the 3′ UTR IRAlus of mRNAs augments their export and/or translation.
Figure 2.
STAU1 coimmunoprecipitates with 3′ UTR IRAlus in competition with p54nrb. (A) Western blotting of lysates of formaldehyde cross-linked HEK293T cells (1.8 × 107 per 150-mm dish), which had been transiently transfected with 15 μg of pcDNA3-HA (−) or pcDNA3-STAU155-HA3 (+), 5 μg of pmRFP, and 5 μg of the specified pEGFP reporter, before (−) or after immunoprecipitation using anti-HA (Supplemental Table S5). (IP) Immunoprecipitation. (B) Histograms of RT-qPCR analyses of RNA from lysates analyzed in A. (Top) EGFP mRNA levels before immunoprecipitation were normalized to the level of GAPDH mRNA, and EGFP mRNA levels after immunoprecipitation were normalized to the level of LACZ RNA (immunoprecipitated samples were spiked with Escherichia coli RNA prior to RNA extraction). Subsequently, the normalized level of EGFP mRNA after immunoprecipitation is presented as a ratio of its normalized level before immunoprecipitation, and this ratio is defined as 1 for samples transfected with pEGFP + pcDNA3-STAU155-HA3 (Supplemental Fig. S2A). (Bottom) As in the top panel but analyzing cellular mRNAs, where the ratio of c-JUN mRNA after immunoprecipitation/before immunoprecipitation is defined as 1 (Supplemental Fig. S2B). (C–E) HEK293T cells (1.8 × 107 per 150-mm dish) were transiently transfected with 25 μg of pcDNA3-HA (−) or 15 μg (+) or 25 μg (++) of pcDNA3-STAU155-HA3, 5 μg of the designated pEGFP reporter, and 5 μg of pmRFP. Cells were then formaldehyde cross-linked, and half was immunoprecipitated using anti-p54nrb, while the other half was immunoprecipitated using anti-HA. (C) Western blotting of lysates before or after immunoprecipitation using anti-p54nrb or gIgG (Supplemental Table S5). (D) Histograms of RT-qPCR quantitations (Supplemental Fig. S2C) of RNA from samples shown in C, where the level of each mRNA was normalized to the level of GAPDH mRNA and LACZ RNA, respectively, before and after anti-p54nrb immunoprecipitation. Normalized values are provided as after immunoprecipitation/before immunoprecipitation, and the ratio in pcDNA3-HA-transfected cells is defined as 100. (E) Histograms of RT-qPCR quantitations (Supplemental Fig. S2D) are as in D except that each mRNA was normalized to the level of GAPDH mRNA before anti-HA immunoprecipitation or LACZ RNA after anti-HA immunoprecipitation. (F) Western blotting of lysates of HEK293T cells (1.8 × 107 per 150-mm dish), which were generated in the presence (+) or absence (−) of RNase A before or after immunoprecipitation, essentially as in C. All results are representative of three independently performed experiments. (*) P < 0.05; (**) P < 0.01; n ≥ 3.
STAU1 and p54nrb compete for binding to 3′ UTR IRAlus
To determine whether STAU1 binding to 3′ UTR IRAlus inhibits the binding of p54nrb, which mediates nuclear retention, we again turned to STAU1 dosage using STAU155-HA3, which can be readily immunoprecipitated. HEK293T cells were transiently transfected with (1) either pcDNA3-HA or one of two concentrations of the STAU155-HA3 expression vector and (2) one of the three pEGFP reporters. After formaldehyde cross-linking, cells were divided into two halves. One half was immunoprecipitated using anti-p54nrb (Fig. 2C,D) or, as a negative control, goat immunoglobulin G (gIgG), and the other half was immunoprecipitated using anti-HA (Fig. 2E).
Upon increasing the level of STAU155-HA3 expression, the amounts of EGFP-IRAlus-NICN1, EGFP-IRAlus-LIN28, NICN1, and CYCS mRNAs progressively decreased in anti-p54nrb immunoprecipitations (Fig. 2D; Supplemental Fig. S2C) and increased in the anti-HA immunoprecipitation (Fig. 2E; Supplemental Fig. S2D). Thus, STAU1 appears to compete with p54nrb for binding to IRAlus. To further confirm these results, cells were transfected with (1) control siRNA or p54nrb siRNA, which down-regulated p54nrb to ∼30% of its cellular level (Supplemental Fig. S2E), and (2) the STAU155-HA3 expression vector. Immunoprecipitations using anti-HA showed that down-regulating p54nrb increased the levels of EGFP-IRAlus mRNAs and cellular NICN1 and CYCS mRNAs that were bound by STAU155-HA3 (Supplemental Fig. S2F), again indicating that STAU1 competes with p54nrb for binding to IRAlus. In control experiments, the coimmunoprecipitation (co-IP) of p54nrb and hNEAT1 RNA was essentially unchanged by variations in the level of STAU155-HA3 (Supplemental Fig. S2G).
Both STAU155-HA3 and ADAR1 proteins coimmunoprecipitated with anti-p54nrb, and increased levels of STAU155-HA3 were accompanied by a modest decrease in the level of ADAR1 protein in anti-p54nrb immunoprecipitations (Fig. 2C; Supplemental Fig. S2H). This suggests that STAU1, ADAR1, and p54nrb may constitute the same mRNP, the composition of which depends on their relative abundance. To gain additional insight into proteins bound to 3′ UTR IRAlus mRNAs, lysates of untransfected HEK293T cells were immunoprecipitated using anti-p54nrb or gIgG in the absence or presence of RNase A, which quantitatively degraded cellular RNA (data not shown). The immunoprecipitation of STAU1 or ADAR1 using anti-p54nrb was largely RNase A-sensitive, whereas the immunoprecipitation of PSF with anti-p54nrb was RNase A-insensitive (Fig. 2F), consistent with their functioning as a heterodimer (Shav-Tal and Zipori 2002). These results were corroborated with the findings that both ADAR1 and p54nrb coimmunoprecipitate with STAU155-HA3 and that their co-IP is largely RNase A-sensitive (Supplemental Fig. S2I). We conclude that the decision to retain a 3′ UTR IRAlus mRNA in the nucleus or transport it to the cytoplasm is influenced by the relative abundance of p54nrb, ADAR1, and STAU1, which can comprise the same mRNP.
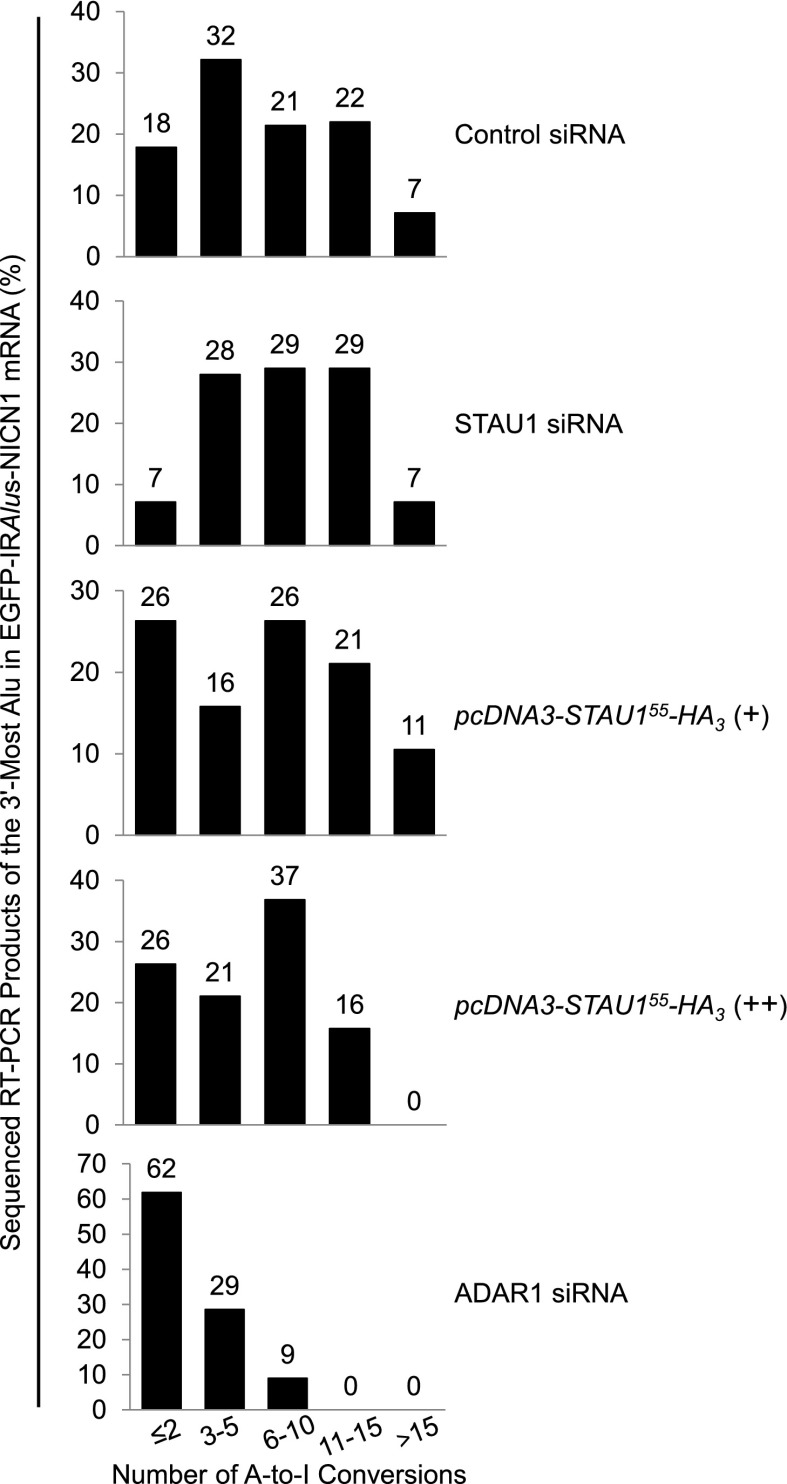
STAU1 does not significantly affect ADAR1-mediated A-to-I editing of 3′ UTR IRAlus
STAU1 competition with p54nrb for binding to IRAlus (Fig. 2D,E) might be via STAU1 inhibiting ADAR1-mediated A-to-I editing, especially since p54nrb has been reported to preferentially bind to edited sites, at least in vitro (Zhang and Carmichael 2001), and we detect modest competition between STAU1 and ADAR1 in the immunoprecipitation of p54nrb (Fig. 2C). To test this possibility, we sequenced (Fig. 3) the 3′-most 3′ UTR Alu element of EGFP-IRAlus-NICN1 mRNA using samples analyzed in Figure 1E and, as a positive control, ADAR1 siRNA-treated samples from Figure 4, A and B. As reported (Kim et al. 2004), editing was preferentially at A*-G, T-A*, and A-A* dinucleotides and TA*G trinucleotides and toward the middle of the duplex (Supplemental Fig. S3). A Mann-Whitney rank-sum test revealed that only in ADAR1 siRNA-treated cells was the degree of editing significantly different (P = 0.00045) than in control siRNA-treated cells (Fig. 3). Thus, varying the HEK293T cell concentration of STAU1 from ∼10-fold below normal to approximately threefold above normal was sufficient to influence the N/C ratio of 3′ UTR IRAlus mRNAs that are normally largely nuclear (Fig. 1E) despite a lack of statistically significant effects on the degree of A-to-I editing (Fig. 3). Additionally, our sequencing (Fig. 3) and co-IP (Fig. 2C,D) data suggest that ADAR1-mediated A-to-I editing might precede the step where STAU1 and p54nrb compete for binding to IRAlus.
Figure 3.
Expressing STAU1 does not significantly change the level of A-to-I editing within the 3′ UTR IRAlus of EGFP-IRAlus-NICN1 mRNA. RT–PCR products of EGFP-IRAlus-NICN1 mRNA that derived from samples analyzed in Figure 1E or 4B were cloned, and plasmid DNA from ≥20 individual clones was sequenced. Numbers above the bars specify the percent of sequenced clones that harbored the specified number of A-to-I conversions in the 3′-most Alu of EGFP-IRAlus-NICN1 mRNA. See also Supplemental Figure S3.
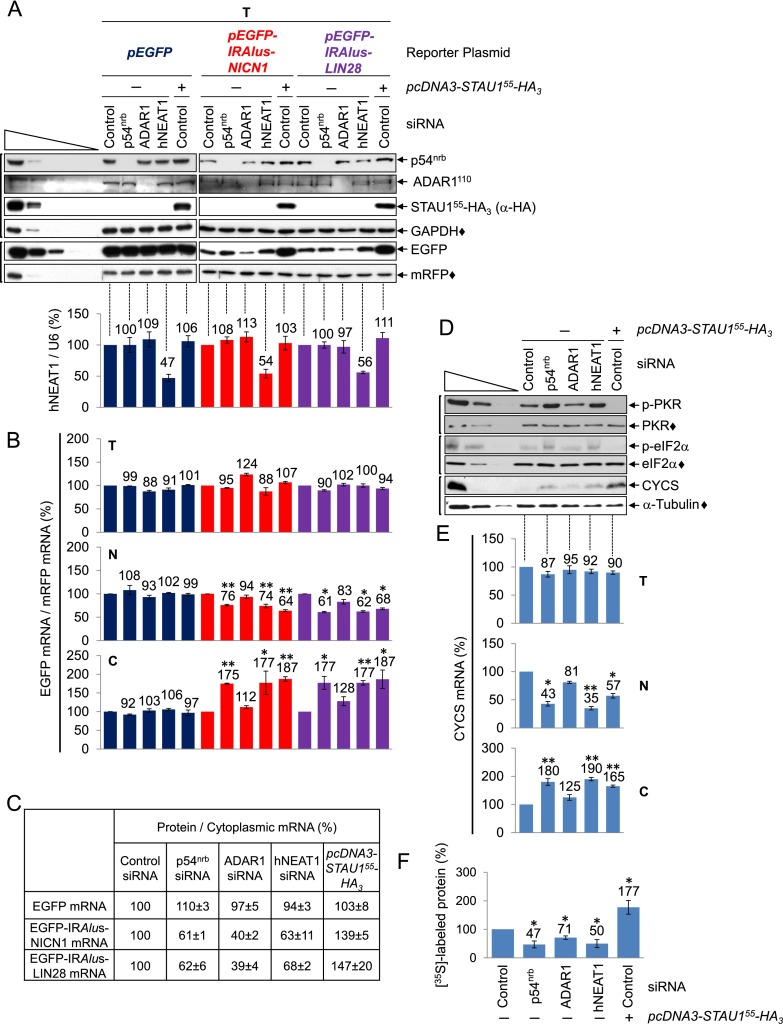
Figure 4.
STAU1 binding to 3′ UTR IRAlus mRNAs augments their export to the cytoplasm and relieves their translational repression. (A, top) Western blotting of lysates of HEK293T cells (1.8 × 107 per 150-mm dish) that had been transiently transfected with 100 nM control siRNA or 50 nM p54nrb siRNA or ADAR1 siRNA, each together with 50 nM control siRNA or 100 nM hNEAT1 siRNA and, 2 d later, 15 μg of pcDNA3-STAU155-HA3 (+) or pcDNA3-HA (−), 5 μg of pmRFP, and 5 μg of the specified pEGFP reporter. See Supplemental Table S3 for quantitations. (Bottom) Histograms of RT-qPCR quantitations of samples analyzed in the top panel. The level of hNEAT1 RNA was normalized to the level of U6 snRNA, and the normalized level in the presence of control siRNA + pcDNA3-HA is defined as 100. (B) Histograms of RT-qPCR quantitations (Supplemental Fig. S4A) of samples analyzed in A but using total (T), nuclear (N), or cytoplasmic (C) cell fractions, where the level of the specified EGFP mRNA in cells treated with control siRNA + pcDNA3-HA, normalized to mRFP mRNA, is defined as 100 for each cell fraction. (C) Tabulation of data from A and B, where the ratio of EGFP protein to its cytoplasmic mRNA in cells treated with control siRNA + pcDNA3-HA is defined as 100. (D) Western blotting of lysates from cells analyzed in A. (p-PKR) phospho-PKR; (p-eIF2α) phospho-eIF2α. (E) Histograms of RT-qPCR quantitations (Supplemental Fig. S4B) as in B except that the level of CYCS mRNA was normalized to the level of GAPDH in total cell and cytoplasmic RNA or to U6 snRNA in nuclear RNA. (F) HEK293T cells (8 × 106 per 100-mm dish) were transfected as in A and incubated with 0.1 mCi of [35S]-Met + [35S]-Cys for 30 min. The amount of [35S] incorporation into cellular protein in the presence of control siRNA + pcDNA3-HA is defined as 100 (Supplemental Fig. S4C). All results are representative of three independently performed experiments. (*) P < 0.05; (**) P < 0.01; n ≥ 3.
STAU1 binding to 3′ UTR IRAlus augments the translation of exported mRNAs
STAU1 has been reported to enhance mRNA translation when tethered to mRNA 5′ UTRs (Dugré-Brisson et al. 2005). Thus, some of the increase in EGFP protein production observed in the presence of increased levels of STAU155-HA3 could be attributable to not only increased EGFP-IRAlus mRNA export, but also increased translation of exported EGFP-IRAlus mRNA.
To test this, HEK293T cells were transfected with control siRNA, p54nrb siRNA, ADAR1 siRNA, or hNEAT1 siRNA and, 2 d later, pcDNA3-STAU155-HA3 or pcDNA3-HA, a pEGFP reporter, and pmRFP. Total cell, nuclear, and cytoplasmic fractions were generated. p54nrb siRNA and ADAR1 siRNA reduced the level of their target protein to nondetectable levels (Fig. 4A, top), STAU155-HA3 was expressed at approximately twofold the level of cellular STAU1 (Fig. 4A, top; data not shown), and hNEAT1 siRNA reduced the level of hNEAT1 RNA to ∼50% the level in the presence of control siRNA (Fig. 4A, bottom).
Compared with in the presence of control siRNA, the level of total cell EGFP protein produced in the presence of p54nrb siRNA, ADAR1 siRNA, hNEAT1 siRNA, or STAU155-HA3 was, respectively, ∼110%, 50%, 115%, or 270% for both EGFP-IRAlus NICN1 and EGFP-IRAlus LIN28 mRNAs (Fig. 4A, top). However, normalizing the level of EGFP mRNA to the level of mRFP mRNA in nuclear and cytoplasmic fractions revealed comparable levels of cytoplasmic EGFP-IRAlus mRNAs in the presence of p54nrb siRNA, hNEAT1 siRNA, or STAU155-HA3 (Fig. 4B; Supplemental Fig. S4A). We conclude from the ratio of EGFP protein to either cytoplasmic EGFP-IRAlus mRNA (Fig. 4C) that 3′ UTR IRAlus mRNAs that are exported to the cytoplasm as a result of STAU155-HA3 expression are translationally active, while those exported as a result of down-regulating a paraspeckle component are translationally repressed. ADAR1 siRNA, unlike p54nrb siRNA, hNEAT1 siRNA, or STAU155-HA3 expression, failed to appreciably augment the export of either EGFP-IRAlus mRNA (Fig. 4B; Supplemental Fig. S4A), which included the significantly underedited EGFP-IRAlus-NICN1 mRNA (Fig. 3). Thus, a failure to A-to-I edit does not significantly promote IRAlus mRNA export. The results obtained for EGFP-IRAlus reporter proteins and mRNAs were recapitulated by endogenous CYCS protein and mRNA (Fig. 4D,E; Supplemental Fig. S4B).
Down-regulating p54nrb or hNEAT1 increases PKR and eIF2α phosphorylation and inhibits translation, whereas expressing STAU155-HA3 has the opposite effects
Why does ablation of paraspeckle components, which results in the nuclear export of IRAlus mRNAs, result in translational repression, while increasing STAU1 dosage, which similarly results in the nuclear export of IRAlus mRNAs, has the opposite effect; i.e., translational induction? We speculated that translational repression was attributable to cytoplasmic PKR. Cytoplasmic dsRNAs from viruses can elicit an innate immune response that involves the PKR-mediated phosphorylation of eIF2α, resulting in a global cell inhibition of translation initiation (Sonenberg and Hinnebusch 2009). Since IRAlus are probably the biggest cellular reservoir of dsRNAs (Kim et al. 2004), it is possible that (1) down-regulating a constituent of the nuclear retention pathway would result in the activation of PKR by autophosphorylation followed by translational repression via the PKR-mediated phosphorylation of eIF2α, and (2) STAU1 promotes translation by attenuating the PKR response via competition with PKR for binding to IRAlus. In support of these ideas, using samples analyzed in Figure 4A, the ratio of phosphorylated PKR (phospho-PKR) to total cell PKR in the presence of p54nrb siRNA, ADAR1 siRNA, hNEAT1 siRNA, or STAU155-HA3 was, respectively, 216% ± 41%, 95% ± 17%, 197% ± 35%, or 25% ± 13% the ratio in the presence of control siRNA, whereas the ratio of phospho-eIF2α to total cell eIF2α was 164% ± 14%, 90% ± 11%, 168% ± 14%, or 56% ± 11% (Fig. 4D). These data suggest that IRAlus mRNAs that are exported to the cytoplasm due to down-regulation of a paraspeckle component are translationally repressed, at least in part, because of PKR activation.
To determine whether repression extends to global cell protein synthesis, the transfections were repeated, and the incorporation of [35S]-Met + [35S]-Cys into newly synthesized protein was quantitated. Results indicate that STAU155-HA3 increased cellular translation ∼1.8-fold, whereas p54nrb siRNA, ADAR1 siRNA, or hNEAT1 siRNA decreased global cell translation, respectively, approximately twofold, ∼1.5-fold, or approximately twofold (Fig. 4F; Supplemental Fig. S4C). It is noteworthy that the effects of siRNA and STAU155-HA3 expression on the translation of EGFP mRNA but not EGFP-IRAlus mRNAs were largely masked when an equal amount of mRFP protein was loaded in all lanes (Fig. 4A). To further examine the role of PKR in the shutdown of global cell translation by p54nrb siRNA, ADAR1 siRNA, or hNEAT1 siRNA, we included PKR siRNA in the transfections using p54nrb siRNA, ADAR1 siRNA, or hNEAT1 siRNA. PKR siRNA, which down-regulated the level of PKR to ∼50% of normal (Supplemental Fig. S4D), largely reversed the inhibition of cellular translation attributable to p54nrb siRNA or hNEAT1siRNA but not ADAR1 siRNA (Supplemental Fig. S4E). We conclude that down-regulating p54nrb or hNEAT1 results in a global PKR-mediated shutdown of translation, whereas expressing STAU1 has the opposite effect. In both cases, the translational effect is stronger on IRAlus mRNA than on other mRNAs.
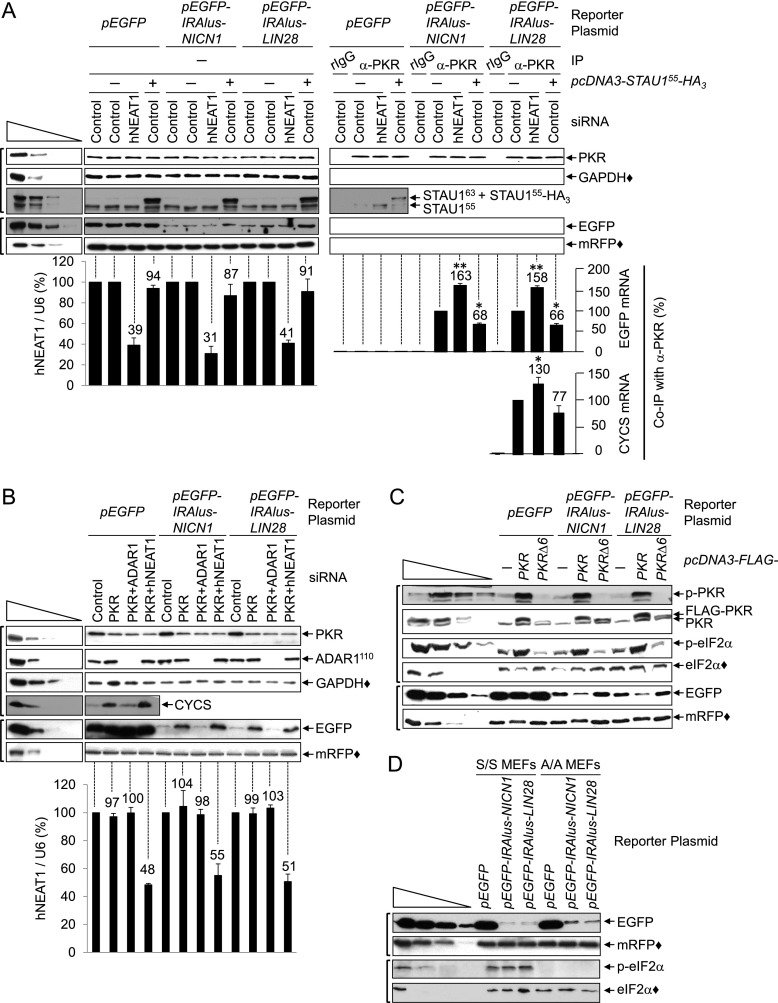
3′ UTR IRAlus bind PKR
Down-regulating a paraspeckle component increases the levels of cytoplasmic 3′ UTR IRAlus mRNAs, which increases PKR and eIF2α phosphorylation so as to inhibit cellular translation (Fig. 4). To test whether these effects involve PKR binding to 3′ UTR IRAlus, HEK293T cells were transfected with either control siRNA or hNEAT1 siRNA and, 2 d later, either pcDNA3-HA or pcDNA3-STAU155-HA3 and a pEGFP reporter. Cells were formaldehyde cross-linked, and lysates were analyzed before or after immunoprecipitation using anti-PKR or rabbit IgG (rIgG). STAU155-HA3 was expressed at approximately twofold the level of cellular STAU1, and hNEAT1 siRNA reduced its target to ∼35% of normal (Fig. 5A). STAU1 could be detected in the anti-PKR immunoprecipitation, and, interestingly, down-regulating hNEAT1 increased the level of STAU1 in this immunoprecipitation ∼1.5-fold. Notably, the co-IP of PKR with STAU1 is largely RNase A-sensitive (Supplemental Fig. S5A), indicating that the STAU1-mediated inhibition of PKR phosphorylation (Fig. 4D) is not primarily due to the direct binding of STAU1 to PKR.
Figure 5.
Translation of 3′ UTR IRAlus mRNAs is repressed by PKR-mediated eIF2α phosphorylation. (A) Western blotting (top) or histograms of RT-qPCR quantitations (bottom) of lysates of formaldehyde cross-linked HEK293T cells (1.8 × 107 per 150-mm dish), which had been transiently transfected with 100 nM control siRNA or hNEAT1 siRNA and, 2 d later, either 15 μg of pcDNA3-STAU155-HA3 (+) or pcDNA3-HA (−), 5 μg of pmRFP, and 5 μg of the specified pEGFP reporter, before (−) or after immunoprecipitation using α-PKR (Supplemental Table S5) or rIgG. The levels of EGFP mRNA and CYCS mRNA were normalized to the level of GAPDH mRNA in preimmunoprecipitation samples and to the level of LACZ RNA in immunoprecipitation samples. Normalized levels after immunoprecipitation/before immunoprecipitation in the presence of control siRNA + pcDNA3-HA are defined as 100 (Supplemental Fig. S5B). (B, top) Western blotting using lysates of HEK293T cells (8 × 106 per 100-mm dish), which had been transiently transfected with 150 nM control siRNA, 50 nM PKR siRNA, 50 nM PKR siRNA + 50 nM ADAR1 siRNA, or 50 nM PKR siRNA + 100 nM hNEAT1 siRNA (where control siRNA was added so transfected siRNAs totaled 150 nM) and, 2 d later, 2.5 μg of pmRFP and 2.5 μg of the specified pEGFP reporter. See Supplemental Table S3 for quantitations. (Bottom) Histograms of RT-qPCR quantitations using samples analyzed in the top panel, where the level of hNEAT1 RNA was normalized to the level of U6 snRNA, and the normalized level in the presence of control siRNA is defined as 100. (C) Western blotting as in B except that along with each pEGFP reporter, cells were transfected with 10 μg of pcDNA3, pcDNA3-Flag-PKR, or pcDNA3-Flag-PKRΔ6. See Supplemental Table S3 for quantitations. (D) Western blotting of lysates of S/S or A/A MEFs (3 × 106 per 100-mm dish), which had been transiently transfected with 5 μg of pmRFP and 5 μg of the specified pEGFP reporter. All results are representative of three independently performed experiments. (*) P < 0.05; (**) P < 0.01; n ≥ 3.
EGFP-IRAlus-NICN1 and EGFP-IRAlus-LIN28 mRNAs were present in the anti-PKR immunoprecipitations, unlike EGFP mRNA, indicating that PKR binds to 3′ UTR IRAlus (Fig. 5A; Supplemental Fig. S5B). The amount of each EGFP-IRAlus mRNA that coimmunoprecipitated with PKR was increased ∼1.6-fold or decreased approximately twofold in the presence of, respectively, hNEAT1 siRNA or STAU155-HA3 (Fig. 5A; Supplemental Fig. S5B). The same pattern of co-IP was obtained for CYCS mRNA (Fig. 5A; Supplemental Fig. S5B). Competition between STAU1 and PKR for binding to cytoplasmic IRAlus mRNAs becomes evident considering that the cytoplasmic abundance of these mRNAs was increased approximately twofold when hNEAT1 RNA was down-regulated or STAU155-HA3 was expressed (Fig. 4B). Also notable, since PKR dimerization and autophosphorylation require at least 33 bp and optimally 80–90 bp (Manche et al. 1992), it follows that when the level of STAU1 is elevated, the observed decrease in the level of 3′ UTR IRAlus in an anti-PKR immunoprecipitation (Fig. 5A) would be smaller than the observed decrease in the level of PKR autophosphorylation (Fig. 4D).
To corroborate that STAU1 and PKR compete for binding to 3′ UTR IRAlus, cells were transfected with PKR siRNA and, subsequently, a pEGFP-IRAlus reporter and the STAU155-HA3 expression vector. An anti-HA immunoprecipitation revealed that down-regulating PKR increased the levels of EGFP-IRAlus and CYCS mRNAs that were bound by STAU155-HA3, confirming that STAU1 and PKR compete for binding to IRAlus (Supplemental Fig. S5C). We conclude that a global shutdown of translation is obviated when 3′ UTR IRAlus mRNAs are retained within nuclei or bound by STAU1, both of which preclude cytoplasmic PKR binding.
PKR represses 3′ UTR IRAlus mRNA translation more effectively than 3′ UTR IRAlus-free mRNA translation via the canonical eIF2α phosphorylation pathway
To gain more insight into the PKR-mediated repression of 3′ UTR IRAlus mRNA translation, HEK293T cells were transiently transfected with (1) control siRNA, PKR siRNA, PKR + ADAR1 siRNAs, or PKR + hNEAT1 siRNAs and, 2 d later, (2) a pEGFP reporter and (3) pmRFP. After normalization to the level of mRFP protein, the level of EGFP production from either EGFP-IRAlus-NICN1 or EGFP-IRAlus-LIN28 mRNA in the presence of PKR siRNA, PKR + ADAR1 siRNAs, or PKR + hNEAT1 siRNAs was, respectively, ∼200%, 75%, or 295% the level in the presence of control siRNA (Fig. 5B). In contrast, none of the siRNAs significantly affected the production of EGFP protein from EGFP mRNA (Fig. 5B).
These results confirm that the PKR-mediated inhibition of translation is more efficient for EGFP-IRAlus mRNAs than for EGFP mRNA. Moreover, the higher levels of cytoplasmic IRAlus mRNAs due to the down-regulation of hNEAT1 (Fig. 4B) allows for more IRAlus mRNA translation when PKR is down-regulated. Similar results were obtained for cellular CYCS mRNA (Fig. 5B). Additionally, PKR siRNA does not promote IRAlus mRNA translation in the presence of ADAR1 siRNA (Fig. 5B). Although it has been reported that ADAR1 and PKR compete for the same dsRNA sequences (e.g., see Pfaller et al. 2011), our finding that ADAR1 siRNA does not promote the phosphorylation of PKR or eIF2α under the conditions used here suggests that the opposing effects of ADAR1 siRNA and PKR siRNA on IRAlus mRNA translation are attributable to different mechanisms.
Next, HEK293T cells were transiently transfected with a pEGFP reporter and pcDNA3-Flag-PKR, pcDNA3-Flag-PKRΔ6 (Koromilas et al. 1992), or pcDNA3 (Fig. 5C). PKRΔ6 manifests dsRNA-binding activity but fails to undergo autophosphorylation or mediate eIF2α phosphorylation. Flag-PKR and Flag-PKRΔ6 were expressed at, respectively, ∼1.5-fold or the same level as cellular PKR (Fig. 5C). Flag-PKR increased the amount of PKR and eIF2α phosphorylation to approximately threefold the amount in the presence of pcDNA3. In contrast, Flag-PKRΔ6 reduced PKR and eIF2α phosphorylation to, respectively, an undetectable level and about half the level in the presence of pcDNA3 (Fig. 5C). Flag-PKR or Flag-PKRΔ6, respectively, decreased or increased the level of mRFP protein and the short-lived cellular protein c-JUN, reflecting their consequences to global cell translation (Supplemental Fig. S5D). When an equal amount of mRFP protein was loaded in each lane (Fig. 5C), Flag-PKR was found to reduce EGFP production from each EGFP-IRAlus mRNA to ∼45% the level in the presence of pcDNA3, while Flag-PKRΔ6 increased EGFP production to ∼120%–150%, and neither Flag-PKR nor Flag-PKRΔ6 altered EGFP production from EGFP mRNA (Fig. 5C). As observed above (Figs. 4, 5B), translational repression was stronger for the IRAlus mRNAs than for mRNAs that lack IRAlus; i.e., EGFP and mRFP mRNAs (Fig. 5C). We suggest that PKR most effectively represses the translation of mRNAs to which it binds.
To further investigate the role of eIF2α phosphorylation in the PKR-mediated translational repression of 3′ UTR IRAlus mRNAs, mouse embryonic fibroblasts (MEFs) (Scheuner et al. 2001) in which both eIF2α alleles (1) harbor a Ser51 → Ala mutation (A/A) that precludes eIF2α phosphorylation upon, e.g., exposure to endoplasmic reticulum stress induced by thapsigargin or (2) are wild type (S/S) (Supplemental Fig. S5E) were transiently transfected with a pEGFP reporter. EGFP produced from each pEGFP-IRAlus reporter in A/A MEFs was ∼195%–275% the level in S/S MEFs (Fig. 5D). These results provide further evidence that PKR-mediated translational repression of IRAlus mRNAs is mediated, at least in part, by eIF2α phosphorylation.
Regulation of LIN28 mRNA via its 3′ UTR IRAlus is critical for myogenesis
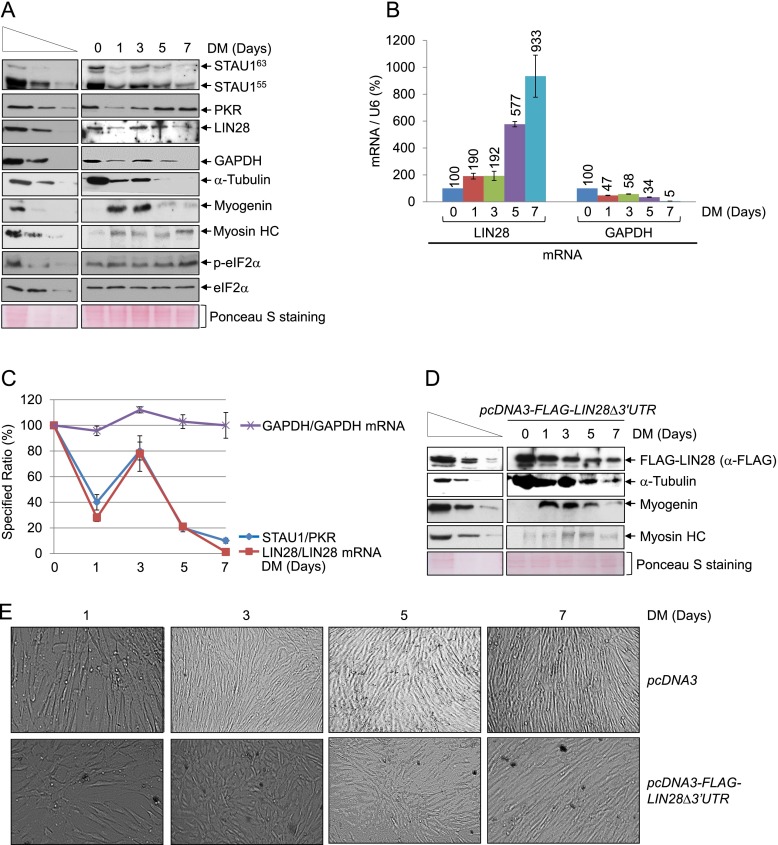
Our data suggest that changes in the STAU1 to PKR ratio impact cellular physiology via effects on IRAlus mRNA metabolism. To examine this, we measured the levels of STAU1 and PKR in primary human skeletal muscle (hSKMc) MBs and myotubes (MTs) (Supplemental Fig. S6A). Compared with the level in MBs, the level in MTs of STAU1 decreased approximately fourfold, PKR increased ∼1.5-fold, and LIN28 protein decreased approximately twofold despite an ∼15-fold increase in the level of LIN28 mRNA that was not accompanied by a loss of its 3′ UTR IRAlus by, e.g., alternative 3′ end formation (Supplemental Fig. S6B–D). As expected, down-regulating STAU1 or PKR in MBs resulted in an approximately threefold decrease or increase, respectively, in the level of LIN28 protein without a concomitant change in the level of its mRNA (Supplemental Fig. S6E,F).
Since LIN28 protein is a potent oncogene and plays a critical role during development and the myogenesis of C2C12 mouse skeletal muscle cells (Polesskaya et al. 2007; Thornton and Gregory 2012), we aimed to determine how STAU1 and PKR regulate the translation of LIN28 mRNA. Thus, we normalized the level of LIN28 protein to LIN28 mRNA during the myogenesis of hSkMc cells after 0, 1, 3, 5, and 7 d in differentiation medium. As reported (Polesskaya et al. 2007), during myogenesis, the level of α-tubulin decreased, the level of myogenin increased and then decreased, and the level of myosin heavy chain increased (Fig. 6A). While the levels of STAU1 and PKR varied during differentiation, remarkably, the levels of LIN28 protein/LIN28 mRNA and eEF2K protein/eEF2K mRNA—unlike the level of GAPDH protein/GAPDH mRNA or c-JUN protein/c-JUN mRNA—increased when the ratio of STAU1/PKR increased and the degree of eIF2α phosphorylation decreased (Fig. 6A–C; Supplemental Fig. S6G; data not shown). Since the translation of LIN28, eEF2K, GAPDH, and c-JUN mRNAs is regulated by the degree of eIF2α phosphorylation, but only LIN28 and eEF2K mRNAs contain a 3′ UTR IRAlus, and since a 3′ UTR IRAlus augments STAU1- and PKR-mediated regulatory events, including translational repression by eIF2α phosphorylation (Fig. 6A–C), these data indicate that during myogenesis, STAU1 and PKR regulate the translation of LIN28 and eEF2K mRNAs via their 3′ UTR IRAlus.
Figure 6.
Changes in the ratio of STAU1 and PKR during hSkMc cell myogenesis alters LIN28 mRNA translation via its 3′ UTR IRAlus, which is key to myogenesis. (A) Western blotting of hSkMc cell lysates. Cells (2 × 106 per 150-mm dish) were propagated in growth medium to 80% confluency. An aliquot was harvested (0 d in differentiation medium [DM]), and the remaining cells were propagated in DM and harvested on the specified day. (Myosin HC) Myosin heavy chain. Ponceau S-staining demonstrated that equal amounts of lysates were analyzed. (B) Histogram representation of RT-qPCR quantitations (Supplemental Fig. S6G) of RNA from samples analyzed in A. The level of each mRNA was normalized to the level of U6 snRNA, and the normalized level in MBs (day 0) is defined as 100. (C) Line graph representation of the denoted ratios using samples analyzed in A and B. Each ratio in MBs (day 0) is defined as 100. (D) Western blotting of lysates of hSkMc MBs (2 × 106 per 150-mm dish), which had been transiently transfected with 1 μg of pcDNA3-Flag-LIN28Δ3′UTR and, 3 d later, cultured for the specified number of days in DM. (E) Differential interference contrast microscopy of hSkMc cells analyzed in A and D. All results are representative of three independently performed experiments.
To test the importance of LIN28 protein and the 3′ UTR IRAlus of LIN28 mRNA to hSkMc myogenesis, LIN28 protein was expressed from pcDNA3-Flag-LIN28Δ3′UTR (Heo et al. 2008) at approximately threefold its cellular level (Supplemental Fig. S6H) throughout the differentiation process (Fig. 6D). LIN28 expression from the pcDNA3-Flag-LIN28Δ3′UTR delayed myogenesis, as evidenced by disregulation of myogenin production and slower rates of (1) α-tubulin down-regulation, (2) myosin heavy chain up-regulation, and (3) the fusion of MBs to MTs (Fig. 6D,E). Our results reveal how STAU1 and PKR coregulate the myogenic process through the efficiency of IRAlus mRNA translation.
Discussion
We show here that STAU1 binding to 3′ UTR IRAlus within nuclei competes with the binding of p54nrb and, to a lesser extent, ADAR1 to inhibit the nuclear retention of 3′ UTR IRAlus mRNAs (Figs. 2, 3, 7). As a consequence, STAU1 increases the nuclear export of 3′ UTR IRAlus mRNAs and inhibits PKR binding to 3′ UTR IRAlus in the cytoplasm so as to preclude the consequential cascade of PKR phosphorylation followed by eIF2α phosphorylation (Figs. 1, 2, 4, 5, 7). This then augments 3′ UTR IRAlus mRNA translation and, to a lesser extent, global cell translation (Figs. 4, 5, 7). STAU1 binding to 3′ UTR IRAlus appears to be responsible for the nuclear export of the fraction of IRAlus mRNAs detected in the cytoplasm under physiological conditions, since there is a linear relationship between the cellular abundance of STAU1 and the cytoplasmic concentrations of those 3′ UTR IRAlus mRNAs examined (Fig. 1; Supplemental Fig. S1). Consistent with this view, a fraction of 3′ UTR IRAlus mRNAs is normally detectable in the cytoplasm associated with polysomes (Chen et al. 2008; Hundley and Bass 2010). Variability among different cell lines in the relative concentrations of STAU1 and p54nrb—and probably other dsRNA-binding proteins—is expected to result in different N/C distributions of IRAlus mRNAs and thus levels of protein produced from 3′ UTR IRAlus mRNAs. The lack of strict sequence- and position-dependent binding of dsRNA-binding proteins allows for their competitive binding to what can be as long as 300 imperfectly base-paired IRAlus, which might account for the variable extent and patterns of editing detected on different transcripts from the same gene (Fig. 3; Supplemental Fig. S3; Chen et al. 2008).
Figure 7.
Model for how STAU1 enhances the nuclear export and translation of 3′ UTR IRAlus mRNAs. In nuclei, p54nrb binds to 3′ UTR IRAlus largely independently of ADAR1-mediated A-to-I editing, resulting in their recruitment to and retention in hNEAT1-containing paraspeckles. STAU1 competes with p54nrb for 3′ UTR IRAlus binding so as to inhibit their nuclear retention and enhance their nuclear export. It is possible (Fig. 2), but not diagrammed here, that nuclear-retained 3′ UTR IRAlus could be bound by both p54nrb and STAU1. In the cytoplasm, IRAlus capable of binding PKR result in PKR autophosphorylation/activation, which leads to eIF2α phosphorylation and the inhibition of IRAlus mRNA translation, and, to a lesser extent, global cell translation. When bound to cytoplasmic 3′ UTR IRAlus so as to disallow PKR binding, STAU1 stimulates IRAlus mRNA translation and obviates a global shutdown of translation. Notably, our data do not exclude the possibility that STAU1 inhibits PKR activation by other pathways in addition to competing for binding to 3′ UTR IRAlus.
It is worth noting that while the prevalence of IR SINEs that form a duplex in the 3′ UTRs of mouse mRNAs is significantly less than in primate mRNAs (Krayev et al. 1982; Neeman et al. 2006), two studies, when taken together, indicate that STAU1 binding to 3′ UTR IR SINEs might also promote mRNA export and translation in rodents. The first study describes two transcripts that derive from the same gene: One transcript contains the 3′ UTR IR SINEs, is nuclear-enriched, and localizes to paraspeckles; the other transcript lacks the repeats and, as a consequence, is exported to the cytoplasm and produces cationic amino acid transporter 2 protein (Prasanth et al. 2005). The second study demonstrates that STAU1 indeed binds dsRNA formed by SINE base-pairing (Wang et al. 2013).
We do not know why STAU1 binding to those 3′ UTR IRAlus studied fails to trigger SMD (Supplemental Fig. S1J,K). Furthermore, we cannot conclude that all IRAlus mRNAs are immune to SMD. However, microarray data also indicate that 3′ UTR IRAlus mRNAs are generally not up-regulated upon STAU1 down-regulation (Kim et al. 2007). Possibly, STAU1 bound to at least some 3′ UTR IRAlus is not configured to support SMD. Notably, there is reason to believe that STAU2 also binds 3′ UTR IRAlus, since, like STAU1, it has been shown to bind single 3′ UTR Alu elements (Park et al. 2013).
Our finding that the degree of A-to-I editing does not significantly correlate with the extent of nuclear retention is consistent with reports that 3′ UTR IRAlus reduce polysome loading on and translational efficiency of mRNAs independent of the degree of A-to-I editing (Hundley and Bass 2010; Capshew et al. 2012). Furthermore, the fact that cytoplasmic 3′ UTR IRAlus inhibit the translation of 3′ UTR IRAlus mRNAs and, to a lesser extent, the bulk of cellular mRNAs adds a new dimension to studies demonstrating that duplexes in the 5′ UTR of interferon-γ (IFN-γ) mRNA, which have been shown to bind and activate PKR, result in the translational repression of only IFN-γ mRNA (Ben-Asouli et al. 2002; Cohen-Chalamish et al. 2009). Extended dsRNA structures within the 3′ UTRs of tropomyosin, troponin, and cardiac actin mRNAs (Nussbaum et al. 2002), like the expanded CUG repeats that typify the 3′ UTRs of mRNAs from patients with myotonic dystrophy type 1 (DM1) (Tian et al. 2000; Huichalaf et al. 2010), have also been shown to activate PKR so as to inhibit global protein synthesis in rabbit reticulocyte lysates, consistent with our results. In fact, elevated levels of STAU1 in skeletal muscles of DM1 patients and DM1 mouse models result in STAU1 binding to CUG repeat-containing mRNAs (Ravel-Chapuis et al. 2012). Binding not only promotes the export of these mRNAs, but also alleviates global cell splicing defects that are attributable to the sequestration of RNA-binding proteins, including muscleblind and CUG-binding protein 1, within nuclei at the repeats (Ravel-Chapuis et al. 2012).
Even though extended dsRNA structures were thought to only rarely exist in the cytoplasm, our data indicate that they do exist and are tightly controlled by STAU1 binding, which precludes a shutdown of global cell translation. Consistent with the negative impact of 3′ UTR IRAlus on cellular translation, while 11.5% of those genes analyzed contain one or more 3′ UTR IRAlus, only 2.4% of poly(A) site-containing sequencing reads could be annotated to a 3′ UTR IRAlus transcript of these genes (Supplemental Table S4).
The 3′ UTR IRAlus-mediated effect on mRNA translation both in cis and in trans via bound STAU1 and/or PKR (Figs. 5–7) constitutes a new regulatory mechanism. Precedents already exist for distinct effects of a protein on the translation of a specific subset of mRNAs compared with the bulk of cellular mRNAs. For example, during megakaryocytic differentiation, eIF2α phosphorylation inhibits global cell translation and also stimulates the translation of cellular mRNAs that contain an internal ribosome entry site (Gerlitz et al. 2002).
Our finding that down-regulating the IRAlus nuclear retention factors p54nrb or hNEAT1 induces eIF2α phosphorylation is the first to implicate nuclear retention in paraspeckles as a means for cells to obviate IRAlus mRNA-mediated innate immunity. Human embryonic stem (hES) cells lack hNEAT1 and paraspeckles but avoid induction of innate immunity via an attenuated response to cytoplasmic dsRNA in a different mechanism: Even though they express PKR, they lack Toll-like receptor 3 and melanoma differentiation-associated protein 5, which are two major cytoplasmic dsRNA sensors (Chen et al. 2010). hES cell differentiation to trophoblasts is accompanied by hNEAT1 expression, paraspeckle formation, and a response to cytoplasmic dsRNAs (Chen et al. 2010).
Our studies underscore the importance of dsRNA-binding proteins that compete for the same or overlapping RNA-binding sites to cellular homeostasis via post-transcriptional effects that reach far beyond the RNAs they bind.
Materials and methods
Deep sequencing of mRNA and data analysis
For deep sequencing of mRNA and data analysis, see the Supplemental Material.
Plasmid construction
For plasmid construction, see the Supplemental Material.
Human cell transfections, formaldehyde cross-linking, and lysis
HEK293T cells were propagated in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS) and, when specified, transiently transfected with plasmid DNA using Lipofectamine 2000 transfection reagent (Invitrogen) and/or siRNA using RNAiMAX transfection reagent (Invitrogen). A/A and S/S MEFs were propagated in DMEM supplemented with 10% FBS and 1× nonessential amino acids (Gibco) (Scheuner et al. 2001) and transiently transfected with plasmid DNA using Lipofectamine LTX with Plus reagent (Invitrogen). Where specified, cells were cross-linked using 0.75% formaldehyde (Sigma) in 1× phosphate-buffered saline (PBS) (pH 7) for 10 min at 25°C and subsequently quenched with 1 M glycine for 5 min at room temperature prior to lysis (Gong et al. 2009). Total cells were pelleted for RNA purification or lysed using RIPA buffer (Sefton 2005) for the analysis of protein. Hypotonic Gentle Lysis buffer (Roche) was used for immunoprecipitations. Nuclear and cytoplasmic lysates were generated using NE-PER Nuclear and Cytoplasmic Extraction reagents (Thermo Scientific). RNA was purified from cell pellets and lysates using TRIzol reagent (Invitrogen).
Primary hSkMc MBs (Cell Application, Inc.) were propagated in Skeletal Muscle Cell Growth medium (PromoCell) and differentiated to MTs in Skeletal Muscle Cell Differentiation medium (Promocell). When specified, MBs were transfected with plasmid using Fugene HD transfection reagent (Promega) or reverse-transfected with siRNA using RNAiMAX transfection reagent (Invitrogen). hSkMc cells were lysed using RIPA buffer.
All siRNAs are described in the Supplemental Material.
IRAlus RNA sequencing
RNA was extracted from pEGFP-IRAlus-NICN1 transfected HEK293T cells using TRIzol reagent and treated twice with RQ1 RNase-Free DNase (Promega). cDNA was then prepared using 1 μg of total cell RNA, SuperScript III, and the EGFP-specific primer 5′-TCTACAAATGTGGTATGGCTG-3′, which anneals downstream from the IRAlus. PCR was performed using Vent DNA Polymerase (New England Biolabbs) and the primer pair 5′-CTTCTGCCATCTGACATAGC-3′ (sense) and 5′-ATGCTGAGCATATCTCTGG-3′ (antisense), which amplify sequences flanking the 3′-most Alu element. The resulting PCR products were inserted into the PCR-Blunt II-TOPO vector using the Zero Blunt TOPO PCR cloning kit (Invitrogen). Plasmid DNA from at least 20 colonies was sequenced.
Immunoprecipitations
Samples for the analysis of protein and RNA were generated before and after immunoprecipitation in the presence or absence of RNase A (Sigma) as reported (Gong et al. 2009). Immunoprecipitations were performed in 150 mM NaCl in the absence of cross-linking and in 500 mM NaCl when using lysates from formaldehyde cross-linked cells. Immunoprecipitations used gIgG (Santa Cruz Biotechnology), rIgG (Santa Cruz Biotechnology), or antibody that reacts with HA (Roche), p54nrb (Santa Cruz Biotechnology), or PKR (Cell Signaling). See Supplemental Table S5 for cell equivalents analyzed before and after immunoprecipitation and for immunoprecipitation efficiencies.
Western blotting
Proteins were electrophoresed in 6%–15% polyacrylamide and transferred to a nitrocellulose or PVDF membrane (Amersham Biosciences). Blots, which were probed using antibodies described in the Supplemental Material, were quantified using ImageQuant (Molecular Dynamics). See Supplemental Table S3 for standard deviations.
RT-qPCR
Total cell cDNA was synthesized using SuperScript III (Invitrogen) and random hexamers, Anchored Oligo(dT)20 Primer (Invitrogen), or cDNA-specific primers. Individual mRNAs were quantitated by amplifying the corresponding cDNA using cDNA-specific primer pairs (Supplemental Table S2), the 7500 Fast Real-Time PCR System (Applied Biosystems), and Fast SYBR Green Master Mix (Applied Biosystems) as specified (Gong et al. 2009). P-values were calculated using the two-tailed Student's t-test.
Fluorescent in situ hybridization (FISH) and microscopy
For FISH, see the Supplemental Material.
Acknowledgments
We thank L.-L. Chen and G.G. Carmichael for pEGFP-IRAlus reporters; M. Hatzoglou for MEFs and PKR vectors; S. de Lucas and J. Ortín for anti-STAU1; R.H. Singer for pmRFP; N. Kim for pcDNA3-Flag-LIN28; K.W. Nehrke for microscope use; L. Callahan at the University of Rochester Medical Center Conventional-Confocal Microscopy core for help with the FISH analysis; L.-L. Chen, M. Hatzoglou, and A. Almudevar for advice; and M.W.-L. Popp for comments on the manuscript. This work was supported by NIH R01 GM074593 to L.E.M. and NIH R01 GM084089 to B.T.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.220962.113.
References
- Athanasiadis A, Rich A, Maas S 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL 2002. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71: 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R 2002. Human interferon-γ mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell 108: 221–232 [DOI] [PubMed] [Google Scholar]
- Berger A, Strub K 2011. Multiple roles of Alu-related noncoding RNAs. Prog Mol Subcell Biol 51: 119–146 [DOI] [PubMed] [Google Scholar]
- Bond CS, Fox AH 2009. Paraspeckles: Nuclear bodies built on long noncoding RNA. J Cell Biol 186: 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY 2002. A monomeric red fluorescent protein. Proc Natl Acad Sci 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capshew CR, Dusenbury KL, Hundley HA 2012. Inverted Alu dsRNA structures do not affect localization but can alter translation efficiency of human mRNAs independent of RNA editing. Nucleic Acids Res 40: 8637–8645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG 2008. Gene regulation by SINES and inosines: Biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle 7: 3294–3301 [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol Cell 35: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, DeCerbo JN, Carmichael GG 2008. Alu element-mediated gene silencing. EMBO J 27: 1694–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Yang L, Carmichael GG 2010. Molecular basis for an attenuated cytoplasmic dsRNA response in human embryonic stem cells. Cell Cycle 9: 3552–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Chalamish S, Hasson A, Weinberg D, Namer LS, Banai Y, Osman F, Kaempfer R 2009. Dynamic refolding of IFN-γ mRNA enables it to function as PKR activator and translation template. Nat Chem Biol 5: 896–903 [DOI] [PubMed] [Google Scholar]
- Deininger P 2011. Alu elements: Know the SINEs. Genome Biol 12: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugré-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L 2005. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res 33: 4797–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick T, Huang S 2012. 3′-UTR-located inverted Alu repeats facilitate mRNA translational repression and stress granule accumulation. Nucleus 3: 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furic L, Maher-Laporte M, DesGroseillers L 2008. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA 14: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz G, Jagus R, Elroy-Stein O 2002. Phosphorylation of initiation factor-2α is required for activation of internal translation initiation during cell differentiation. Eur J Biochem 269: 2810–2819 [DOI] [PubMed] [Google Scholar]
- Gleghorn ML, Gong C, Kielkopf CL, Maquat LE 2013. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nat Struct Mol Biol 20: 515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE 2009. SMD and NMD are competitive pathways that contribute to myogenesis: Effects on PAX3 and myogenin mRNAs. Genes & Dev 23: 54–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN 2008. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 32: 276–284 [DOI] [PubMed] [Google Scholar]
- Huichalaf C, Sakai K, Jin B, Jones K, Wang GL, Schoser B, Schneider-Gold C, Sarkar P, Pereira-Smith OM, Timchenko N, et al. 2010. Expansion of CUG RNA repeats causes stress and inhibition of translation in myotonic dystrophy 1 (DM1) cells. FASEB J 24: 3706–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley HA, Bass BL 2010. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci 35: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A 2004. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res 14: 1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Furic L, DesGroseillers L, Maquat LE 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120: 195–208 [DOI] [PubMed] [Google Scholar]
- Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE 2007. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J 26: 2670–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N 1992. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science 257: 1685–1689 [DOI] [PubMed] [Google Scholar]
- Krayev AS, Markusheva TV, Kramerov DA, Ryskov AP, Skryabin KG, Bayev AA, Georgiev GP 1982. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res 10: 7461–7475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Manche L, Green SR, Schmedt C, Mathews MB 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol 12: 5238–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Macchi P, Furic L, Kiebler MA, DesGroseillers L 2006. Staufen1 is imported into the nucleolus via a bipartite nuclear localization signal and several modulatory determinants. Biochem J 393: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Dugre-Brisson S, Boulay K, Breton B, Lapointe G, Armando S, Trepanier V, Duchaîne T, Bouvier M, DesGroseillers L 2010. Multimerization of Staufen1 in live cells. RNA 16: 585–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeman Y, Levanon EY, Jantsch MF, Eisenberg E 2006. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA 12: 1802–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum JM, Gunnery S, Mathews MB 2002. The 3′-untranslated regions of cytoskeletal muscle mRNAs inhibit translation by activating the double-stranded RNA-dependent protein kinase PKR. Nucleic Acids Res 30: 1205–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Gleghorn ML, Maquat LE 2013. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci 110: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller CK, Li Z, George CX, Samuel CE 2011. Protein kinase PKR and RNA adenosine deaminase ADAR1: New roles for old players as modulators of the interferon response. Curr Opin Immunol 23: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A 2007. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes & Dev 21: 1125–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL 2007. Eukaryotic regulatory RNAs: An answer to the ‘genome complexity’ conundrum. Genes & Dev 21: 11–42 [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL 2005. Regulating gene expression through RNA nuclear retention. Cell 123: 249–263 [DOI] [PubMed] [Google Scholar]
- Ravel-Chapuis A, Belanger G, Yadava RS, Mahadevan MS, DesGroseillers L, Cote J, Jasmin BJ 2012. The RNA-binding protein Staufen1 is increased in DM1 skeletal muscle and promotes alternative pre-mRNA splicing. J Cell Biol 196: 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 7: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Sefton BM 2005. Labeling cultured cells with 32Pi and preparing cell lysates for immunoprecipitation. Cur Protocols Mol Biol 40: 18.2.1–18.2.8 [DOI] [PubMed] [Google Scholar]
- Sen SK, Han K, Wang J, Lee J, Wang H, Callinan PA, Dyer M, Cordaux R, Liang P, Batzer MA 2006. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet 79: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Zipori D 2002. PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett 531: 109–114 [DOI] [PubMed] [Google Scholar]
- Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, et al. 2007. Bright far-red fluorescent protein for whole-body imaging. Nat Methods 4: 741–746 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB, et al. 2010. The solution structure of the ADAR2 dsRBM–RNA complex reveals a sequence-specific readout of the minor groove. Cell 143: 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Gregory RI 2012. How does Lin28 let-7 control development and disease? Trends Cell Biol 22: 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, White RJ, Xia T, Welle S, Turner DH, Mathews MB, Thornton CA 2000. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA 6: 79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RD, Kugel JF, Goodrich JA 2009. InvAluable junk: The cellular impact and function of Alu and B2 RNAs. IUBMB Life 61: 831–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gong C, Maquat LE 2013. Control of myogenesis by rodent SINE-containing lncRNAs. Genes & Dev 27: 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham L, Duchaîne T, Luo M, Nabi IR, DesGroseillers L 1999. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol 19: 2220–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG 2001. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106: 465–475 [DOI] [PubMed] [Google Scholar]