Abstract
In a recently introduced transgenic multicolor labeling strategy called “Brainbow,” Cre-lox recombination is used to create a stochastic choice of expression among fluorescent proteins (XFPs), resulting in the indelible marking of mouse neurons with multiple, distinct colors. This method has been adapted to non-neuronal cells in mice and to neurons in fish and flies, but has yet to realize its full potential in the mouse brain. Here, we present several new lines of mice that overcome limitations of the initial lines and an adaptation of the method for use in adeno-associated viral (AAV) vectors. We also provide technical advice about how best to image Brainbow transgenes.
INTRODUCTION
The discovery that recombinant jellyfish green fluorescent protein (GFP) fluoresces when expressed in heterologous cells1 has led to a vast array of powerful methods for marking and manipulating cells, subcellular compartments and molecules. The discovery or design of numerous spectral variants2–13 (together called XFPs; ref. 14), expanded the scope of the “GFP revolution” by enabling discrimination of nearby cells or processes labeled with contrasting colors. At least in the nervous system, however, two or three colors are far too few, because each axon or dendrite approaches hundreds or thousands of other processes in the crowded neuropil of the brain.
Several years ago, we developed a transgenic strategy called Brainbow15 that addresses this problem by marking each neuron with a different color. In this method, three or four XFPs are incorporated into a transgene, and the Cre-loxP recombination system16 is used to make a stochastic “choice” of a single XFP to be expressed from the cassette. Because multiple cassettes are integrated at a single genomic site, and choice within each cassette is made independently, combinatorial expression can endow individual neurons with one of ~100 colors, endowing nearby neurons with distinct spectral identities.
If Cre recombinase is expressed transiently, descendants of the initially marked cell inherit the color of their progenitor. Accordingly, the Brainbow method has been adapted for use in lineage analysis in non-neural tissues of mice17–21. In addition, it has been adapted for analyzing neuronal connectivity, cell migration and lineage in fish22, 23 and flies24, 25. In disappointing contrast, the method has been little used in the mouse nervous system26. We believe that the main reasons are limitations of the initial set of Brainbow transgenic mice. These include suboptimal fluorescence intensity, failure to fill all axonal and dendritic processes, and disproportionate expression of the “default” (i.e., non-recombined) XFP in the transgene. We have now addressed several of these limitations, and present a new set of Brainbow reagents here. In addition, we provide guidelines for imaging Brainbow tissue.
RESULTS
Design of Brainbow 3.0 transgenes
As a first step in improving Brainbow methods, we sought XFPs with minimal tendency to aggregate in vivo, high photostability, and maximal stability to fixation with paraformaldehyde. Because some XFPs that ranked highly in cultured cells performed poorly in vivo, we therefore generated transgenic lines from 15 XFPs (Supplementary Table 1 and refs. 2–13). Of the XFPs tested in this way, 7 were judged suitable: mTFP1, EGFP, EYFP, mOrange2, TagRFPt, tdTomato, and mKate2.
From these XFPs, we chose three, based on the criteria of minimal spectral overlap and minimal sequence homology. These were mOrange2 from a coral (Excitation peak [Ex]549nm, Emission peak [Em]565nm), EGFP from a jelly fish (Ex488nm, Em507nm), and mKate2, from a sea anemone (Ex588nm, Em635nm)2, 9, 11. Our reason for minimizing sequence homology was to ensure that the XFPs would be antigenically distinct, in contrast to spectrally distinct but antigenically indistinguishable jellyfish variants (EBFP, ECFP, EGFP, and EYFP). Exploiting this property, we generated antibodies to them in different host species (rabbit anti-mCherry/mOrange2/tdTomato, chicken anti-EGFP/EYFP/ECFP and guinea pig anti-mKate2/TagBFP/TagRFP; Supplementary Table 1). Tests in transfected cultured cells confirmed lack of cross-reactivity (data not shown).
Next, we addressed the need to fill all parts of the cell evenly. Unmodified XFPs labeled somata so strongly that nearby processes were difficult to resolve, whereas palmitoylated derivatives, which targeted the XFPs to the plasma membrane, were selectively transported to axons and labeled dendrites poorly15. We therefore generated farnesylated derivatives27, which were trafficked to membranes of all neurites (see below).
Based on these results, we generated “Brainbow 3.0” transgenic lines incorporating farnesylated derivatives of mOrange2, EGFP, and mKate2. We retained the “Brainbow 1” format15 in which incompatible wild-type and mutant lox sites are concatenated, so that Cre recombinase yields a stochastic choice among XFPs (Fig. 1a,b). We also retained two other features of the Brainbow 1 strategy. First, we used neuron-specific regulatory elements from the thy1 gene14 because it promotes high levels of transgene expression in many, although not all, neuronal types; other promoter-enhancer sequences that we tested support considerably lower levels of expression21. Second, we generated transgenic lines by injection into oocytes because this method leads to integration of multiple copies of the cassette and thus a broad spectrum of outcomes15, 28; by contrast, “knock-in” lines generated by homologous recombination contain one or two copies of the cassette (as heterozygotes or homozygotes, respectively), and thus a smaller number of possible color combinations17–20.
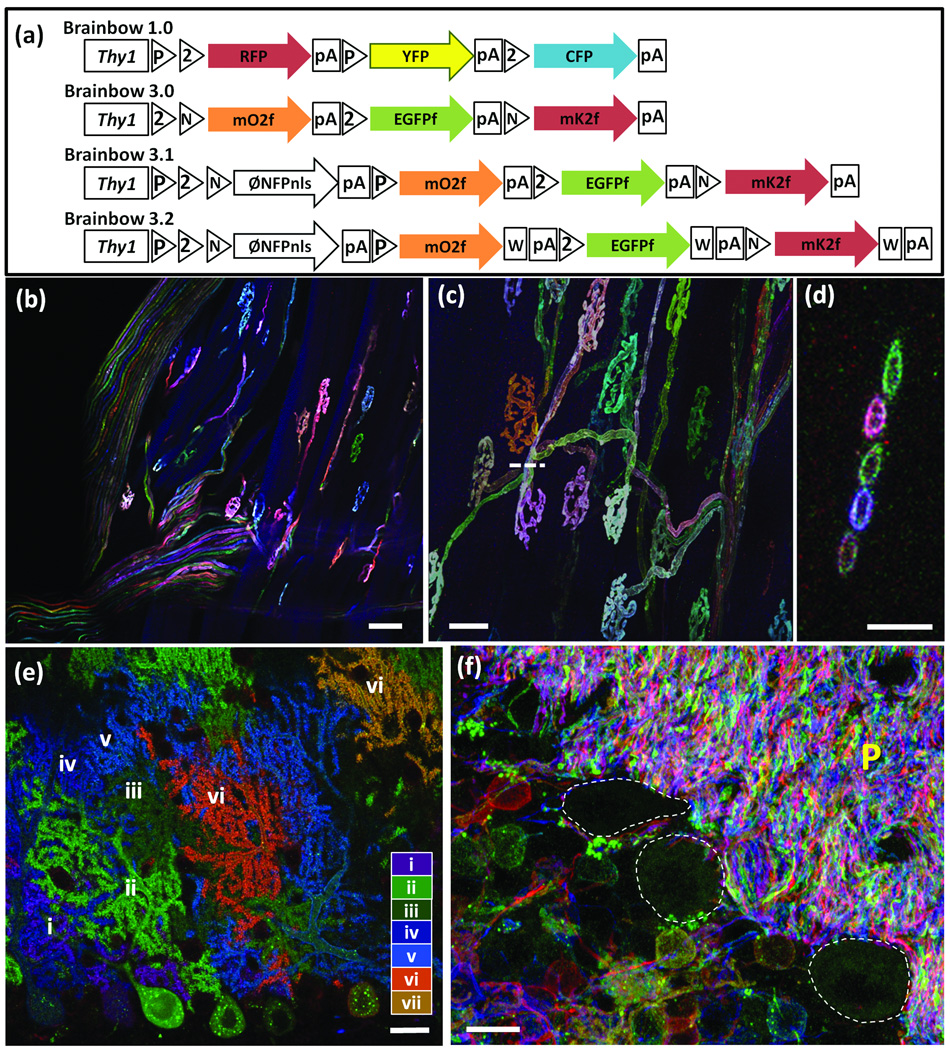
Figure 1. Brainbow 3 transgenic mice.
(a–d) Steps in generation of Brainbow 3 transgenic mice. (a) Brainbow 1.0 (described in ref. 15). (b) Brainbow 3.0 incorporates farnesylated, antigenically distinct XFPs (mOrange2f, EGFPf, and mKate2f) for membrane labeling and antibody amplification. (c) Brainbow 3.1 incorporates a nuclear-targeted non-fluorescent XFP (PhiNFPnls) in the first (default) position to limit fluorescence to Cre-expressing cells while retaining the ability to screen lines with a fourth antibody in the absence of Cre. (d) Brainbow 3.2 incorporates a WPRE into Brainbow 3.1 to increase XFP levels. P, LoxP; 2, Lox2272; N, LoxN; W, WPRE; pA, polyadenylation sequence.
(e–g) Low (e) and high (f) power views of muscles from Brainbow 3.0 (line D); Islet-cre mouse, showing terminal axons and neuromuscular junctions in extraocular muscle. (g) Rotated image along dashed bar in (f) to show 5 motor axons labeled in distinct colors. The open circles show that farnesylated XFPs mark plasma membranes more than cytoplasm.
(h) Cerebellum from Brainbow 3.1 (line 3); L7-Cre mouse. The 10 Purkinje cells in this field are labeled by at least 7 distinct colors (antibody amplified and numbered i–vii). Because Cre is selectively expressed by Purkinje cells in the L7-cre line, no other cell types are labeled.
(i) Cerebellum from Brainbow 3.2 (line 7); CreER mouse showing granule native fluorescence in red, pink, yellow, green, cyan, blue and brown. P, parallel fibers in molecular layer. Purkinje cell bodies, which are unlabeled, are outlined.
Bars are 50µm in e, 20µm in f and h, 5µm in g, 10µm in i.
Design of Brainbow 3.1 and 3.2 transgenes
In Brainbow 1, 2 and 3.0 (Fig. 1a,b), one XFP is expressed “by default” in Cre-negative cells. The presence of a “default” XFP has both drawbacks and advantages. In cases of limited Cre expression this XFP is expressed in a majority of cells, reducing spectral diversity among recombined neurons. On the other hand, incorporation of a “default” XFP allows one to screen numerous lines in the absence of a Cre reporter to assess the the number and types of cells within which XFPs could be expressed following recombination.
To eliminate the default XFP while retaining the ability to assess expression in the absence of Cre we adopted the following strategy. First, we incorporated three rather than two pairs of incompatible lox sites, which allowed insertion of a “stop cassette”29 into a first position. With this modification, the three XFPs were expressed only in Cre-positive neurons giving more spectral diversity. Second, we inserted a fourth XFP, Phi-YFP (from the hydrozoa, Phialidium)4, which is antigenically distinct from the other three, into the stop cassette. We mutated Phi-YFP to eliminate its endogenous fluorescence (PhiYFP Y65A), fused it to a nuclear localization sequence and generated antibodies to it in rat. In “Brainbow 3.1” mice generated from this cassette (Fig. 1c), one can screen sections with rat-anti-Phi-YFP prior to Cre recombination (Supplementary Fig. 1).
Finally, we inserted a sequence that stabilizes mRNAs, called a woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) to the 3’ untranslated sequences following each XFP. The WPRE has been used in many cases to increase protein levels produced by viral vectors and transgenes30, 31. We call lines incorporating the WPRE “Brainbow 3.2” (Fig. 1d).
Characterization of Brainbow 3 mice
We generated 31 lines of transgenic mice from the Brainbow 3.0, 3.1 and 3.2 cassettes. Offspring were crossed to several Cre transgenic lines32–37, leading to multicolor spectral labeling of neuronal populations in numerous regions including cerebral cortex, brainstem, cerebellum, spinal cord and retina (Fig. 1e–i, Supplementary Table 2, Supplementary Fig. 2 and Supplementary Movies 1 and 2). Intensity of expression varied markedly among lines, making quantitative comparison of dubious value but the strongest expressing lines were those that incorporated the WPRE (Brainbow 3.2).
Analysis of these lines confirmed their advantages over Brainbow 1 and 2 lines15. First, the use of farnesylated XFPs led to concentration of the XFPs at the plasma membrane (Fig. 1g). As a consequence, somata were less intensely labeled in Brainbow 3 than in Brainbow 1 mice, so processes could be visualized without saturating somata (Fig. 2a,b and Supplementary Fig. 3). Use of farnesylated XFPs also improved labeling of fine processes and dendritic spines (Fig. 2a,b insets). Second, the ability to immunostain all three XFPs enabled enhancement of the intrinsic fluorescence without losing color diversity (Fig. 2c,d and Supplementary Fig. 4). Third, XFP fluorescence is only visible in Cre-positive cells, correcting the color imbalance caused by “default” XFP expression in cells that were Cre-negative or expressed Cre at low levels in Brainbow 1 and 2 lines (Fig. 2e–i and Supplementary Fig. 5). Thus, Brainbow 3 lines are likely to be more useful than Brainbow 1 and 2 lines for multicolor labeling.
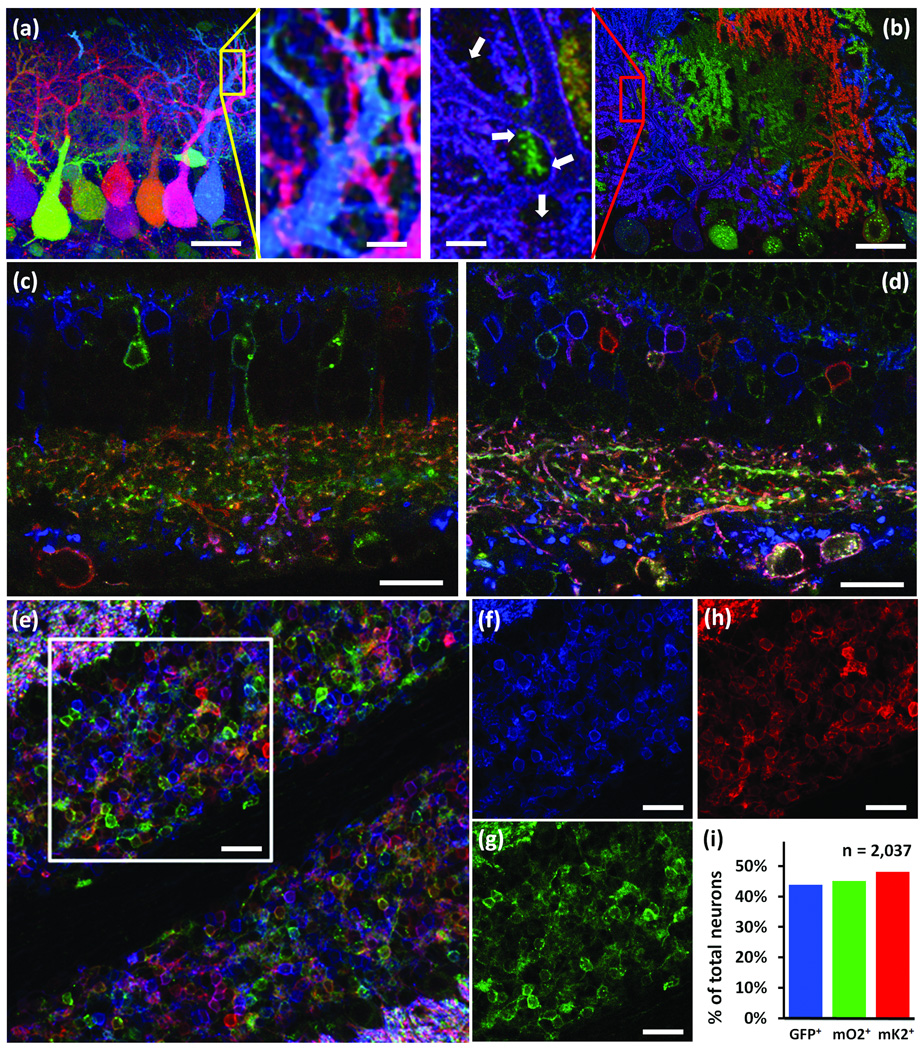
Figure 2. Improved visualization of neurons in Brainbow 3 mice.
(a,b) Cerebellum fromf Brainbow 1.0 (a) and Brainbow 3.1 (line3); L7Cre (b) mice. Left and right panels in the middle show high magnification views of boxed regions in a and b, respectively. The farnesylated XFPs clearly label the fine processes and dendritic spines (arrows, middle right), which are missing in the cytoplasmic labeling (middle left).
(c,d) Retina from a Brainbow 3.0 (line D); Islet-cre mouse expressing EGFP (blue), mOrange2 (green) and mKate2 (red). c shows Intrinsic XFP fluorescence and d shows a nearby section immunostained with chicken-anti-GFP, rabbit-anti-mOrange2 and guinea pig-anti-mKate2. Immunostaining enhances fluorescence and the number of labeled neurons and processes without compromising color diversity.
(e–i) Immunostained cerebellum section from Brainbow 3.2 (line 7);PV-cre mouse. Separate channels of region boxed in e are shown in f–h. The fraction of all labeled neurons that express EGFP (f), mOrange2 (g) or mKate2 (h) is indicated in I (2037 neurons from 15 regions).
Bars are 40µm in a and b, 20µm in c and d, 25µm in e–g, 10µm in inserts.
Brainbow with self-excising cre recombinase
In Brainbow 1–3 lines, the cassette encodes XFPs separated by lox sites; Cre recombinase is supplied from a separate transgene. For analysis of connectivity in mouse mutants, breeding two transgenes (Brainbow and Cre) into an already complex background is cumbersome. We therefore attempted to combine XFPs and Cre in a single cassette. In this transgene, called Autobow, we substituted a self-excising Cre recombinase38 for the “STOP” sequences in Brainbow 3.1 (Fig. 3a). The Thy1 regulatory elements lead to expression of Cre in differentiated neurons; Cre then simultaneously activates an XFP and excises itself.
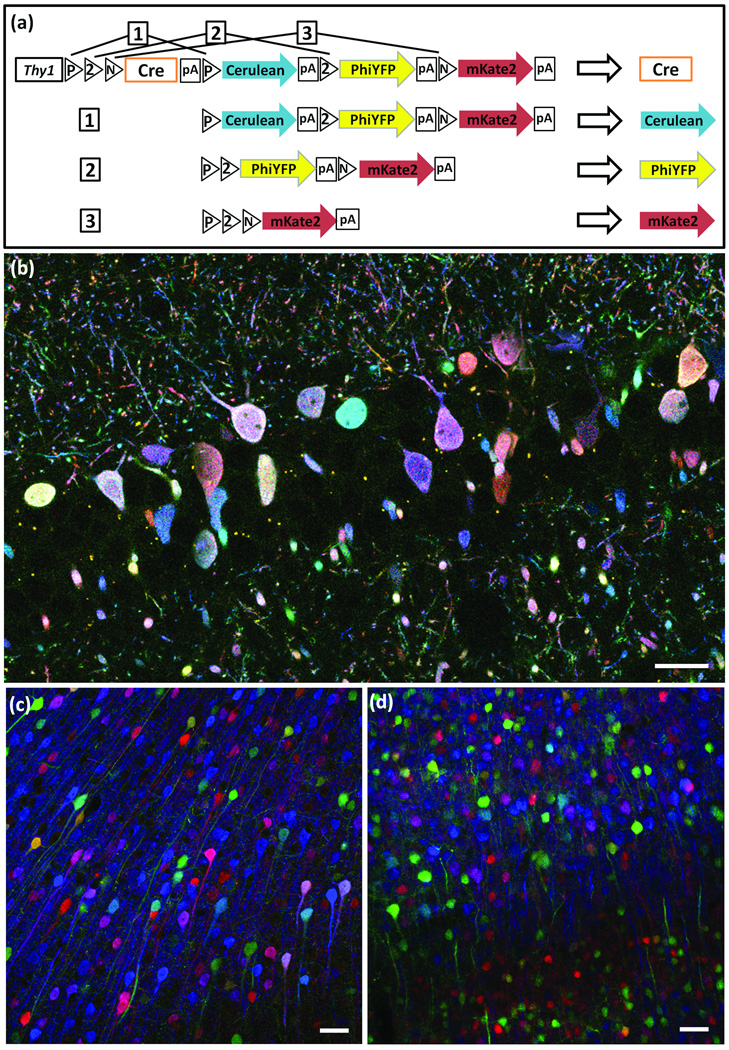
Figure 3. Autobow.
(a) In Autobow, a self-excising Cre recombinase cDNA is placed in the first (default) position. Once expressed, Cre can lead to expression of any of the 3 following XFPs (outcomes 1–3), but is itself excised in the process, terminating recombination.
(b) Labeling of hippocampal neurons by Autobow founder 3. The 20 large neurons (diameter <5µm) in this section are labeled in 20 distinct colors. Antibody amplified Cerulean, PhiYFP and mKate2 are in blue, green and red, respectively.
(c,d) Cortical neurons of an Autobow mouse line show similar color palette in the second (c) and sixth (d) generations. Antibody amplified Cerulean, PhiYFP and mKate2 are in blue, green and red, respectively. Bars are 50µm.
We generated three founders using this construct. Two were sacrificed as adults, and both expressed combinatorial expression of XFPs in multiple neurons (Fig. 3b and Supplementary Fig. 3e). We were concerned that precocious Cre activation in the germ line might lead to loss of the cassette. We therefore established a line from the third founder, and examined mice in the second and sixth generations. Color range was limited in this line, perhaps because only a few copies had been integrated into the genome, but the variety of colors and level of expression were similar in both generations (Fig. 3c,d). Thus, Autobow transgenes can be stably maintained.
Brainbow using Flp recombinase and Frt sites
A second recombination system, orthogonal to Cre-Lox, could be used to independently control expression of distinct XFPs in, for example, excitatory and inhibitory neurons. In this way, the color of a neuron could denote cell identity, a feature lacking in currently available Brainbow lines15, 17–20. We therefore tested a second recombination system, in which Flp recombinase acts on Frt sites. The Flp-Frt system has been used in conjunction with Cre-Lox in mice16, and in Brainbow-like transgenes in Drosophila24, 25.
We tested previously described mutant Frt sites39, 40 to find incompatible sets (Supplementary Fig. 6) and used these sets to construct “Flpbow” lines (Fig. 4a,b). In one of the cassettes, we fused the XFPs to an epitope tag41, allowing discrimination of cells labeled by Cre- and Flp-driven cassettes (Supplementary Fig. 7). When mated to Flp-expressing mice42,43, multicolor labeling was observed (Fig. 4c,d). Although the few lines tested to date exhibit narrow expression patterns, these results demonstrate that Flp- and Cre-based Brainbow systems can be used in combination.
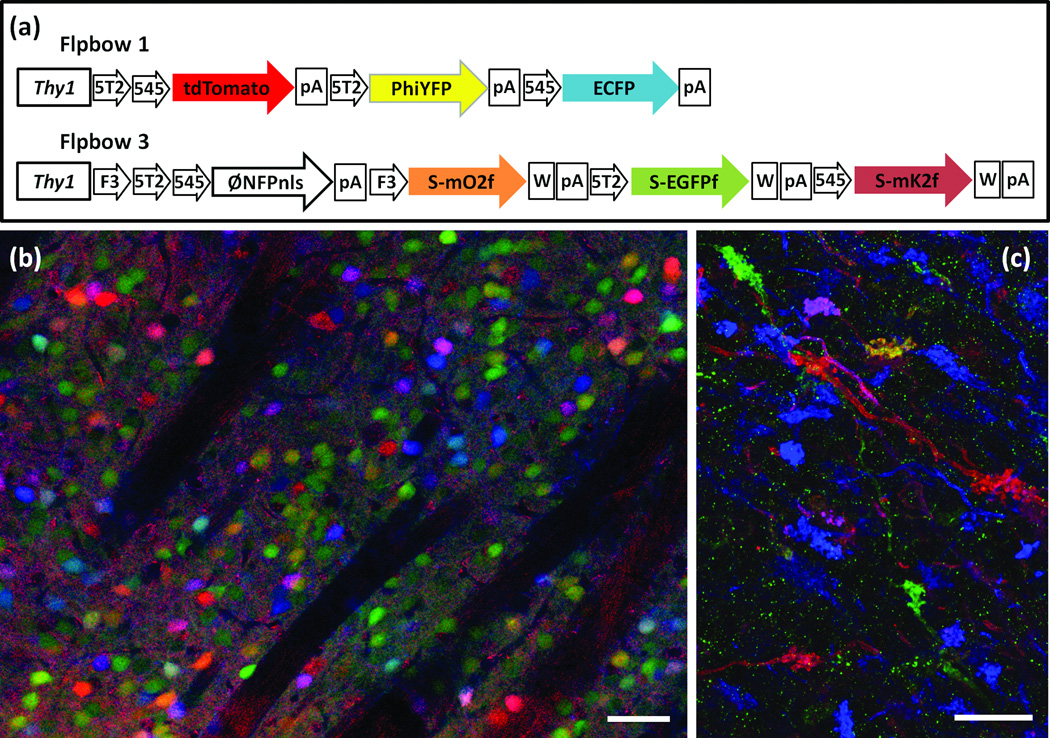
Figure 4. Flpbow.
(a,b) Flpbow transgenes incorporate Frt sites, which are substrates for Flp recombinase, instead of Lox sites, which are substrates for Cre recombinase. In Flpbow 1 (a), the Lox sites of Brainbow 1.0 were replaced by incompatible Frt sites Frt5T2 and Frt545. In Flpbow 3 (b), the Lox sites of Brainbow 3.2 were replaced by incompatible Frt sites F3, Frt5T2 and Frt545. In addition, XFPs in this construct were fused to SUMO-Star, providing an epitope tag.
(c) Labeling of neurons in caudate putamen of Flpbow 1; Wnt-flp double transgenic. Neurons are labeled by at least nine colors (red, orange, yellow, yellow-green, green, cyan, blue, purple, pink). Native fluorescence of ECFP, tdTomato and antibody amplified PhiYFP are in blue, red and green, respectively.
(d) Labeling of mossy fibers in Flpbow3; Wnt-flp double transgenic. Fluorescence of antibody amplified EGFP, mOrange2 and mKate2 are in blue, red and green, respectively.
Bars are 50µm in c, 20µm in d.
Brainbow adeno-associated viral vectors
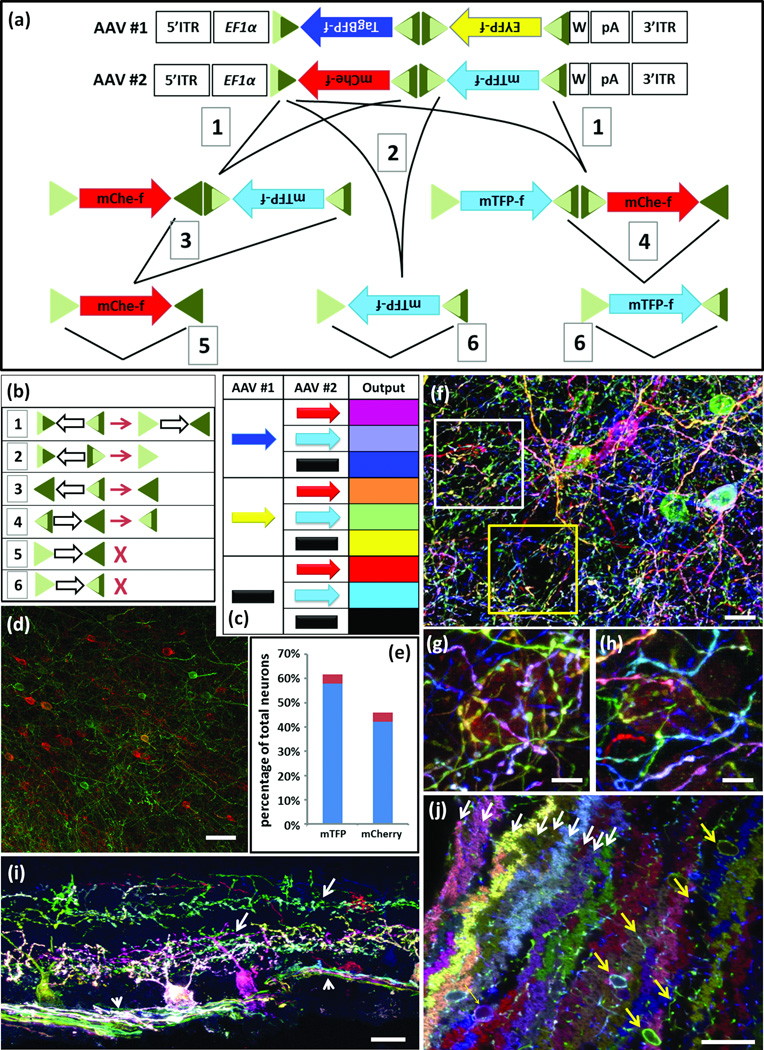
In parallel to generation of Brainbow transgenic lines, we generated adeno-associated viral (AAV) vectors to provide spatial and temporal control over expression and to make the method applicable to other species. Because the Brainbow 3.1 cassette described above is >6kB but the capacity of AAV vectors is <5kB, we reengineered the cassette. Based on results from initial tests illustrated in Supplementary Fig. 8, we devised a scheme in which lox sites with left or right element mutations44 were used for unidirectional Cre-dependent inversion (Fig. 5a,b). Farnesylated XFPs were positioned in reverse orientation to prevent Cre-independent expression, and WPRE elements were added to increase expression. In this design, recombination can lead to three outcomes from two XFPs: XFP1, XFP2 or neither. We generated two AAVs with 2 XFPs each, such that co-infection would lead to a minimum of 8 hues (3 × 3 −1; Fig. 5c). Because AAV can infect cells at high multiplicity, the number of possible colors is >>8. An additional feature is that excision of the non-expressed XFP in a second step (#3 and #4 in Fig. 5a) enhances and equalizes expression of the remaining XFP (Fig. 5d,e).
Figure 5. Brainbow AAV.
(a) AAV Brainbow constructs and recombination scheme. Farnesylated TagBFP and EYFP or mCherry and mTFP were placed in reverse orientation between mutant Lox sites. EF1α, regulatory elements from elongation factor 1α gene; W, WPRE; triangles, Lox mutants and their recombination products; dark and light sectors indicate wild-type and mutant portions of Lox sites, respectively; 1–6, recombination events.
(b) Outcomes of recombination events numbered in a. Open arrows, direction of intervening cDNA; X, fully mutant (light green) lox site cannot serve as substrate for Cre.
(c) Eight color outcomes resulting from pairs of Brainbow AAVs following recombination as shown in a. This is a minimum value, because it does not account for differences in relative intensity of the four XFPs
(d,e) Test of color balance. The mTFPf-mCherryf AAV vector was injected at low titer into the cortex of a Thy1-Cre mouse and neurons of each color were counted. Similar fractions of neurons expressed mTFPf (58%) and mCherryf (42%; n=1523 neurons in 4 sections of three mice; red shows standard deviation.)
(f–h) AAV injected into cortex of PV-cre mice predominantly labels interneurons. g and h, high magnification views of boxed regions in f.
(i) AAV injected into retina of mouse expressing Cre in retinal ganglion cell subset shows labeling of ganglion cell somata, dendrites (arrow) and axons (arrowhead).
(j) AAV injected into cerebellum of PV-Cre mouse labels Purkinje cells (fan shaped dendrites, white arrows) and interneurons (cell bodies and axons, yellow arrows).
In f–j antibody amplified mTFP and EYFP were in green; TagBFP was in blue and mCherry was in red.
Bars are 20µm in f, 10µm in g and h, 50µm in i and j
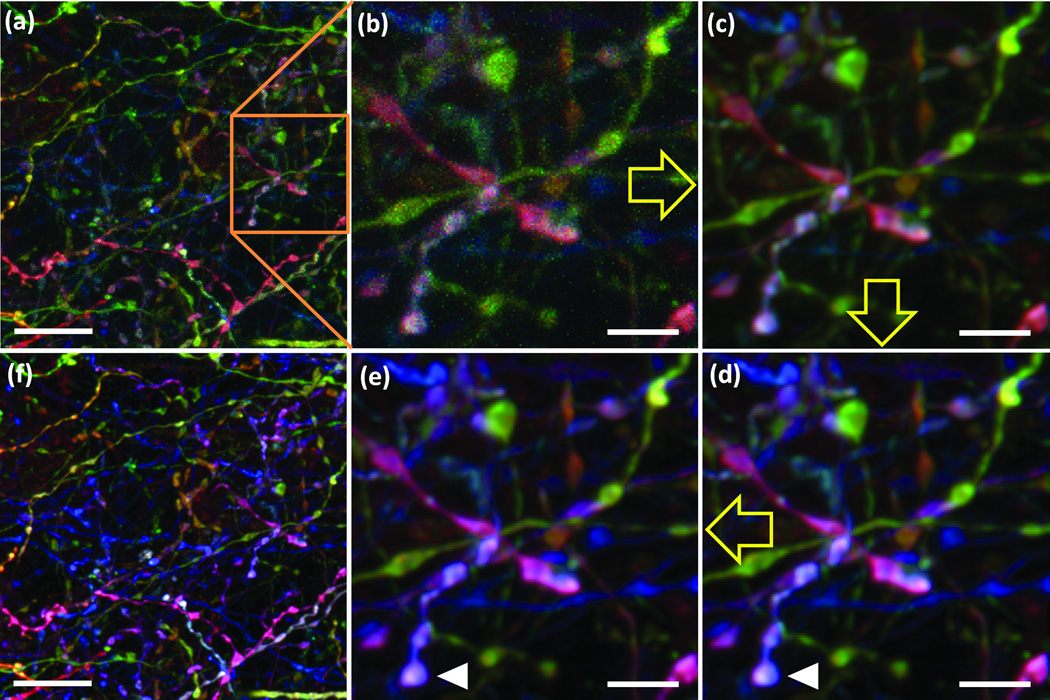
We infected cortex, cerebellum and retina of Cre transgenic mice with these vectors. When examined 3–5 weeks later, neurons were labeled in multiple colors (Fig. 2f–j and Supplementary Movie 3). Near injection sites, high levels of infectivity led to coexpression of all XFPs in single cells and “gray-white” labeling. The variety of colors increased with distance from these sites, then decreased again in sparsely injected regions, presumably because each labeled neuron received only one virion (Supplementary Fig. 9).
Methods to optimize Brainbow imaging
Obtaining high quality images from of tissues expressing Brainbow transgenes is challenging. Because colors are derived by mixing images of multiple fluorophores over a wide range of concentrations, factors that differentially affect the labels degrade the final image. In addition, it is usually necessary to image a tissue volume rather than a single section, so methods for taking image stacks must be optimized. Here, we summarize guidelines for imaging Brainbow tissue:
Sample preparation
To minimize background, section thickness should be less than the working distance of the objective, generally <100µm for high numerical aperture [NA] lenses. It is also important to match the refractive indices of the immersion medium and the sample, because chromatic aberrations caused by mismatches between these values lead to spatial offsets between color channels (Supplementary Fig. 10).Commercial antifade mountants such as VECTASHIELD®, or ProLong® Gold that have refractive indices of ~1.47 are optimal for objectives that use glycerin as the immersion medium. Polyvinyl alchohol mountants (such as Mowiol® 4–88) provide a better match for oil immersion (~1.52) objectives.
Confocal laser scanning microscopy
Epifluoresence microscopy can be used for imaging thin sections (<10µm) or monolayer cultures but confocal microscopes are preferable for thick specimens because they avoid contamination by light from outside the plane of focus. Newly developed 2 photon multi-XFP imaging techniques are also useful45–47. Apochromatic or fluorite microscope objectives that are corrected for three or more colors are strongly recommended.Most lens manufacturers specify preferred oils and coverslips. Using the wrong oil or coverslip degrades sharpness of focus and increases chromatic aberration.
Fluorophores with overlap in the excitation or emission spectra should be imaged sequentially rather than simultaneously to minimize fluorescence crosstalk and thereby optimize color separation. Laser power should be set as low as possible for several reasons. First, all planes are bleached as each image plane is scanned so generation of stacks leads to gradual bleaching and decreased signal through the stack. Second, because each fluorophore bleaches at a different rate, colors may shift during imaging. Third, linear signaling requires that fluorophores emit photons at submaximal rates; at higher excitation intensities only the out of focus signal is increased48. Fourth, if one fluorophore species is saturated but another is not, small changes in laser power will affect their intensity differently, leading to color change. With high NA objectives, laser power of just a few milliwatts is saturating; this is generally a small percentage of the total power the laser can provide.
Having adjusted the laser power to a low level, the photmuliplier tubes (PMT) voltages and digital gains must be set to relatively high values. In some confocal microscopes it is possible to compensate for signal loss from deep layers by automatic adjustment of laser power, PMT voltage or digital gain as a function of depth. Imaging parameters can be adjusted to obtain images with similar signal ranges throughout the stack.
Image processing
Brainbow images must be post-processed to maximize color information, but care is needed to avoid introducing artifacts. Often, one begins by reducing noise. Because confocal laser scanning of multicolor stacks is generally done at speeds of ~1 µs per pixel or less to save time, the small number of photons collected for each pixel gives rise to sufficient shot noise to cause perceptible local color differences. This problem can be minimized by slower scanning or averaging of multiple scans, but when this is infeasible, simple filtering and deconvolution methods are helpful (Fig. 6a–c). For example, median or Gaussian filters with 0.5–2 pixel radius reduces color noise, but at the expense of resolution. Deconvolution algorithms (see Online Methods) are more challenging to use than simple filters but can remove color noise without compromising spatial resolution (Supplementary Fig. 11).
Figure 6. Processing a Brainbow image.
(a) Original image. (b) Region boxed in a. (c) Deconvolution, resulting in decreased noise without sacrificing spatial resolution. (d) Intensity normalization, expanding perceptible color range. (e) Color shift correction (see also Supplemenatry Fig. 10). (f) Fully processed image.
Yellow arrows indicate sequence of the image processing. White arrowheads indicate corresponding objects in the original and color-shift corrected images. Bars are 10µm in a and f, 3µm in b–e.
Subsequent processing steps can expand the detectable color range and correct for color shifts (Fig. 6 d,e). To obtain easily perceived color differences, pixel intensity values for each channel in each image are normalized to the same minimum and maximum intensity values for that color in the whole image stack. This linearly stretches all channels and images to the full dynamic range. Color shifts also arise because illumination strength is generally uneven across the imaging field and differs among lasers. This effect can be attenuated by intensity or shading correction for each channel46, 49. The resulting composite RGB images provide maximum color separation for viewing by eye (Fig. 6f).
DISCUSSION
The goal of the work reported here was to design, generate and characterize improved reagents for multicolor “Brainbow” imaging of neurons in mice. First, we generated new transgenic lines that overcome some limitations of the Brainbow 1 and 2 lines that are currently available15. Improvements were substitution of XFPs (especially red and orange fluorescent proteins) that are more photostable and less prone to aggregation than those used initially; use of XFPs with minimal sequence homology so they could be immunostained separately; farnesylation of the XFPs for even staining of somata and the finest processes; insertion of a “stop” cassette to increase color variety by eliminating broad expression of a “default” XFP; inclusion of a non-fluorescent marker in the default position to facilitate screening of multiple lines; and insertion of a WPRE to boost expression (Figs. 1 and 2). These lines under control of regulatory elements from the Thy-1 gene enables marking of many but not all neuronal types. To date, elements tested other than those from the Thy1 gene do not support the high expression levels needed to image Brainbow 1 and 2 material. The ability to immunostain provided by Brainbow 3 cassettes may allow weaker promoters to be used.
Second, we designed two additional transgenes and performed initial tests to demonstrate that they can be used effectively in vivo. One, called Autobow, incorporates a self-excising Cre recombinase. Autobow lacks the temporal and spatial control afforded by use of specific Cre lines or ligand-activated Cre (CreER). However, because it does not require generation of double transgenics, it may be useful for rapid screening of neuronal morphology in mutant mice or mice submitted to various experimental interventions (e.g., drug treatments). The other novel transgene, Flpbow, replaces Lox sites by Frt sites, so that recombination can be controlled by Flp recombinase rather than Cre recombinase. Flpbow 3 also incorporates an epitope tag so that XFPs in Flpbow can be distinguished immunohistochemically from XFPs in Brainbow. By using Cre and Flp transgenic lines with distinct, defined specifities, it should be possible to map separate sets of neurons in a single animal.
Finally, we generated Brainbow AAV vectors. These, along with recently described Brainbow herpes viral vectors50 may be more useful than Brainbow transgenic mice in some situations. Like Autobow, they avoid the need for double transgenic animals. Because time of infection can be varied, they provide an alternative to use of CreER for temporal control. Moreover, localized delivery of AAV enables tracing of connections from known sites to multiple targets and discrimination of long-distance inputs from local connections.
The three most broadly useful Brainbow 3 lines (Brainbow 3.0 line D, Brainbow 3.1 line 3, Brainbow 3.1 line 18 and Brainbow 3.2 line 7) have been provided to Jackson Laboratories (www.jax.org) for distribution. Their stock numbers are xxx, xxx and xxx. The two AAVs shown in Fig. 5 can be obtained from the University of Pennsylvania Vector Core (www.med.upenn.edu/gtp/vectorcore/). Plasmids used to generate Brainbow 3.0, 3.1, and 3.2, Autobow, and Flpbow 1.1 and 3.1 mice, as well as bacterial expression vectors used to produce immunogens, are available through Addgene, (www.addgene.org).
METHODS
Brainbow Constructs
cDNA encoding the following fluorescent proteins were used: EGFP1, EYFP2, ECFP2, mCerulean3, PhiYFP4 (Evrogen), mTFP5(Allele Biotechnology), TagBFP6 (Evrogen), EBFP27 (Addgene), Kusabira-Orange8, TagRFPt9, mOrange29, tdTomato10, mCherry10, mKate211 (Evrogen) and eqFP65012 (Evrogen). A HRAS farnesylation sequence51 was used to tether XFPs to the cell membrane. A nuclear localization signal (NLS, APKKKRKV) was added to the N-terminal end of PhiYFP(Y65A). WPRE sequence was used to stabilize mRNA and enhance nuclear mRNA export30, 31. Polyadenylation signals were from the SV40 t antigen for mouse transgenes and from the human Growth Hormone gene for AAV. Brainbow constructs were assembled by standard cloning methods. A cloning scaffold containing concatenate Lox mutant sequences and unique restriction digestion sites was synthesized (DNA2.0, Inc., Menlo Park, CA) to facilitate cloning.
Brainbow modules were cloned into the pCMV- N1 mammalian expression vector (Clontech) for transient mammalian cell expression. Brainbow mouse constructs were cloned into a unique XhoI site in a genomic fragment of Thy1.2 containing neuron-specific regulatory elements14. Brainbow AAV constructs were cloned into vectors provided by the University of Pennsylvania Virus Core. Constructs were tested by expression in HEK293 cells (ATCC) prior to generation of mice or AAV.
Mice
Transgenic mice were generated by pronuclear injection at the Harvard Genome Modification Core. Mice were maintained on C57B6 or CD-1 backgrounds. Brainbow mice were crossed to mice that expressed Cre or Flp recombinases, including PV-Cre32, Islet-Cre33, CreER34, L7-Cre35, ChAT-Cre36, Thy1-cre37, Wnt-flp43 and Actin-flp42. All experiments conformed to NIH guidelines and were carried out in accordance with protocols approved by the Harvard University Standing Committee on the Use of Animals in Research and Teaching.
AAV
Two Brainbow AAVs were mixed to equal titer (7.5E12 genome copy/mL) before injection. For retina injection, adult mice were anesthetized with ketamine-xylazine by intraperitoneal injection. A small hole was made in the temporal eye by puncturing the sclera below the cornea with a 30 1/2 G needle. Using a Hamilton syringe with a 33G blunt-ended needle, 0.5–1 µL of AAV virus was injected intravitreally. After injections, animals were treated with Antisedan (Zoetic) and monitored for full recovery. For cortex injection, adult mice were anesthetized with isoflurane via continuous delivery through a nose-cone and fixed to a stereotaxic apparatus Surgery took place under sterile conditions with the animal lying on a heating pad. 1 µL of 1:5 saline diluted AAV mix (1.5E12 genome copy/mL) was injected over 10 minutes. The head wound was sutured at the end of the experiment. One injection of the non-steroidal anti-inflammatory agent Meloxicam was given at the end of the surgery and mice were kept on a heating pad with accessible moistened food pellets and/or HydroGel until fully recovered. The mice were given another dose of Meloxicam one day later and examined 4–6 weeks infection.
Antibodies
Expression vectors were constructed to produce His tag fusions of XFPs in E. coli. Proteins were produced in bacterial, purified using His Fusion Protein Purification Kits (Thermo Scientific), concentrated to > 3 mg/ml, and used as immunogens to produce rat anti-mTFP, chicken anti-EGFP, rat anti-PhiYFP, rabbit anti-mCherry and guinea pig anti-mKate2. (Covance, Inc.). Chicken anti-EGFP IgY was purified from chicken egg yolks using Pierce Chicken IgY Purification Kit (Thermo Scientific). Other sera were used without purification. Other antibodies used were: rabbit anti-GFP (ab6556, Abcam), rabbit anti-PhiYFP (#AB604, Evrogen), chicken anti-SUMOstar (#AB7002, LifeSensors Inc), Dylight 405 conjugated goat anti-rat (Jackson ImmunoResearch Laboratories, Inc.) and Alexa fluorescent dye conjugated goat include anti-rat 488, anti-chicken 488 and 594, anti-rabbit 514 and 546, and anti-guinea pig 647 (Life Technologies Corporation).
Histology
Mice were anesthetized with sodium pentobarbital before intracardiac perfusion with 2–4% paraformaldehyde in PBS. Brains were sectioned at 100 µm using a Leica vt1000s vibratome. Muscle and retina were sectioned at 20 µm in a Leica CM1850 cryostat or processed as whole mounts. For immunostaining, tissues were permeablized by 0.5% Triton X-100 with 0.02% sodium azide in StartingBloc (Thermo Scientific) at room temperature for 2 hours, then incubated with combinations of anti-XFPs (see above) for 24–48 hours at 4 °C. After extensive washing in PBST (0.01 M PBS with 0.1% Triton X-100), all secondary antibodies (1:500) were added for 12 hours at 4 °C. Finally, sections and tissues were mounted in Vectashield mounting medium (Vector Laboratory) and stored at −20 °C until they were imaged.
Antibody combinations used in figures are as following: In Figure 1e–h, Figure 2b and d–g, Figure 4d, Supplementary Figure 2a–d, Supplementary Figure 4b and Supplementary Figure 5, primary antibodies are chicken anti-GFP (1:2,000), rabbit anti-mCherry (1:1,000, for mOrange2) and guinea pig anti-mKate2 (1:500). Secondary antibodies are Alexa dye conjugated goat anti-chicken488, anti-rabbit546, and anti-guinea pig647. In Figure 3b–d and Supplementary Figure 2e, primary antibodies are chicken anti-GFP (for ECFP), rabbit anti-PhiYFP (1:1,000) and guinea pig anti-mKate2. Secondary antibodies are Alexa dye conjugated goat anti-chicken488, anti-rabbit546, and anti-guinea pig647. In Figure 4c, rabbit anti-PhiYFP and Alexa dye conjugated goat anti-rabbit514 are used. In Figure 5d, primary antibodies are rat anti-mTFP and rabbit anti-mCherry. Secondary antibodies are Alexa dye conjugated goat anti-rat488 and anti-rabbit546. In Figure 5f–j and Supplementary Figure 9, primary antibodies are guinea pig anti-mKate2 (for TagBFP), rat anti-mTFP (1:1,000), chicken anti-GFP (for EYFP) and rabbit anti-mCherry. Secondary antibodies are Alexa dye conjugated goat anti-rat488, anti-chicken488, anti-rabbit546, and anti-guinea pig647. In Supplementary Figure 1, primary antibodies are rat anti-PhiYFP (1:1,000) and rabbit anti-mCherry (1:1,000, for mOrange2). Secondary antibodies are Alexa dye conjugated goat anti-rat488 and anti-rabbit546. In Supplementary Figure 7, primary antibodies are rabbit anti-EGFP (1:1,000, for ECFP) and chicken anti-SUMOstar (1:1,000). Secondary antibodies are Alexa dye conjugated goat anti-rabbit514 and anti-chicken594.
Imaging
Fixed brain and muscle samples were imaged using a Zeiss LSM710 confocal microscope. Best separation of multiple fluorophores was obtained by using a 405 photodiode laser for TagBFP and Dylight405, a 440 nm photodiode laser for mTFP, a 488 nm Argon line for EGFP and Alexa488, a 514 nm Argon line for EYFP and Alexa514, a 561 nm photodiode for mOrange2 and Alexa546, a 594 nm photodiode for mCherry, mKate2 and Alexa594 or a 633 nm photodiode for Alexa647. Images were obtained with 16× (0.8 NA), and 63× (1.45 NA) oil objectives. Confocal image stacks for all channels were acquired sequentially, and maximally or 3D view projected using ImageJ (NIH). Intensity levels were uniformly adjusted in ImageJ.
Optimal imaging for Brainbow 3 tissue used a Zeiss LSM710 with fixed dichroic mirror combinations of DM455+514/594 in order to reduce lag time between the two sequential scans. EGFP and mKate2 are excited by 458 nm and 594 nm lasers simultaneously; and fluorescence is collected at 465–580 nm in Channel 1 and 605–780 nm in Channel 2, respectively. In a subsequent scan, 514 nm laser is used to excite mOrange2; and fluorescence is collected at 545–600 nm in Channel 2. In the antibody amplified samples, conjugated Alexa dyes normally produce much stronger fluorescence signal than XFPs. The Zeiss microscope we used is optimized for imaging the Alexa488/546/647 combination. The fixed dichroic mirror is DM488/561/633. Alexa488 and Alexa647 are excited by 488 nm and 633 nm lasers simultaneously; and fluorescence is collected at 495–590 nm in Channel 1 and 638–780 nm in Channel 2, respectively. In the subsequent scan, 561 nm laser is used to excite mOrange2; and fluorescence is collected at 566–626 nm in Channel 2.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from National Institute of Health (5U24NS063931), the Gatsby Charitable Foundation, and Collaborative Innovation Award #43667 from Howard Hughes Medical Institute. We thank S. Haddad for assistance with mouse colony maintenance, X. Duan, L. Bogart and J. Lefebvre for testing Brainbow mice and AAV, R. W. Draft for valuable discussions and advice, R. Y. Tsien (University of California, San Diego) for mOrange2 and TagRFPt, and D. M. Chudakov (IBCH RAS) for TagBFP, PhiYFP, mKate2 and eqFP650.
Footnotes
AUTHOR CONTRIBUTIONS
D.C., K.B.C., and T.L. performed experiments. D.C., J.W.L. and J.R.S. designed experiments, interpreted results and wrote the manuscript.
REFERENCES
- 1.Tsien RY. The green fluorescent protein. Annual review of biochemistry. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Current biology : CB. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 3.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nature biotechnology. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 4.Shagin DA, et al. GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Molecular biology and evolution. 2004;21:841–850. doi: 10.1093/molbev/msh079. [DOI] [PubMed] [Google Scholar]
- 5.Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. The Biochemical journal. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subach OM, et al. Conversion of red fluorescent protein into a bright blue probe. Chemistry & biology. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ai HW, Shaner NC, Cheng Z, Tsien RY, Campbell RE. Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry. 2007;46:5904–5910. doi: 10.1021/bi700199g. [DOI] [PubMed] [Google Scholar]
- 8.Karasawa S, Araki T, Nagai T, Mizuno H, Miyawaki A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. The Biochemical journal. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaner NC, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nature methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaner NC, et al. Improved monomeric, red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature biotechnology. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 11.Shcherbo D, et al. Far-red fluorescent tags for protein imaging in living tissues. The Biochemical journal. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shcherbo D, et al. Near-infrared fluorescent proteins. Nature methods. 2010;7:827–829. doi: 10.1038/nmeth.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 14.Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 15.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 16.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Developmental cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 17.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by developmental reprogramming of venous cells. Nature. 2010;464:549–553. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476:409–413. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schepers AG, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 21.Tabansky I, et al. Developmental bias in cleavage-stage mouse blastomeres. Current biology : CB. 2013;23:21–31. doi: 10.1016/j.cub.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta V, Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;484:479–484. doi: 10.1038/nature11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan YA, Livet J, Sanes JR, Lichtman JW, Schier AF. Multicolor Brainbow imaging in zebrafish. Cold Spring Harbor protocols. 2011;2011 doi: 10.1101/pdb.prot5546. pdb prot5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hampel S, et al. Drosophila Brainbow: a recombinase-based fluorescence labeling technique to subdivide neural expression patterns. Nature methods. 2011;8:253–259. doi: 10.1038/nmeth.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hadjieconomou D, et al. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nature methods. 2011;8:260–266. doi: 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- 26.Lang C, Guo X, Kerschensteiner M, Bareyre FM. Single collateral reconstructions reveal distinct phases of corticospinal remodeling after spinal cord injury. PloS one. 2012;7:e30461. doi: 10.1371/journal.pone.0030461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Badaloni A, et al. Transgenic mice expressing a dual, CRE-inducible reporter for the analysis of axon guidance and synaptogenesis. Genesis. 2007;45:405–412. doi: 10.1002/dvg.20307. [DOI] [PubMed] [Google Scholar]
- 28.Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nat Rev Neurosci. 2008;9:417–422. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakso M, et al. Targeted oncogene activation by site-specific recombination in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paterna JC, Moccetti T, Mura A, Feldon J, Bueler H. Influence of promoter and WHV post-transcriptional regulatory element on AAV-mediated transgene expression in the rat brain. Gene therapy. 2000;7:1304–1311. doi: 10.1038/sj.gt.3301221. [DOI] [PubMed] [Google Scholar]
- 31.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hippenmeyer S, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS biology. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo C, Yang W, Lobe CG. A Cre recombinase transgene with mosaic, widespread tamoxifen-inducible action. Genesis. 2002;32:8–18. doi: 10.1002/gene.10021. [DOI] [PubMed] [Google Scholar]
- 35.Zhang XM, et al. Highly restricted expression of Cre recombinase in cerebellar Purkinje cells. Genesis. 2004;40:45–51. doi: 10.1002/gene.20062. [DOI] [PubMed] [Google Scholar]
- 36.Rossi J, et al. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell metabolism. 2011;13:195–204. doi: 10.1016/j.cmet.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campsall KD, Mazerolle CJ, De Repentingy Y, Kothary R, Wallace VA. Characterization of transgene expression and Cre recombinase activity in a panel of Thy-1 promoter-Cre transgenic mice. Developmental dynamics : an official publication of the American Association of Anatomists. 2002;224:135–143. doi: 10.1002/dvdy.10092. [DOI] [PubMed] [Google Scholar]
- 38.Bunting M, Bernstein KE, Greer JM, Capecchi MR, Thomas KR. Targeting genes for self-excision in the germ line. Genes & development. 1999;13:1524–1528. doi: 10.1101/gad.13.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLeod M, Craft S, Broach JR. Identification of the crossover site during FLP-mediated recombination in the Saccharomyces cerevisiae plasmid 2 microns circle. Molecular and cellular biology. 1986;6:3357–3367. doi: 10.1128/mcb.6.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 41.Peroutka RJ, Elshourbagy N, Piech T, Butt TR. Enhanced protein expression in mammalian cells using engineered SUMO fusions: secreted phospholipase A2. Protein science : a publication of the Protein Society. 2008;17:1586–1595. doi: 10.1110/ps.035576.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- 43.Awatramani R, Soriano P, Rodriguez C, Mai JJ, Dymecki SM. Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nature genetics. 2003;35:70–75. doi: 10.1038/ng1228. [DOI] [PubMed] [Google Scholar]
- 44.Araki K, Okada Y, Araki M, Yamamura K. Comparative analysis of right element mutant lox sites on recombination efficiency in embryonic stem cells. BMC biotechnology. 2010;10:29. doi: 10.1186/1472-6750-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Entenberg D, et al. Setup and use of a two-laser multiphoton microscope for multichannel intravital fluorescence imaging. Nat Protoc. 2011;6:1500–1520. doi: 10.1038/nprot.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahou P, et al. Multicolor two-photon tissue imaging by wavelength mixing. Nature methods. 2012;9:815–818. doi: 10.1038/nmeth.2098. [DOI] [PubMed] [Google Scholar]
- 47.Wang K, et al. Three-color femtosecond source for simultaneous excitation of three fluorescent proteins in two-photon fluorescence microscopy. Biomedical optics express. 2012;3:1972–1977. doi: 10.1364/BOE.3.001972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conchello JA, Lichtman JW. Optical sectioning microscopy. Nature methods. 2005;2:920–931. doi: 10.1038/nmeth815. [DOI] [PubMed] [Google Scholar]
- 49.Ducros M, et al. Efficient large core fiber-based detection for multi-channel two-photon fluorescence microscopy and spectral unmixing. Journal of neuroscience methods. 2011;198:172–180. doi: 10.1016/j.jneumeth.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 50.Card JP, et al. A dual infection pseudorabies virus conditional reporter approach to identify projections to collateralized neurons in complex neural circuits. PloS one. 2011;6:e21141. doi: 10.1371/journal.pone.0021141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Reference for methods
- 51.Hancock JF, Cadwallader K, Paterson H, Marshall CJ. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. The EMBO journal. 1991;10:4033–4039. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.