The mammalian circadian timing system consists of a master pacemaker in the suprachiasmatic nucleus (SCN) in the hypothalamus. Saini et al. use a device consisting of a cylindrical cage with reflecting conical walls that channel photons toward a photomultiplier tube to record bioluminescence emitted in real time by hepatocytes expressing circadian luciferase reporter genes in freely moving mice. This revealed that hepatocyte oscillators of SCN-lesioned mice synchronized more rapidly to feeding cycles than hepatocyte clocks of intact mice.
Keywords: circadian gene expression, real-time bioluminescence monitoring, liver oscillators, resetting cues, SCN lesion
Abstract
The mammalian circadian timing system consists of a master pacemaker in the suprachiasmatic nucleus (SCN) in the hypothalamus, which is thought to set the phase of slave oscillators in virtually all body cells. However, due to the lack of appropriate in vivo recording technologies, it has been difficult to study how the SCN synchronizes oscillators in peripheral tissues. Here we describe the real-time recording of bioluminescence emitted by hepatocytes expressing circadian luciferase reporter genes in freely moving mice. The technology employs a device dubbed RT-Biolumicorder, which consists of a cylindrical cage with reflecting conical walls that channel photons toward a photomultiplier tube. The monitoring of circadian liver gene expression revealed that hepatocyte oscillators of SCN-lesioned mice synchronized more rapidly to feeding cycles than hepatocyte clocks of intact mice. Hence, the SCN uses signaling pathways that counteract those of feeding rhythms when their phase is in conflict with its own phase.
Virtually all mammalian physiology is subject to daily oscillations governed by a circadian timekeeping system. This system is composed of a master pacemaker in the suprachiasmatic nucleus (SCN) and subsidiary clocks in most peripheral cell types. The molecular oscillators operative in SCN neurons and peripheral cell types are thought to consist of two coupled feedback loops in the expression of specific core clock genes, including the transcriptional activators CLOCK, BMAL1, RORα, RORβ, and RORγ and the repressors PER1, PER2, CRY1, CRY2, REV-ERBα, and REV-ERBβ (Preitner et al. 2002; Jetten 2009; Relogio et al. 2011; Takeda et al. 2011; Bugge et al. 2012; Cho et al. 2012; for review, see (Dibner et al. 2010). The rhythmic output of the SCN, whose phase is entrained to daily light–dark (LD) cycles, synchronizes the oscillators in peripheral organs by a variety of direct and indirect signaling pathways. Feeding–fasting cycles, generated by rest–activity rhythms, have been shown to be dominant Zeitgebers (timing cues) for circadian clocks in the liver and other organs (Damiola et al. 2000; Yamazaki et al. 2000; Le Minh et al. 2001). However, endocrine (Balsalobre et al. 2000; Le Minh et al. 2001) and neuronal (Vujovic et al. 2008) outputs as well as body temperature oscillations (Brown et al. 2002; Kornmann et al. 2007; Reinke et al. 2008; Buhr et al. 2010; Saini et al. 2012) may also contribute to the phase resetting in these tissues.
The synchronization of peripheral clocks is obviously of utmost importance, since it is a prerequisite for the generation of overt cyclic outputs in physiology and behavior. While a bewildering array of signals can reset the clocks in cultured cells and explanted tissue sections (Stratmann and Schibler 2006), only few of the corresponding signaling pathways have been shown to be relevant in vivo. Owing to the functional redundancy of multiple pathways, the steady-state phase of circadian gene expression is quite resilient to the disruption of a particular pathway and therefore cannot be used as a reliable readout for the implication of the examined pathway in the synchronization process (Kornmann et al. 2007). In contrast, kinetics of phase shifting of circadian gene expression after the inversion of feeding rhythm has been shown to be quite sensitive to the disruption of a given pathway (Le Minh et al. 2001; Kornmann et al. 2007; Asher et al. 2010). Unfortunately, such experiments require the quantification of mRNA or proteins from hundreds of mice, since the phase of circadian gene expression must be tracked during several consecutive days (Asher et al. 2010). Moreover, this procedure yields low-resolution data at best, and the results may be confounded by individual variations in phase-shifting kinetics between different animals. While Shibata and coworkers (Tahara et al. 2012) have recently succeeded in imaging circadian bioluminescence emitted by the liver, kidney, and submandibular gland of individual PER2∷luciferase mice, these experiments were performed during only 1 d on mice that were anesthetized for each image acquisition. Clearly, a long-term, real-time recording procedure applicable to unconstrained, nonanesthetized animals would greatly facilitate the recording of phase-shifting kinetics of peripheral clocks.
Here we report on the development of a device that records bioluminescence in real-time, dubbed the RT-Biolumicorder, which enabled us to monitor circadian liver gene expression in freely moving mice at a high temporal resolution during many consecutive days. We explored the RT-Biolumicorder technology to determine the relative importance of the SCN and feeding cycles in the synchronization of hepatocyte clocks and observed that, upon the inversion of feeding cycles, the hepatocyte oscillators of SCN-lesioned mice adapted more rapidly to the new feeding regimen than those of wild-type mice.
Results
Visualization of circadian liver gene expression by whole-body bioluminescence imaging in anaesthetized mice
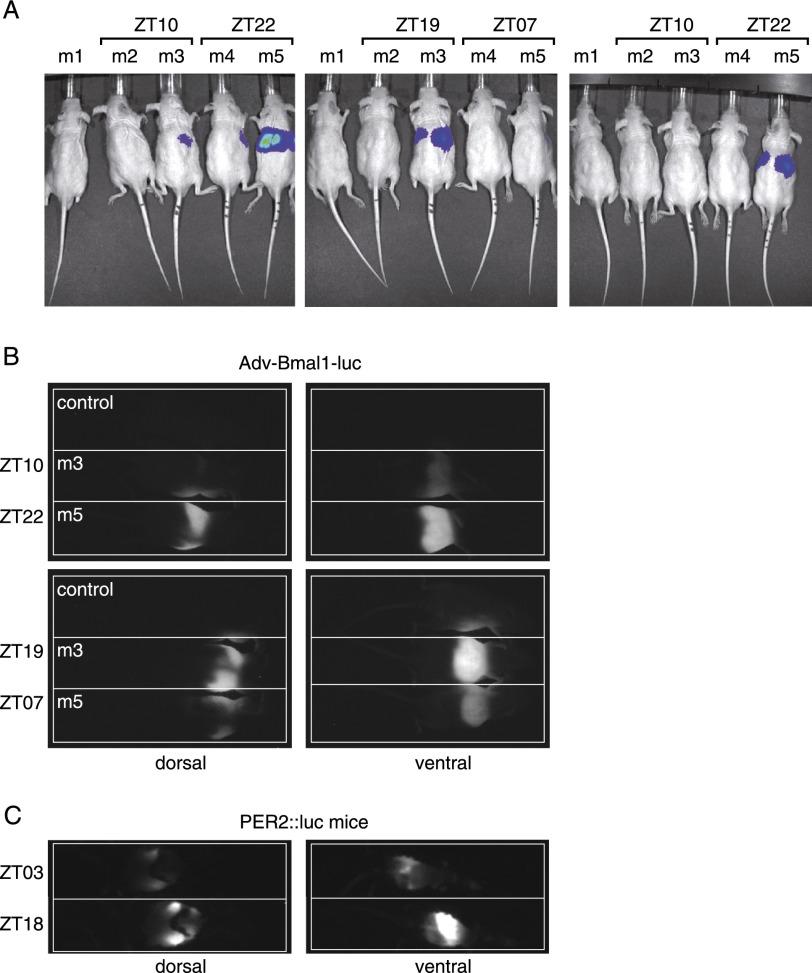
The activity of genes can be visualized in real-time by the recording of transgenic cells or organisms expressing fluorescent proteins or luciferase. Both of these techniques have been successfully adapted to whole-body imaging of mice (Hoffman and Yang 2006; Sandhu et al. 2010). Fluorescence measurements involve the excitation of fluorescent proteins with bright light, which would phase-shift the SCN (Albrecht et al. 2001). Therefore, bioluminescence monitoring appeared to be a more judicious method for tracking rhythmic gene expression in animals. By inserting an optical fiber, Yamaguchi et al. (2001) have succeeded in recording bioluminescence emitted by the SCN of freely moving mice harboring a Per1-luciferase transgene in real time. However, this has not yet been accomplished for peripheral organs. In order to examine the feasibility of monitoring bioluminescence in the liver in real time, we first conducted a few pilot experiments with anesthetized mice. Liver cells of hairless mice (SKH1-E, immunocompetent) were transduced with an adenoviral vector, Adv-Bmal1-luc, harboring a Bmal1-luciferase reporter gene via tail vein injection (Supplemental Fig. S1; Prill et al. 2011). Such adenoviral vector particles are known to primarily transduce hepatocytes. The mechanism is based on the fact that they bind to blood coagulation factor X upon injection into the blood stream. Factor X (bound to the vector particles) mediates a bridging to heparan surface proteoglycans (HSPGs) on the hepatocyte surface, and this triggers the uptake of viral particles by hepatocytes (Waddington et al. 2008; Alba et al. 2009; Bradshaw et al. 2010; Duffy et al. 2011). The bioluminescence recording of freely moving mice can only be performed on the dorsal side of animals, since the cage bottom is covered with nontransparent litter. Figure 1A displays dorsal bioluminescence images of hairless mice, which were injected with Adv-Bmal1-luc particles and luciferin. Three days to 4 d after Adv-Bmal1-luc transduction, the animals were anesthetized at the indicated Zeitgeber times (ZTs; ZT00 and ZT12 are the times when lights were switched on and off, respectively), injected intraperitoneally with a luciferin solution, and prepared for bioluminescence imaging using an IVIS-200 imaging station (Xenogen). As shown in Figure 1A, the livers of mice m3 and m5 (injected with the higher Adv-Bmal1-luc dose) yielded dorsal bioluminescence signals that were low and high when the endogenous Bmal1 gene was expressed at nadir and zenith levels, respectively. Hoping that freely moving animals could be imaged as well, we built a round lightproof cylindrical cage (211 mm) equipped with a hypersensitive EM-CCD camera (C900-13, Hamamatsu). We first determined whether bioluminescence signals could also be acquired for anesthetized mice with this homemade device. Again, strong, daytime-dependent signals of luminescent livers were obtained from both sides at the time of maximal Bmal1 expression (Fig. 1B). Similar results were obtained with PER2∷luc mice, in accordance with the observations recently reported by Tahara et al. (2012). PER2∷luc mice were engineered by Yoo et al. (2004) by inserting an ORF for luciferase downstream from and in phase with the Per2 ORF by homologous recombination in embryonic stem cells. These investigators showed that the PER2∷luciferase fusion protein performed all functions of the wild-type PER2 protein. Since PER2∷luciferase is expressed in most cell types (see Supplemental Fig. S2), we enriched the bioluminescence signal of the liver by depilating the mice on only a small region covering this organ. As the hair coat strongly absorbs photons, this yielded bioluminescence pictures of which a large part of the photons were contributed by the liver (Fig. 1C).
Figure 1.
Imaging of Bmal1-luciferase and PER2∷luciferase reporter gene expression in the livers of anesthetized mice. (A) The livers of four hairless mice were transduced with an adenoviral Bmal1-luciferase expression vector (Adv-Bmal1-luc) by the injection of 200 μL of PBS containing 2.5 × 109 (mice m2 and m4) and 2.5 × 1010 (mice m3 and m5) infectious particles into the tail vein. The mice were exposed during 10 d to light/dark (LD) cycles (m2 and m3: lights on and off at 11:00 pm and 11:00 am, respectively; m4 and m5: lights on and off at 11:00 am and 11:00 pm, respectively), before pictures were taken in an IVIS-200 imaging station (Xenogen) 3 and 4 d after Adv-Bmal1-luc transduction at the indicated ZTs. Fifty minutes before imaging, they were injected intraperitoneally with a luciferin solution (150 μg/g of body weight). They were under isoflurane (2%) anesthesia during image acquisition. The exposure time was 30 sec. (B) Mice m3 and m5 from A were injected intraperitoneally with a solution containing luciferin (as in A) and an anesthetic Ketarom (10 μL/g of body weight) (see the Material and Methods); placed in a homemade, lightproof cage (head toward the right side); and imaged dorsally and ventrally at the indicated ZTs 5 d after Adv-Bmal1-luc transduction. Images were taken 15 min after luciferin injection with an exposure time of 120 sec. (C) A PER2∷luc mouse was entrained during 10 d to LD cycles (lights on and off at 3:00 pm and 3:00 am, respectively), depilated in a zone surrounding the liver, and imaged at the indicated ZTs 30 min after luciferin injection with an exposure time of 30 sec.
Recording of circadian liver gene expression in real-time and in unconstrained mice
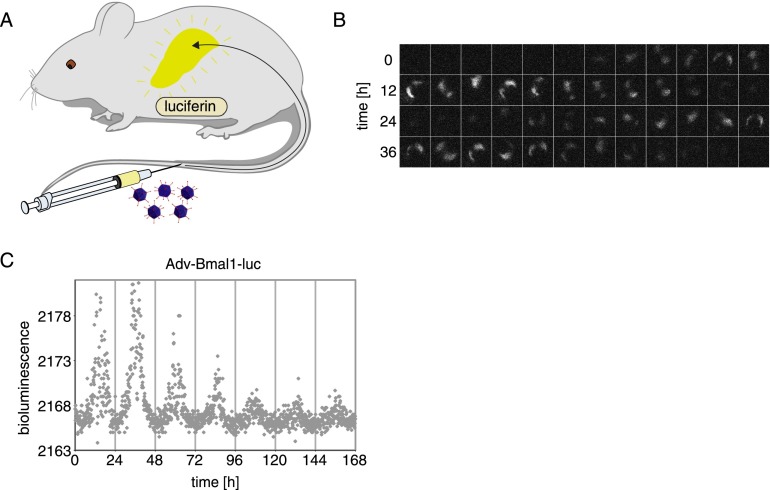
Next, we used a freely moving mouse expressing the Adv-Bmal1-luc reporter in the liver for a live-imaging movie. The animal was intraperitoneally injected with a luciferin solution (as above), placed in the lightproof cage when Bmal1-luc expression was expected to be maximal, and bioluminescence images were taken every 0.5 sec. Supplemental Movie M1, recorded over a period of 4.167 min, shows that readily discernible signals were produced from the liver of this unconstrained animal. For long-term recording, a constant supply of luciferin was afforded by the intraperitoneal or subcutaneous implantation of osmotic minipumps (Alzet) (cartoon in Fig. 2A). The luciferin concentrations reached in such animals were lower than those achieved by intraperitoneal injection, necessitating longer exposures. Nonetheless, it was thus possible to reconstruct time series of images during several days for mice expressing Adv-Bmal1-luc in the liver (Fig. 2B).
Figure 2.
Real-time recording of hepatic Adv-Bmal1-luc expression with a CCD camera. (A) Cartoon showing the preparation of a mouse for real-time bioluminescence imaging. The mouse was injected with 1 × 1011 infectious Adv-Bmal1-luc particles into the tail vein, and a osmotic minipump (model 2001, Alzet) containing a concentrated luciferin solution (90 mg/mL) was implanted into the intraperitoneal cavity (see the Materials and Methods). (B) Real-time imaging of bioluminescence emitted from the liver. A movie (exposure time, 30 sec) was taken during 48 h. The frames were inspected manually, and a picture was selected for each hour in which the mouse was immobile during the exposure time. The intervals between the selected frames were between 20 and 100 min. (C) Gray dots indicate time-integrated bioluminescence photon counts (30-sec EM-CCD exposure time) averaged over 7.5-min intervals as a function of time. Data were corrected for isolated high-intensity signal outliers (i.e., cosmic rays) (see the Materials and Methods).
Once the source of bioluminescence—the liver in this case, as shown in Figure 1A—is known, it suffices to count all photons emitted by a mouse, irrespective of whether it is moving or not. We hence designed a program (see the Materials and Methods) for the processing of total photon counts acquired by the CCD camera per time unit. The diagram in Figure 2C exhibits photon counts recorded during 1 wk. For all recordings presented in this study, bioluminescence monitoring was started at the beginning of nighttime. Presumably due to a gradual loss of luciferase-expressing cells and/or active Adv-Bmal1-luc vector molecules per cell (see below), the signals were somewhat weaker during the last days of recording, but circadian Bmal1-luc expression with the expected phase could be visualized by this technique during multiple days. While this method can be used for the long-term monitoring of circadian liver gene expression, it has two drawbacks. As seen in Figure 2C, the signal to background ratio is quite low when total photon counts are determined, and EM-CCD cameras are very expensive.
Development of the RT-Biolumicorder, a sensitive real-time bioluminescence recording device
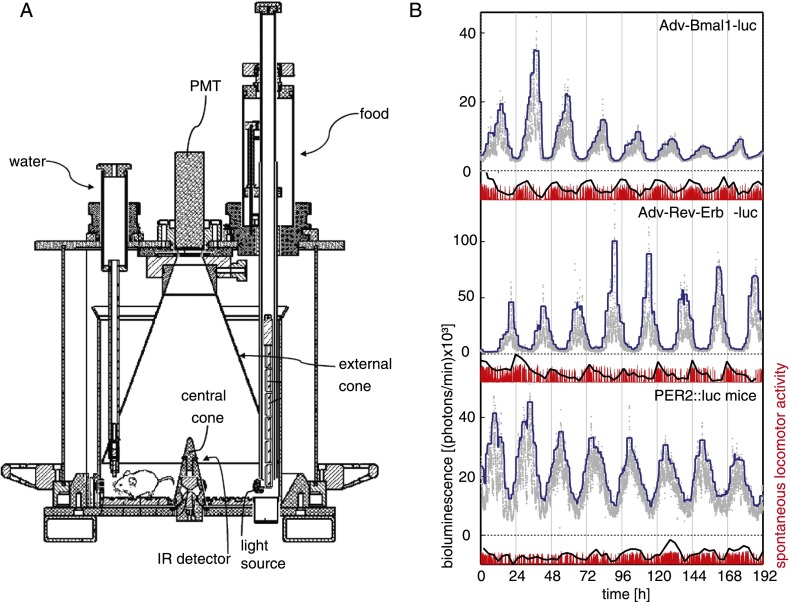
In cultured cells and tissue explants, circadian bioluminescence can be conveniently monitored by using photomultiplier tubes (PMTs) (Yamazaki et al. 2000; Nagoshi et al. 2004; Yoo et al. 2004). Since the dark current of PMTs is low and their efficiency in detecting photons is high, we adapted the lightproof recording cage to PMT technology and dubbed it the RT-Biolumicorder (Fig. 3A). Thus, the CCD camera was replaced by a PMT, and the stainless-steel walls of the cage, which conically converge to the counting surface of the PMT, were polished in order to reflect photons more efficiently. External and central cones increased photon counts by a factor of 1.5 and diminished the differences measured for a light source placed at different positions on the cage floor, respectively (Supplemental Fig. S3). The cage was also equipped with a device allowing the programming of feeding rhythms, a water delivery system, and an infrared sensor that monitors spontaneous locomotor activity. Finally, the most recent version of the RT-Biolumicorder (available from Lesa-Technology) (see the Materials and Methods) contains a programmable lighting system through which the SCN master pacemaker of experimental animals can be synchronized by skeleton photoperiods (i.e., two light pulses at 12-h intervals) (Oishi et al. 2002). Figure 3B displays bioluminescence data of 1-wk recordings obtained with animals in which Adv-Bmal1-luc and Adv-Rev-erbα-luc vectors (see the Supplemental Material; Supplemental Fig. S1) were transduced into hepatocytes and with a PER2∷luc transgenic mouse. Animals were fed ad libitum, and the corresponding locomotor activity profiles are shown at the bottom of each panel of Figure 3. The circadian profiles of Bmal1-luc, Rev-erbα-luc, and PER2∷luc could easily be visualized by this technique during multiple days, and the expression of these three reporters oscillated with phases corresponding to those observed for the endogenous Bmal1, Rev-erbα, and Per2 genes. We noticed in all experiments that the magnitude of Adv-Rev-erbα-luciferase expression remained high throughout the recording periods, whereas this parameter progressively decreased after a few days for the Adv-Bmal1-luciferase expression. It thus appears that the Bmal1-luc reporter gene, but not the Rev-erbα-luc reporter gene, loses activity over time. Conceivably, this difference is due to the difference in regulatory sequences driving luciferase expression in these two reporters. The genome-wide sequencing of DNase1-hypersensitive sites (DHSs) in mouse liver chromatin indeed revealed that the genomic Rev-erbα fragment used for the reporter gene expression (−3.5-kb flanking region + first intron) (see the Materials and Methods) encompasses all DHS regions associated with the Rev-erbα gene. In contrast, the Bmal1 genomic fragment only contains the promoter DHS region but lacks two intronic DHS regions (T Andersin, J Sobel, I Krier, U Schibler, and F Naef, unpubl.). As DHS regions are reliable marks for cis-acting regulatory sequences in promoters, enhancers, and locus control regions, we speculate that the Rev-erbα-luc reporter gene, but not the Bmal1-luc reporter gene, encompasses all DNA elements required for the long-term maintenance of an active chromatin configuration.
Figure 3.
Recording of hepatic Bmal1-luc, Rev-erbα-luc, and PER2∷luc expression using PMT technology. (A) Schematic representation of the RT-Biolumicorder (Lesa-Technology). The RT-Biolumicorder consists of a cylindrical cage equipped with a PMT (installed at the center above the cage), a feeding machine (right side, filled with food granules), a water flask (left side), and photon-reflecting walls. In addition, the device contains a large reflecting cone on top of the cage (external cone) that projects photons to the PMT and a small reflecting cone in the center of the floor (central cone) that projects photons to the reflecting walls. An infrared sensor built into the small cone records the locomotor activity of the mice. (B) One-week recordings of hairless mice transduced with Adv-Bmal1-luc vector (top panel) and Adv-Rev-erbα-luc vector (middle panel) and a PER2∷luc mouse (bottom panel) that was depilated in the dorsal region above the liver area. All mice had implanted osmotic minipumps (model 2001, Alzet), ensuring continuous luciferin delivery. Photons were accumulated during 1-min intervals (gray dots). The continuous blue line is a guide to the eye representing the 95% percentile of values associated with a smoothing window of 200 data points (see the Materials and Methods). Spontaneous locomotor activity profiles monitored simultaneously are shown below the bioluminescence tracings. Raw data of activity (red) were integrated over 200 data point intervals to help highlight the periodic oscillations in the signal (black line) (see the Materials and Methods). These three recordings were obtained with a prototype of the RT-Biolumicorder that was not yet equipped with reflecting cones.
Different clock genes are differentially responsive to food- and SCN-derived signals
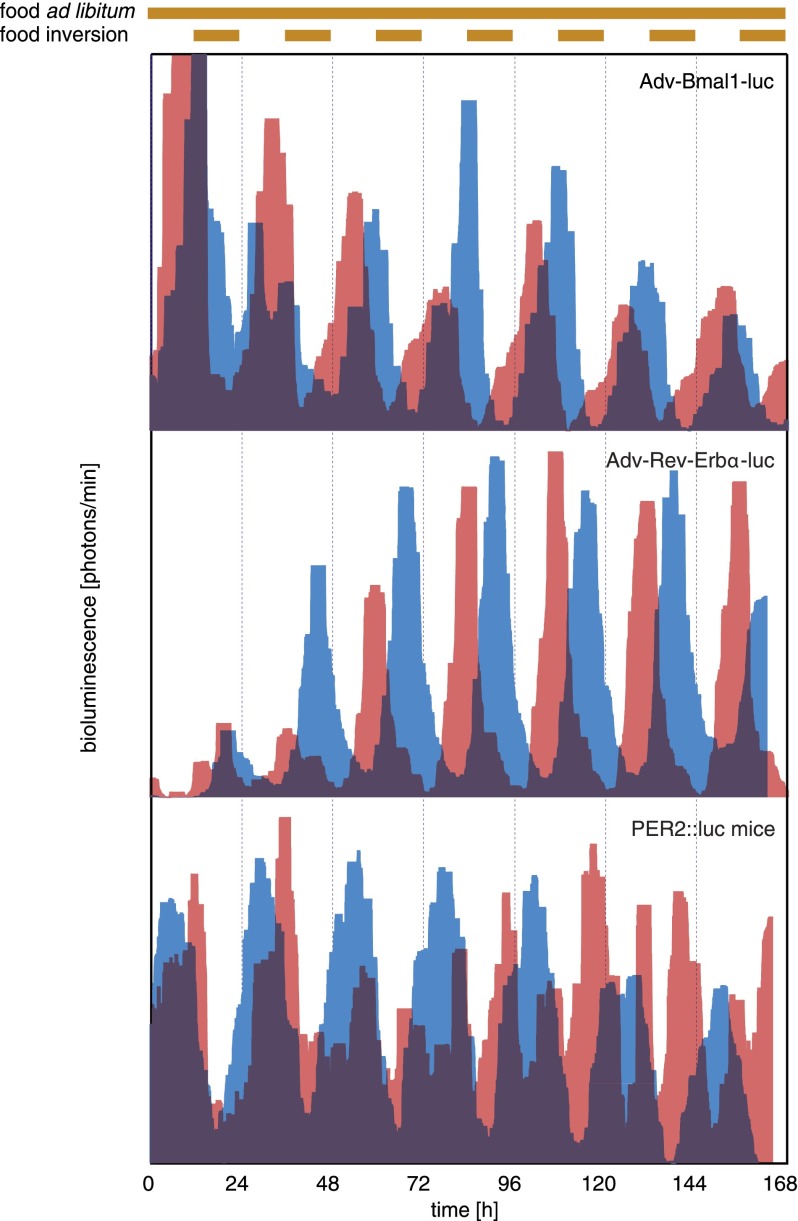
Feeding cycles, driven by rest–activity rhythms governed by the SCN, are dominant Zeitgebers for the oscillators in several peripheral organs, including the liver (Damiola et al. 2000; Stokkan et al. 2001; van der Veen et al. 2006). However, the SCN is also thought to use more direct pathways to synchronize peripheral clocks (e.g., circadian glucocorticoid signaling) (see Balsalobre et al. 2000; Le Minh et al. 2001; Reddy et al. 2007). The arguments in favor of this hypothesis rely on the kinetics of phase shifting of circadian gene expression after the inversion of feeding rhythms. Thus, when animals were switched from nighttime to daytime feeding, the phase of clock gene expression gradually changed and reached the new, inverted steady-state phase only after several days. However, when food was offered ad libitum again, the phase observed before inversion of the feeding regimen was rapidly resumed (Damiola et al. 2000). Moreover, hepatocytes of mice with a liver-specific glucocorticoid receptor gene disruption adapted their phase more rapidly to daytime feeding than hepatocytes proficient for glucocorticoids signaling (Le Minh et al. 2001). Since the phase of the SCN was found to be sensitive to neither feeding rhythms nor glucocorticoids signaling (Balsalobre et al. 2000; Damiola et al. 2000; Le Minh et al. 2001), it was argued that, upon the inversion of feeding rhythms, the SCN counteracted phase shifting in the liver and other organs by more direct signals; for example, by controlling the hypothalamic–pituitary–adrenal axis and thereby diurnal glucocorticoid signaling. Although this scenario seems plausible, it has never been examined by monitoring food-induced phase-shifting kinetics in peripheral organs of animals containing or not containing a functional SCN. Figure 4 and Supplemental Figure S4 display phase-shifting kinetics of the expression of Bmal1-luciferase, Rev-erbα-luciferase, and PER2∷luciferase in the livers of mice upon inversion of the feeding regimen. As expected, the phases of these expression profiles were gradually changed. Interestingly, while the phase angle of Bmal1-luciferase expression was shifted by only ∼60° 7 d after the restricted feeding regimen had been imposed, that of Rev-erbα-luciferase expression was already advanced by ∼120° at that time (Fig. 4; Supplemental Fig. S4). The bioluminescence profiles of PER2∷luciferase expression during this phase transition was a bit more difficult to interpret, since a double peak was observed during several days of the phase transition period. Conceivably, one of these peaks was contributed by nonhepatic tissues, which change the phase less rapidly upon restricted feeding (see Damiola et al. 2000; Stokkan et al. 2001). Nevertheless, 7 d after restricted feeding, the main peak of PER2∷luciferase expression was already advanced by ∼150°. These results indicated that different circadian genes responded with different kinetics to food entrainment and that the phase relationships between core clock components were uncoupled during the transition period. These observations were in accordance with the recent study by Asher et al. (2010) and suggested that different clock genes were differentially responsive to food- and SCN-derived signals. Kornmann et al. (2007) previously reported that circadian gene expression in the liver can be driven by both local cues and systemic signals. As the phases of Bmal1-luciferase expression were adjusted more slowly to inverted feeding rhythms, the transcription of this clock gene was probably strongly influenced by cues from the SCN, whose phase is not significantly affected by feeding rhythms. In contrast, Rev-erbα-luciferase and PER2∷luciferase appeared to be more sensitive to food-derived signals, suggesting that Rev-erbα and Per2 can act as immediate early genes responding to systemic signals. Hence, to evaluate the impact of SCN-derived phase-shifting signals, we used the Bmal1-luciferase reporter gene in subsequent experiments.
Figure 4.
Food-induced phase shifting of hepatic Bmal1-luc, Rev-erbα-luc, and PER2∷luc expression. One-week bioluminescence recordings of hairless mice transduced with Adv-Bmal1-luc (top panel) or Adv-Rev-erbα-luc (middle panel) and a PER2∷luc mouse depilated in a region over the liver (bottom panel). Before monitoring, animals were housed under LD cycles (lights on and off at 6:00 pm and 6:00 am, respectively) and obtained food ad libitum. As soon as the bioluminescence monitoring was started, food (brown bars above the panels) was offered exclusively during the subjective days (red profiles) or ad libitum (blue profiles). Note that 7 d (168 h) after the feeding regimen had been inverted, the phase of Bmal1-luciferase expression was shifted by only 6–7 h. In contrast, the phases of Rev-Erbα-luciferase and PER2∷luciferase expression were almost completely inverted (by 12 h) at this time. For the purpose of clarity, only filtered data (see the Materials and Methods) are shown, and the values on the Y-axis are not depicted. These values were between 2000 and 30,000 photons per minute for the Bmal1-luciferase reporter, 5000 and 250,000 photons per minute for the Rev-Erbα-luciferase reporter, and 2000 and 70,000 photons per minute for PER2∷luciferase mice. The background of the PMT is <1000 photons per minute. These recordings were obtained with a prototype of the RT-Biolumicorder that was not yet equipped with reflecting cones.
Circadian Bmal1 expression adapts more rapidly to inverted feeding cycles in SCN-lesioned mice
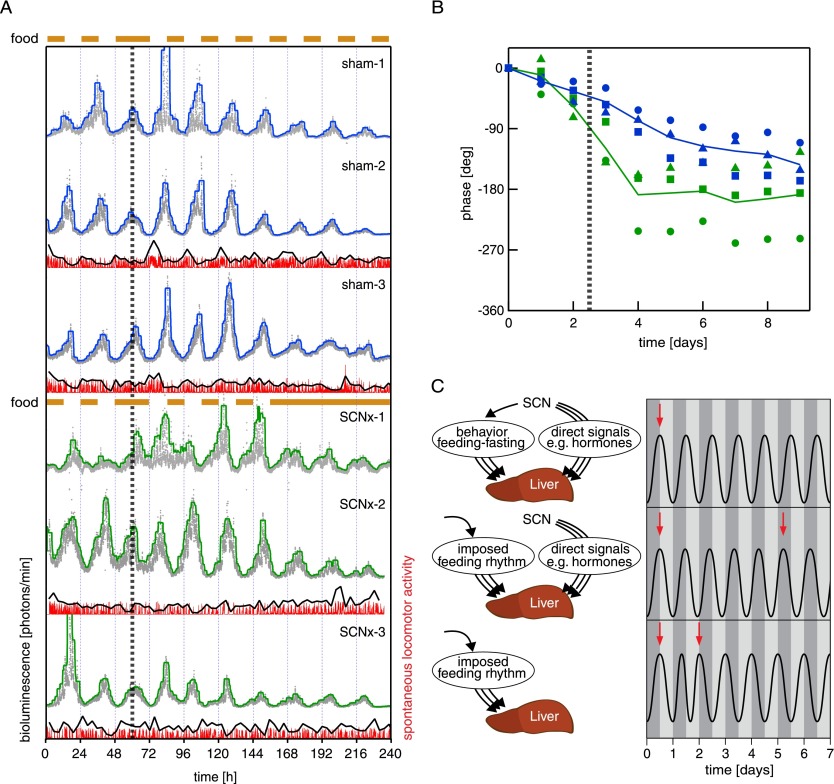
SCN tissues of mice were ablated by stereotaxic lesions (see the Materials and Methods), and actograms of locomotor activities were examined to ensure that the surgical intervention resulted in arrhytmicity (Supplemental Fig. S5). In sham-operated mice, which served as controls, the electrode was inserted into the SCN region, but the electrical current cauterizing the surrounding tissue was not turned on (Supplemental Fig. S5). The animals were kept on a night feeding regimen for 4 wk before transduction with Adv-Bmal1-luc reporter vectors and implantation of osmotic minipumps delivering luciferin. The mice were then placed into the RT-Biolumicorder, and photons were recorded at 1-min intervals. After two additional days of nighttime feeding, the feeding regimen was inverted, and the phase-shifting kinetic was monitored. As shown in Figure 5A (top three panels) the sham-operated mice did not yet reach the new, expected steady-state phase 6 d after the inversion of the feeding regimen, similar to the results obtained previously with intact animals (Fig. 4, top panel). The SCN-lesioned animals exhibited a somewhat delayed phase of circadian Bmal1-luc expression already during nighttime feeding when compared with the sham-operated mouse (Fig. 5A, bottom three panels). Hence, in the latter, the SCN probably contributed to the timing of the steady-state phase of hepatic gene expression not only through controlling feeding rhythms. Strikingly, hepatic Bmal1-luc expression in SCN-lesioned mice was rapidly shifted to the new steady-state phase after the feeding rhythm had been inverted (Fig. 5A,B). Therefore, in SCN-proficient animals, the more direct signals emitted by the SCN slowed down the phase-shifting kinetics of liver clocks when feeding cycles were in conflict with the phase normally established by the SCN. In SCN-lesioned animals, only food-dependent signals were operative, and the phase of the liver was thus rapidly inverted. This scenario is illustrated by the cartoons depicted in Figure 5C. Under normal conditions (Fig. 5C, top panel), the SCN synchronizes hepatocyte oscillators by driving feeding rhythms (through rest–activity cycles) and more direct timing cues (e.g., glucocorticoid signaling). It is likely that multiple feeding-dependent and more direct SCN-dependent molecular signaling pathways (symbolized by arrows in Fig. 5C) are involved in the synchronization of liver clocks, but under normal conditions, all of these pathways act in concert. Upon inversion of the feeding regimen (Fig. 5C, middle panel), the SCN keeps its phase. Hence, the direct signals emanating from the SCN are now in conflict with the signals dependent on feeding (or fasting), and since the latter are dominant, they gradually change the phase of peripheral oscillators in the liver. This may take between 1 and 2 wk, depending on the clock gene analyzed. In SCN-lesioned animals (Fig. 5C, bottom panel), only food-dependent signals are operative, and the phase of the liver is therefore reversed rapidly upon inverting the feeding regimen.
Figure 5.
Phase shifting of hepatic Bmal1-luciferase expression in mice with and without a SCN pacemaker. (A) Mice were exposed to nighttime feeding during 4 wk before monitoring. Two days after the real-time bioluminescence recording was initiated, the feeding regimen (brown bars above the panels) was inverted (vertical dotted line), and the animals only had access to food during the subjective day. Note that the SCN-lesioned mice (SCNx-1, SCNx-2, and SCNx-3; three bottom panels) reached the new steady-state phase of hepatic Bmal1-luciferase expression within 1–2 d after inversion of the feeding rhythm. In contrast, the animals with an intact SCN (sham-1, sham-2, and sham-3; three top panels) only advanced the phase of circadian Bmal1-luciferase expression by ∼6–7 h 7 d after the inversion of the feeding regimen. For clarity purposes, the raw data (gray for bioluminescence and red for locomotor activity) were filtered (blue/green and black, respectively) (see the Materials and Methods). These values on the Y-axis were between 2000 and 30,000 photons per minute. (Fourth panel from top) Note that if food was offered ad libitum again (SCNx-1), the hepatocyte oscillators free-ran with the food-entrained phase for a few days and did not resume the old phase (see also Damiola et al. 2000). Sham-1, Sham-2, Sham-3, and SCNx-1 recordings were obtained with a prototype of the RT-Biolumicorder that was not yet equipped with reflecting cones, while SCNx2 and SCNx3 recordings were acquired using the final version of the RT-Biolumicorder. For sham-1 and SCNx-1 recordings, the prototype of the RT-Biolumicorder was not yet equipped with the infrared detection device. (B) This diagram shows the phase angle changes recorded in A as a function of time after inversion (vertical dotted line) of the feeding regimen (note that in this diagram, the peaks before feeding inversion have been aligned in sham-operated and SCN-lesioned mice) (for calculation, see the Materials and Methods). (C) The three cartoons schematically illustrate our interpretations of the findings illustrated in A and B. See the text for detailed explanations.
Discussion
SCN-controlled signals counteract food-derived signals during the synchronization of liver clocks to feeding rhythms
The RT-Biolumicorder procedure described in this study holds promise for the investigation of gene expression features that were difficult to study thus far. Here we used the technology to examine the impact of signals controlled by feeding rhythms and the SCN on the synchronization of circadian oscillators in the livers of unconstrained freely moving mice. We are confident that the RT-Biolumicorder technology faithfully tracks the phases of rhythmically transcribed gene. Indeed, in all of our experiments, the phases of the recorded bioluminescence cycles closely reflected the known phases of Bmal1, Rev-erbα, and Per2 expression. The results demonstrated that the SCN, whose phase is not sensitive to feeding times, slows down the phase shifting of hepatocyte clocks upon inversion of the feeding regimen. Presumably, it uses signaling pathways, counteracting those related to metabolic cues when the phase of feeding rhythms is in conflict with its own phase. Hence, the RT-Biolumicorder technology will be particularly useful in examining the contribution of molecular signaling pathways to the phase entrainment of peripheral clocks. By using the food-dependent phase inversion paradigm, it should be possible to examine the kinetics of phase shifting in the presence and absence of a given signaling component. The Adv-Bmal1-luc expression vector that we engineered also encodes a Cre recombinase (see Supplemental Fig. S1). Therefore, the transduction of mice with floxed alleles for genes encoding such signaling components should result in the inactivation of the respective gene in the hepatocytes expressing Bmal1-luciferase.
Circadian liver gene expression persists in SCN-lesioned mice
We also examined circadian reporter gene expression in SCN-lesioned mice fed ad libitum, as previous studies have reported somewhat conflicting results. According to Yoo et al. (2004), individual organs still display circadian PER2 expression in peripheral organs of SCN-lesioned PER2∷luc mice, although phase coherence is lost between different organs. Tahara et al. (2012) also observed circadian gene expression in organs such as the liver, kidney, and submandibular gland of a large fraction of SCN-lesioned mice. In contrast, Guo et al. (2006) claimed that phase coherence is lost even between cells of the same organ in SCN-lesioned hamsters. As shown in Supplemental Figure S6A, our results revealed circadian reporter gene expression for several days in freely moving SCN-lesioned mice and thus tend to support the conclusions of Yoo et al. (2004) and Tahara et al. (2012). However, our results must be interpreted with caution, since it is conceivable that the anesthesia and surgical inventions (i.e., tail vein injection and/or implantation of a minipump) reset the phase of desynchronized hepatocyte oscillators. For example, the stress associated with these manipulations might have induced a surge of glucocorticoid hormones, which are strong Zeitgebers for liver clocks (Balsalobre et al. 2000; Le Minh et al. 2001; Reddy et al. 2007). In accordance with the study by Tahara et al. (2012), we noticed that the amplitudes of Bmal1-luciferase expression were somewhat lower in SCN-lesioned mice fed ad libitum as compared with intact mice. This could indicate a partial desynchronization of hepatocyte oscillators in the former (Supplemental Fig. S6B,C).
The RT-Biolumicorder technology opens new avenues in a wide range of biomedical fields for studying gene expression in vivo
The injection of adenoviral vectors into the tail vein results mostly in the transduction of hepatocytes. However, our RT-Biolumicorder technology should also be applicable to tissues other than the liver. For example, transgenic mouse strains could be engineered in which circadian luciferase reporter gene expression is only activated after Cre-mediated site-specific recombination and in which Cre recombinase expression is directed to organs like the kidney, skeletal muscle, heart muscle, pancreas, skin, and brain. Obviously, signal strength may be a limiting factor for tissues containing a small number of cells. However, since in our experiments with Adv-Rev-erbα-luc, the signal obtained for liver is at least 10-fold above the threshold of detection, it probably will be feasible to determine the circadian phase in the aforementioned organs. Moreover, the in vivo bioluminescence monitoring should be readily applicable to measuring circadian gene expression in grafted bioluminescent tumors.
Obviously, our method is by no means limited to circadian biology. Thus, it should readily reveal the kinetics of signaling by hormones; cytokines; neuronal pathways; metabolites such as bile acids, cholesterol, fatty acids, glucose, and oxygen; and medical drugs. Metabolites and drugs are frequently detected by nuclear receptors and other transcription factors, such as family members of the peroxisome proliferator-activated receptors (PPARs) (Tan et al. 2005; Peraza et al. 2006; Grimaldi 2007), farnesoid X receptor (Handschin and Meyer 2005; Kuipers et al. 2007), liver X receptors (LXRs) (Handschin and Meyer 2005), retinoic acid receptors (RARs) (Altucci et al. 2007), sterol regulatory element-binding proteins (SREBPs) (McPherson and Gauthier 2004), carbohydrate response element-binding protein (ChREBP) (Postic et al. 2007), constitutive androstane receptor (CAR) (Handschin and Meyer 2005), pregnane X receptor (PXR) (Handschin and Meyer 2005), aryl hydrocarbon receptor (AHR; also known as dioxin receptor) (Abel and Haarmann-Stemmann 2010), and hypoxia-induced factor (HIF) (Kaelin and Ratcliffe 2008). Thus, we are hopeful that our technology will find wide and fruitful applications in many areas of biomedical research.
Materials and methods
Animal care
Male hairless mice (strain code 477; formerly SKH1 mice with strain code 313) were purchased from Charles River, and PER2∷luc mice were generously provided by Joseph S. Takahashi (University of Texas Medical Center, Dallas, TX) (Yoo et al. 2004). All animals were housed according to standard conditions, and the surgery experiments were approved by the Veterinary Offices of the Cantons of Geneva and Vaud. For the phase entrainment to defined LD cycles, the animals were kept individually in cages placed in homemade, ventilated cabinets with computer-programmable LD cycles (A. Liani and Y.A. Poget, Mechanical Workshop of the Department of Molecular Biology, University of Geneva). When indicated, feeding cycles were imposed before and during bioluminescence recording by a homemade programmable feeding machine (A. Liani and Y.A. Poget).
Locomotor activity recording
Male SKH1 mice (age 3–8 mo) were individually housed with food and water offered ad libitum. Cages were placed in a sound-attenuated and temperature-controlled recording room (25°C). Activity was recorded under 12 h:12 h LD cycles (lights on at 9:00 am, 110 lux) for 10 d before surgery and 3–4 d after surgery. Thereafter, locomotor activity was monitored in constant darkness (DD) for 3 wk to assess the circadian organization of overt behavior under Zeitgeber-free conditions. Spontaneous and voluntary locomotor activities were recorded using passive infrared (PIR) sensors (SPY 4/RTE-A, Visonic) and running wheels, respectively. ClockLab software (Actimetrics) was used for both data acquisition and analyses. The presence or absence of significant circadian rhythmicity in locomotor activity was evaluated using a χ2 periodogram analysis (P < 0.01).
SCN ablation
Bilateral lesion of the SCN was performed stereotaxically (963LS, Kopf Instruments) under ketamine/xylazine anesthesia (intraperitoneal injection, 75 and 10 mg/kg, at a volume of 8 mL/kg). Two electrodes (0.3 mm in diameter) were introduced at the following coordinates: anteroposterior, +0.2 mm using the bregma as the reference; lateral, ±0.2 mm; and depth, −5.7 mm. Electrolytic lesions (1 mA, 5 sec) were made using a DC lesion-making device (3500, Ugo Basile). In sham-operated animals, no electric current was applied. These experiments were performed under the permission of the Veterinary Office of the Canton of Vaud, granted to Paul Franken.
Osmotic minipump implantation and tail vein injection
Micro-osmotic pumps (pump model 2001, blue flow moderator 0002489, Alzet, Charles River) were filled with 90 mg/mL luciferin (306A NF-FF free-acid firefly D-luciferin, Nanolight Technology, Prolume Ltd.) solution under a sterile hood. The luciferin solution was previously filtered with a 0.45-μm filter (Millex-HN SLHN R04 NL, Millipore) to prevent flow moderator obstruction. The pumps were implanted into the intraperitoneal cavity of hairless mice anesthetized by intraperitoneal administration of Ketarom solution (12% ketasol 100 [Dr. E. Graeub, AG Bern] and 8% rompun [Bayer Provet SA]). Experiments in which the osmotic minipumps were implanted subcutaneously yielded similar bioluminescence recordings. To avoid cooling of hairless animals, the operating platform was kept warm (near body temperature) using a homemade heating system (A. Liani and Y.A. Poget). Subsequently, 1 × 1011 adenoviral particles diluted into sterile PBS (final injected volume 200 μL) were injected into the tail vein using a 0.3 × 13-mm 30-gauge needle (304000, BD Microlance) under a P2 hood. To facilitate penetration of the needle, the tail of the animal was maintained for 30 sec in warm water (∼40°C) prior to injection, thereby expanding the vein. The animals were then placed in a cage with a filter cover in a box preheated to 32°C until they woke up from anesthesia. After 30–32 h of recovery, the animals were transferred to the RT-Biolumicorder, and the bioluminescence monitoring was started. During the recovery time, a painkiller (117 mg/L Algifor 400, Vifor Pharma) was added to the drinking water. All of these experimental protocols were approved by the Veterinary Office of the Canton of Geneva. PER2∷luc mice were depilated in a zone surrounding the liver area under anesthesia before pump implantation using depilatory cream (Veet).
In vivo bioluminescence recording and image processing
In experiments with a CCD camera (EM-CCD C900-13, Hamamatsu), bioluminescence recording was performed in a homemade, ventilated, and light-tight cylindrical cage with black walls (A. Liani and Y.A. Poget). For long-term monitoring, the floor of the cage was filled with a layer of nonchemiluminescent litter (Cha-Cha-Cha cat litter, Coop, or mulching film [BIOFILIA]), and food and water were provided ad libitum. Images were acquired using CCD camera software (Hokawoo, Hamamatsu) with exposure time ranging from 30 sec to 2 min depending on the type of analysis (moving or anesthetized mice). For data collected from freely moving animals, CCD images were processed using a homemade LabView-based program (A Liani, unpubl.). Briefly, the software converts TIF images into pixel arrays whose elements are subjected to two filters in a sampling window. A “low filter” eliminates pixels with intensity levels close to the background of the CCD camera, whereas a “high filter” eliminates pixels with a very high intensity probably generated by cosmic rays. The newly created array of pixels was taken into account if it contained a minimum of 200 samples. The average value of the samples was displayed in a graph and stored as TXT file.
For real-time acquisitions of photons by PMT technology, the RT-Biolumicorder technology was established (A. Liani and Y.A. Poget; now available at Lesa-Technology, see http://www.lesa-technology.com). The interior walls of the RT-Biolumicorder are light-reflecting, and the shape of the cage is optimized for a maximum capture of the photons (see Supplemental Fig. S3). Food can be delivered at will by a computer-programmable feeding system, and the food and water levels within the corresponding reservoirs can be read from the outside. The cage is also equipped with an air ventilation system, an infrared sensor for the monitoring of locomotor activity, and a computer-programmable LED light source, allowing for the synchronization of the master circadian pacemaker in the SCN by “skeleton photoperiods.” For long-term monitoring, the floor of the cage (surface of 349.67 cm2) was filled with nonchemiluminescent litter (see above). Photons emitted by the liver of freely moving mice were integrated over 1-min intervals and collected together with activity counts using RT-Biolumicorder data acquisition software (Instrument Control based on the LabView platform).
Processing of bioluminescence and locomotor activity data
As previously mentioned, bioluminescence intensity traces recorded with RT-Biolumicorder technology were acquired at a 1-min sampling rate. The continuous thick line superposed to the raw data (see Figs. 3B, 5A; Supplemental Fig. S6) was a guide to the eye, representing the 95% percentiles of values associated to a smoothing window of 200 data points. Percentile smoothing was preferred to median smoothing (i.e., 50% percentile) to provide a stronger bias for high-intensity values and minimize the sharp signal losses caused by a temporarily poor supply of luciferin or unfavorable animal positions in the cage (parameters that obviously distort the quantitative relationship between photon emission and reporter gene expression). In Figure 4 and Supplemental Figure S4, raw data are not shown; only filtered values are depicted in area-style profiles.
The thick black line superposed to the raw activity data (see Figs. 3B, 5A; Supplemental Fig. S6) was computed taking the integral of the activity traces over intervals of 200 consecutive data points. The latter procedure helped highlight the periodic oscillations in the signal, which were otherwise hindered by the saturation of the activity counts due to the limited sampling rate (60 events per minute).
Calculation of phase angle changes after inversion of the feeding regimen
Figure 5B highlights the phase angle changes recorded in Figure 5A as a function of time after inversion of the feeding regimen. The peaks in the oscillatory bioluminescence response of a given animal were fitted with a Gaussian function to automatically retrieve the positions of their maxima. The time shift with respect to an ideal 24-h daily cycle was computed by subtracting to the nth peak position the position of the first peak and the total number of daily cycles: (n − 1) × 24 h. The phase shift was finally calculated by setting 24 h = 360°.
Generation of Bmal1-luciferase- and Rev-erbα-luciferase-expressing adenoviruses and viral particle production
The preparation of these two adenoviral vectors is schematically depicted in Supplemental Figure S1. The construction of the adenoviral Bmal1-luc expression vector has been published previously (Asher et al. 2008). For the Rev-erbα-luc adenoviral vector, the MluI/PciI fragment from the mRev-erbα locus was cloned in two steps into pGL3-basic (Promega) digested with MluI and NcoI in order to fuse the first 16 amino acids of REV-ERBα with luciferase (previously published) (Stratmann et al. 2010). The MluI–DraI fragment from pGL3-Rev-erbα-luc (encompassing 3493 base pairs [bp] of the 5′ upstream Rev-erbα regulatory region, exon1, intron1, the first 14 bp of exon 2 followed by the luciferase coding sequence, and a polyA signal sequence) was inserted into the pCV100 adenoviral backbone plasmid digested with PacI. Adenoviruses were generated and purified as described previously (Kreppel et al. 2002).
Histochemical procedures and Nissl staining
At the end of the experiment, mice were anaesthetized by ketamine/xylazine (intraperitoneal injection, 75 and 10 mg/kg, at a volume of 8 mL/kg) and subjected to intracardiac perfusion with a PBS solution containing heparin (2 U/mL) and 4% paraformaldehyde (PFA) (pH 7.4). Brains were carefully removed and successively incubated in PBS solutions containing 10% sucrose (1 h), 20% sucrose (1 h), and 30% sucrose (48 h) at 4°C before being embedded in the OCT compound at −20°C until cryostat sectioning.
Coronal brain sections (20 μm) were washed in PBS and then fixed in PBS containing 4% PFA for 10 min at 4°C. Brain sections were then incubated for 10–15 min in Cresyl violet (Sigma-Aldrich) and washed in distilled water. Brain slides were then successively immersed in 95% ethanol (30 sec), 100% ethanol (30 sec), and xylene (three washes, 3–5 min each) before being placed in Eukitt medium (Fluka) and covered with coverslips. Images for chemically stained whole-brain coronal sections were finally acquired using a stereomicroscope MZ16FA (Leica) (see Supplemental Fig. S5).
Detrending of bioluminescence data by moving average transformation and amplitude calculation
To compare bioluminescence values with regard to amplitude (Supplemental Fig. S6), we eliminated variations in the magnitude of the signals resulting from different adenoviral transduction efficiencies or differences in luciferin metabolization. To this end, the oscillation period of each animal was calculated on raw data by Fourier transform. A moving-window algorithm, as previously described (Saini et al. 2012), was then applied to the data set at an interval corresponding to the period calculated for each animal. Each peak of detrended values was fitted by a Gaussian to retrieve amplitude. Amplitude values were plotted as function of time.
Acknowledgments
We thank David Suter for his helpful suggestions, Christophe Bauer and Jérôme Bosset for their help with CCD camera settings, and Nicolas Roggli for the artwork. Work in U.S.'s laboratory was supported by the Swiss National Science Foundation (SNF 31-113565 and SNF 31-128656/1 and the NCCR program grant Frontiers in Genetics), the European Research Council (ERC-AdG-TimeSignal), the Canton of Geneva, and the Louis Jeantet Foundation of Medicine. Work in P.F.'s laboratory was supported by the Swiss National Science Foundation (SNF 31003A-130825), the Marie Curie Intra-European program (IEF-FP7 project no: 221254), the Novartis Foundation, the EUMODIC (European FP6 program: contract no. 037188), and the Canton of Vaud. Work in J.P.W.'s laboratory was supported by the Swiss National Science Foundation National Centers of Competence in Research (NCCR) program grant Molecular Ultrafast Science and Technology (NCCR MUST).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.221374.113.
References
- Abel J, Haarmann-Stemmann T 2010. An introduction to the molecular basics of aryl hydrocarbon receptor biology. Biol Chem 391: 1235–1248 [DOI] [PubMed] [Google Scholar]
- Alba R, Bradshaw AC, Parker AL, Bhella D, Waddington SN, Nicklin SA, van Rooijen N, Custers J, Goudsmit J, Barouch DH, et al. 2009. Identification of coagulation factor (F)X binding sites on the adenovirus serotype 5 hexon: Effect of mutagenesis on FX interactions and gene transfer. Blood 114: 965–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC 2001. MPer1 and mper2 are essential for normal resetting of the circadian clock. J Biol Rhythms 16: 100–104 [DOI] [PubMed] [Google Scholar]
- Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H 2007. RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov 6: 793–810 [DOI] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U 2008. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134: 317–328 [DOI] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U 2010. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 142: 943–953 [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U 2000. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289: 2344–2347 [DOI] [PubMed] [Google Scholar]
- Bradshaw AC, Parker AL, Duffy MR, Coughlan L, van Rooijen N, Kahari VM, Nicklin SA, Baker AH 2010. Requirements for receptor engagement during infection by adenovirus complexed with blood coagulation factor X. PLoS Pathog 6: e1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U 2002. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol 12: 1574–1583 [DOI] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA 2012. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev 26: 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, Takahashi JS 2010. Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330: 379–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. 2012. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485: 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U 2000. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14: 2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U 2010. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol 72: 517–549 [DOI] [PubMed] [Google Scholar]
- Duffy MR, Bradshaw AC, Parker AL, McVey JH, Baker AH 2011. A cluster of basic amino acids in the factor X serine protease mediates surface attachment of adenovirus/FX complexes. J Virol 85: 10914–10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi PA 2007. Peroxisome proliferator-activated receptors as sensors of fatty acids and derivatives. Cell Mol Life Sci 64: 2459–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Brewer JM, Lehman MN, Bittman EL 2006. Suprachiasmatic regulation of circadian rhythms of gene expression in hamster peripheral organs: Effects of transplanting the pacemaker. J Neurosci 26: 6406–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Meyer UA 2005. Regulatory network of lipid-sensing nuclear receptors: Roles for CAR, PXR, LXR, and FXR. Arch Biochem Biophys 433: 387–396 [DOI] [PubMed] [Google Scholar]
- Hoffman RM, Yang M 2006. Whole-body imaging with fluorescent proteins. Nat Protoc 1: 1429–1438 [DOI] [PubMed] [Google Scholar]
- Jetten AM 2009. Retinoid-related orphan receptors (RORs): Critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal 7: e003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG Jr, Ratcliffe PJ 2008. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol Cell 30: 393–402 [DOI] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U 2007. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreppel F, Biermann V, Kochanek S, Schiedner G 2002. A DNA-based method to assay total and infectious particle contents and helper virus contamination in high-capacity adenoviral vector preparations. Hum Gene Ther 13: 1151–1156 [DOI] [PubMed] [Google Scholar]
- Kuipers F, Stroeve JH, Caron S, Staels B 2007. Bile acids, farnesoid X receptor, atherosclerosis and metabolic control. Curr Opin Lipidol 18: 289–297 [DOI] [PubMed] [Google Scholar]
- Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U 2001. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J 20: 7128–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson R, Gauthier A 2004. Molecular regulation of SREBP function: The Insig–SCAP connection and isoform-specific modulation of lipid synthesis. Biochem Cell Biol 82: 201–211 [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U 2004. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119: 693–705 [DOI] [PubMed] [Google Scholar]
- Oishi K, Fukui H, Sakamoto K, Miyazaki K, Kobayashi H, Ishida N 2002. Differential expressions of mPer1 and mPer2 mRNAs under a skeleton photoperiod and a complete light-dark cycle. Brain Res Mol Brain Res 109: 11–17 [DOI] [PubMed] [Google Scholar]
- Peraza MA, Burdick AD, Marin HE, Gonzalez FJ, Peters JM 2006. The toxicology of ligands for peroxisome proliferator-activated receptors (PPAR). Toxicol Sci 90: 269–295 [DOI] [PubMed] [Google Scholar]
- Postic C, Dentin R, Denechaud PD, Girard J 2007. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu Rev Nutr 27: 179–192 [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U 2002. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251–260 [DOI] [PubMed] [Google Scholar]
- Prill JM, Espenlaub S, Samen U, Engler T, Schmidt E, Vetrini F, Rosewell A, Grove N, Palmer D, Ng P, et al. 2011. Modifications of adenovirus hexon allow for either hepatocyte detargeting or targeting with potential evasion from Kupffer cells. Mol Ther 19: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Maywood ES, Karp NA, King VM, Inoue Y, Gonzalez FJ, Lilley KS, Kyriacou CP, Hastings MH 2007. Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology 45: 1478–1488 [DOI] [PubMed] [Google Scholar]
- Reinke H, Saini C, Fleury-Olela F, Dibner C, Benjamin IJ, Schibler U 2008. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev 22: 331–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relogio A, Westermark PO, Wallach T, Schellenberg K, Kramer A, Herzel H 2011. Tuning the mammalian circadian clock: Robust synergy of two loops. PLoS Comput Biol 7: e1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini C, Morf J, Stratmann M, Gos P, Schibler U 2012. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev 26: 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu GS, Solorio L, Broome AM, Salem N, Kolthammer J, Shah T, Flask C, Duerk JL 2010. Whole animal imaging. Wiley Interdiscip Rev Syst Biol Med 2: 398–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M 2001. Entrainment of the circadian clock in the liver by feeding. Science 291: 490–493 [DOI] [PubMed] [Google Scholar]
- Stratmann M, Schibler U 2006. Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythms 21: 494–506 [DOI] [PubMed] [Google Scholar]
- Stratmann M, Stadler F, Tamanini F, van der Horst GT, Ripperger JA 2010. Flexible phase adjustment of circadian albumin D site-binding protein (DBP) gene expression by CRYPTOCHROME1. Genes Dev 24: 1317–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara Y, Kuroda H, Saito K, Nakajima Y, Kubo Y, Ohnishi N, Seo Y, Otsuka M, Fuse Y, Ohura Y, et al. 2012. In vivo monitoring of peripheral circadian clocks in the mouse. Curr Biol 22: 1029–1034 [DOI] [PubMed] [Google Scholar]
- Takeda Y, Kang HS, Angers M, Jetten AM 2011. Retinoic acid-related orphan receptor γ directly regulates neuronal PAS domain protein 2 transcription in vivo. Nucleic Acids Res 39: 4769–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan NS, Michalik L, Desvergne B, Wahli W 2005. Multiple expression control mechanisms of peroxisome proliferator-activated receptors and their target genes. J Steroid Biochem Mol Biol 93: 99–105 [DOI] [PubMed] [Google Scholar]
- van der Veen DR, Minh NL, Gos P, Arneric M, Gerkema MP, Schibler U 2006. Impact of behavior on central and peripheral circadian clocks in the common vole Microtus arvalis, a mammal with ultradian rhythms. Proc Natl Acad Sci 103: 3393–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujovic N, Davidson AJ, Menaker M 2008. Sympathetic input modulates, but does not determine, phase of peripheral circadian oscillators. Am J Physiol Regul Integr Comp Physiol 295: R355–R360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington SN, McVey JH, Bhella D, Parker AL, Barker K, Atoda H, Pink R, Buckley SM, Greig JA, Denby L, et al. 2008. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell 132: 397–409 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kobayashi M, Mitsui S, Ishida Y, van der Horst GT, Suzuki M, Shibata S, Okamura H 2001. View of a mouse clock gene ticking. Nature 409: 684. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H 2000. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288: 682–685 [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. 2004. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci 101: 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]