Abstract
One of the more provocative realizations that have come out of the genome sequencing projects is that organisms possess a large number of uncharacterized or poorly characterized enzymes. This finding belies the commonly held notion that our knowledge of cell metabolism is nearly complete, underscoring the vast landscape of unannotated metabolic and signaling networks that operate under normal physiological conditions, let alone in disease states where metabolic networks may be rewired, dysregulated, or altered to drive disease progression. Consequently, the functional annotation of enzymatic pathways represents a grand challenge for researchers in the post-genomic era. This review will highlight the chemical technologies that have been successfully used to characterize metabolism, and put forth some of the challenges we face as we expand our map of metabolic pathways.
Keywords: activity based protein profiling, proteomics, metabolomics, metabolic flux analysis, cancer metabolism, dysregulated metabolism, metabolomic imaging
Introduction
What is a metabolic network and why do we care about it? A metabolic network comprises the biochemical and regulatory interactions of all of the biochemical processes inside of a cell, tissue, or organism. While in its simplest form, a metabolic network consists of enzymes converting substrate metabolites to products (e.g. hexokinase converts glucose to glucose-6-phosphate), a series of enzymes may catalyze the conversion of one metabolite, through a series of interconversions, to form a penultimate product (e.g. glucose to pyruvate by glycolysis)— a metabolic pathway. These pathways can then interact with other biochemical pathways to generate a plethora of metabolites which can be used for synthesis of cellular building blocks, regulatory metabolites, and signaling molecules [e.g. glycolysis and tricarboxylic acid (TCA) cycle to generate ATP]—a metabolic network. The regulation of enzyme activities within these biochemical networks is subject to multiple levels of regulation, including transcriptional, translational, post-translational and allosteric processes, which in-turn influence metabolite levels or rates of reactions through metabolic pathways, i.e. metabolic flux. How biochemical pathways are wired and how these pathways interact with each other are also tightly regulated, and while we don’t completely understand the underlying regulatory mechanisms, the wiring of metabolic networks are distinct between each tissue and under different (patho)physiological states.
As we will show in this review, many human diseases such as obesity, diabetes, atherosclerosis, cancer, pain, inflammation, and degenerative diseases possess dysregulated, rewired, or even neomorphic networks that directly contribute to disease progression [57,15,30,40,56]. Identifying and manipulating critical nodes in these dysregulated biochemical networks have given rise to enzyme targets that show promise in combating these “metabolic diseases.” However, our efforts in identifying and annotating these dysregulated metabolic pathways in disease, let alone understanding biochemistry in normal physiology, is hindered by our incomplete understanding of enzyme function and the metabolites, pathways, and networks that they control [73]. In our biochemistry courses, we learn of standard metabolic pathways occurring inside of a cell which gives the disarming impression that we know all the individual components of metabolism and how they are interwoven. However, genome sequencing efforts have clearly shown us that there is a large swath of unannotated biochemical pathways [73,69]. Upon further scrutiny, even the seemingly characterized biochemical pathways are nowhere near completely understood in terms of their physiological roles in vivo in living systems.
This brings us to our next question, which will be the focus of this review. How do we interrogate metabolic pathways and integrate them into larger networks? The answer to this question as well as addressing the aforementioned challenges in the field of metabolism lies in the integrated use of modern proteomic and metabolomic approaches coupled with the advancements in chemical tools that have provided penetrating insights into novel aspects of metabolism in (patho)physiology. This review will discuss chemical approaches that have arisen to investigate metabolism and how these technologies have been used to provide insights into enzyme or metabolite functions.
Probing the Functional State of the Proteome
Systems biology approaches such as DNA microarrays and mass spectrometry (MS)-based proteomics have provided in-depth information about key components of metabolic and signaling pathways that are relevant to disease pathogenesis [11,25,78]. However, these technologies have limitations in their scope of characterizing enzymes and metabolites. This is because enzyme activity is regulated not only transcriptionally, but also post-translationally and by endogenous and exogenous inhibitors, which are poorly accounted for by standard gene and protein expression profiling [36]. Furthermore, a large swath of the proteome remains uncharacterized and is therefore difficult to assemble into larger biochemical networks [73]. Also, many enzymes display difficult physicochemical properties that complicate their analysis in biological samples (e.g. low abundance, difficult in enrichment) [37,64].
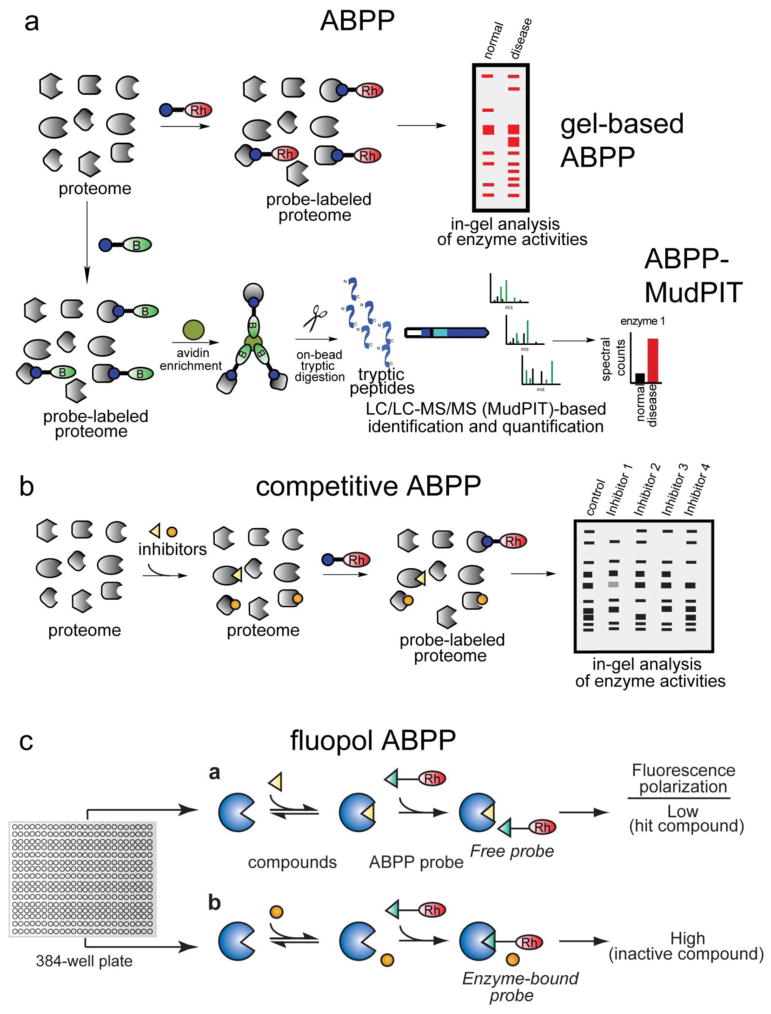
A chemoproteomic approach termed activity-based protein profiling (ABPP) addresses many of the aforementioned challenges. ABPP uses active-site directed chemical probes to directly assess enzyme activities on a proteome-wide scale in complex biological systems (Figure 1) [52,48,22]. These activity-based probes (ABPs) consist of a chemical reactive group for binding and covalently labeling the active or reactive sites of enzymes that share conserved mechanistic and/or structural features; and a reporter tag (e.g. biotin or rhodamine) for the detection, enrichment and identification of labeled enzymes from proteomes. Enzyme activities can be read-out in any biological sample (cell line, tissue, fluid, tumor) by SDS/PAGE and in-gel fluorescence scanning (for ABPs with a fluorescent tag—gel-based ABPP), or through avidin enrichment and MS-based quantitation and identification of tryptic peptides [for ABPs with a biotin tag—ABPP coupled with Multidimensional Protein Identification Technology (ABPP-MudPIT)] [32]. There are currently several ABPs for specific enzyme classes including hydrolases, kinases, oxidoreductases, glycosidases, nitrilases, cytochrome P450s, and glutathione-S-transferases [52]. There are also reactivity-based probes for mapping hyperreactive and functional amino acid residues in the proteome, as has been shown with hyper-reactive cysteines [75,74]. ABPs facilitate enrichment of specific classes of proteins based on shared functional properties and therefore assist in the characterization of low abundance as well as uncharacterized proteins. Importantly, these probes selectively label active enzymes, but not their inactive forms, facilitating characterization of changes in enzyme activities that occur without alterations in protein or transcript expression [32,31].
Figure 1. Activity-based protein profiling (ABPP).
a) ABPP uses active site-directed chemical probes to assess the functional state of large numbers of enzymes directly in complex proteomes. Activity-based probes (ABPs) consist of a reactive group, a spacer arm, and a detection handle [e.g. fluorophore such as a rhodamine (Rh) or biotin (B)]. In a typical ABPP experiment, a proteome is reacted with the ABP and readout either by fluorescence on a 1D-SDS-PAGE gel for rhodamine-ABPs (above), or by avidin enrichment, on-bead tryptic digest, and identification and quantification of peptides by Multidimensional Protein Identification Technology (MudPIT) for biotinylated-ABPs (below). b) ABPP can also be used in a competitive format to assess potency and selectivity of inhibitors in complex proteomes. Inhibitors can compete with the ABP for binding enzyme active sites and enzyme inhibition can be readout by loss of fluorescence on a SDS-PAGE gel (using a rhodamine-ABP) or loss of spectral counts by mass spectrometry (using a biotinylated-ABP). c) Competitive ABPP can be adapted to an HTS format using fluorescent polarization screening (fluopol-ABPP). Assays are conducted in 384-well plates with pure or recombinant protein. Fluorescent polarization is high if the ABP is bound to the active-site of the enzyme, and low if an inhibitor is bound to the enzyme and prevents ABP binding.
ABPP has been used to identify dysregulated metabolic pathways in various diseases such as cancer and obesity [9,54,8,10]. One enzyme class for which ABPP has been used extensively has been the serine hydrolase superfamily, one of the largest and most diverse metabolic enzyme classes in mammalian proteomes, which include esterases, thioesterases, lipases, amidases and proteases [41]. The fluorophosphonate ABPs for the serine hydrolase superfamily has been used to discover several dysregulated enzymes in disease [32,31,54,68]. Serine hydrolases KIAA1363 and monoacylglycerol lipase (MAGL) were found to be highly expressed across multiple types of human aggressive cancer cells and malignant primary tumors, where they drive tumorigenicity and aggressiveness of cancer cells [54,31,32]. Shields and colleagues found that the activity of the serine hydrolase retinoblastoma binding protein 9 (RBBP9) was heightened in pancreatic carcinomas to promote anchorage-independent growth and pancreatic carcinogenesis through overcoming transforming growth factor -β (TGF)-mediated antiproliferative signaling [68].
Several ABPs for proteases have also been used to identify dysregulated enzyme activities in cancer. Upon profiling secreted serine hydrolase activities in highly human breast cancer cells, Jessani et al found that the serine proteases uPA and tPA were secreted from highly tumorigenic and aggressive human breast cancer cells, indicating that these proteolytic activities can contribute to heightened cancer pathogenicity [31]. Bogyo and colleagues have used quenched near-infrared fluorescent ABPs of cysteine proteases including caspases to image these enzymes in tumor xenografts in vivo and ex vivo [10,21]. These probes emit a fluorescent signal only when covalently bound to the active site of the enzyme and can be used to monitor target occupancy in vivo of protease inhibitors or drug efficacy, directly by imaging the tumor [21,10]. The metalloproteinase ABP was used to identify neprilysin activity in aggressive melanoma cells [60].
ABPP can also be used to identify protein-protein interaction partners by embedding photocrosslinkable tags (e.g. benzophenone or diazarine) and bioorthogonal handles (e.g. alkyne). Salisbury et al treated melanoma cancer cells in situ with a suberoylanilide hydroxamic acid (SAHA)-based histone deacetylase (HDAC) ABP (SAHA-BPyne) with a benzophenone and an alkyne, then exposed cells to UV radiation to crosslink HDAC-interacting proteins [63]. This proteome was then subjected to “click chemistry” to append detection handles, such as rhodamine- or biotin-azide to react with the SAHA-BPyne-labeled proteins. Subsequent proteomic analysis of pulled-down proteins not only revealed HDAC enzyme activities, but also members of the HDAC complex, including CoREST, p66β, methyl CpG binding protein 3 (MBD3), and the metastasis-associated proteins MTA1 and MTA2 [63].
While ABPs for specific enzyme classes have been instrumental in identifying and characterizing dysregulated metabolic pathways in diseases, there are several enzyme classes for which there are no cognate ABPs. Weerapana et al recently developed a broader ABPP platform for mapping functional amino acids within the proteome using a reactivity-based probe for mapping hyper-reactive and functional cysteines [75]. This cysteine reactivity-based probe consists of: 1) an iodoacetamide probe to label cysteine residues in proteins; 2) an alkyne handle for “click chemistry” conjugation of probe-labeled proteins that can be appended to 3) an azide-functionalized TEV-protease recognition peptide conjugated to biotin for streptavidin enrichment of probe labeled proteins, and 4) an isotopically coded valine for quantitative MS measurements of iodoacetamide-labeled peptides across multiple proteomes. The authors globally identified hyper-reactive and functional cysteines with a wide range of activities, including nucleophilic and reductive catalysis and sites of oxidative modification. The authors even demonstrated that quantitative reactivity profiling can form the basis for screening and functional assignment of cysteines in computationally designed proteins, where it discriminated catalytically active from inactive cysteine hydrolase designs [75].
Collectively, ABPP complements standard genome sequencing, transcriptomic, and proteomic methodologies in identifying alterations not only in protein expression, but also in enzyme activities in pathological states.
Development of Pharmacological Tools for Dissecting Metabolic Pathways
While ABPP is a useful and important chemoproteomic platform that provides the ability to assess enzyme activities on a global proteomic scale, tools to perturb the activity of these enzymes are required to uncover their metabolic or pathophysiological roles. Genetic tools to knockout, knockdown, mutate, or overexpress enzymes in living systems have been incredibly useful in annotating the biochemical and pathophysiological functions of enzymes. However, there are several drawbacks to genetic perturbation of enzymes in organisms including compensation, lack of temporal control, and toxicity and cell or organismal death that can occur from genetically manipulating the enzyme. Developing pharmacological inhibitors of enzymes are therefore critical in overcoming these challenges, in which enzyme function can be blocked to identify acute versus chronic changes in a temporally controlled manner while avoiding any toxicity or adaptation that can occur from chronic target inactivation. Yet, there is a notable lack of selective chemical inhibitors for the majority of human enzymes, which hinders investigations into the biochemical and (patho)physiological roles of enzymes in physiology and disease.
Because ABPs or reactivity-based probes bind to active sites or functional domains of enzymes, inhibitors can be directly competed with probe labeling (competitive ABPP), and thus can be used in a competitive format for identifying both reversible and irreversible enzyme inhibitors and confirming target occupancy in situ or in vivo (Figure 1) [6,52]. As ABPP globally assesses activities of whole classes of enzymes or reactivities of proteins, inhibitors can be tested for potency as well as selectivity. Enzyme inhibition by small-molecule inhibitors can be read-out by both gel-based and MS-based ABPP methods using fluorescent- and biotin-tagged probes, respectively [39,7,42]. These gel-based and MS-based competitive ABPP platforms have facilitated the discovery and development of highly selective inhibitors for a multitude of metabolic enzymes, which have in-turn been used for in-depth characterization of the (patho)physiological roles of these enzymes in living systems (Figure 2) [42,44,2].
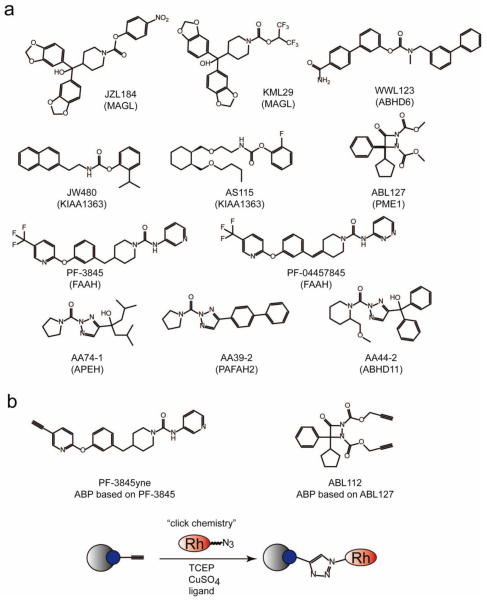
Figure 2. Examples of selective enzyme inhibitors found through competitive ABPP.
a) Structures of selective inhibitors for enzymes (in parentheses) that have been found through competitive ABPP platforms. b) To ascertain selectivity of these inhibitors outside of their cognate enzyme classes, these small-molecules can themselves be turned into activity-based probes by appending a chemoorthogonal handle (such as an alkyne or azide). PF-3845yne and ABL112 are alkyne-appended ABP analogs of PF-3845 and ABL127, respectively. These probes can be reacted with proteomes in vitro, in situ, or even in vivo and proteomes can be subjected to click chemistry with biotin or rhodamine-azide and detected by ABPP-MudPIT or gel-based approaches, respectively.
Considering the serine hydrolase superfamily as an example, gel-based and MS-based competitive ABPP screening has enabled selective inhibitor discovery for numerous serine hydrolases, including metabolic enzymes that break down endogenous cannabinoid signaling lipids. Among these are the relatively characterized enzymes fatty acid amide hydrolase (FAAH) and monoacylglycerol lipase (MAGL), as well as uncharacterized enzymes such as KIAA1363 and alpha/beta hydrolase domain 6 (ABHD6) (Figure 2) [46,16,39,42]. For these specific enzymes, electrophilic scaffolds that specifically target the serine hydrolase catalytic mechanism, such as carbamates, ketones, and ureas, were critical in inhibitor development. For FAAH, which hydrolyzes fatty acid amides, the urea compounds PF-3845 and PF-04457845 were developed as a highly potent, in vivo active, and selective irreversible inhibitors of FAAH [2,3]. Initial scaffolds for MAGL inhibitors were found through a competitive ABPP screen with a structurally diverse library of carbamates. Through medicinal chemistry efforts, JZL184 was developed as the first highly potent and selective, in vivo active and irreversible inhibitor of MAGL [42]. While JZL184 has been used in countless studies to annotate the role of MAGL and its biological functions, it has weak cross-reactivity with FAAH and peripheral carboxylesterases. Recent studies have provided an even better MAGL inhibitor KML29, based on a distinct class of O-hexafluoroisopropyl carbamates that irreversibly inhibit MAGL with superior potency and selectivity [13]. While characterized enzymes can also be assayed by traditional substrate turn-over assays, uncharacterized enzymes are often unamenable to inhibitor discovery, hindering further characterization. However, employing competitive ABPP can circumvent these issues, since ABPs assess enzyme activity based on shared catalytic mechanisms or chemical reactivity rather than the status of enzyme characterization. Highly potent and selective inhibitors of KIAA1363 and ABHD6, both serine hydrolases that were completely uncharacterized but labeled by the serine hydrolase ABP, were developed through screening of carbamate libraries and subsequent optimization. KIAA1363 inhibitors include the initially developed reversible trifluoromethylketone inhibitors and the irreversible carbamate inhibitor AS115, as well as the recently developed highly selective and in vivo active carbamate JW480 [16,14]. ABHD6 inhibitors include carbamates such as WWL123 [39].
Bachovchin et al expanded upon this gel-based ABPP screening approach for enzyme inhibitors by screening a library of structurally diverse inhibitors against a library of serine hydrolases [7]. Using this approach, the authors found lead inhibitors for nearly 40% of greater than 70 serine hydrolases screened, including many poorly characterized serine hydrolases. Adibekian et al recently found 1,2,3-triazole urea compounds as a versatile chemotype for developing serine hydrolase inhibitors. They developed a simple and efficient click-chemistry approach to create substituted triazole ureas, where substituted alkynes were reacted with in situ-formed azidomethanol to yield 4-substituted triazoles, which were then carbamoylated to give triazole urea products. Through this approach, the authors developed, screened, and optimized a triazole urea library and found highly selective in situ and in vivo active inhibitors AA74-1, AA39-2, and AA44-2, for uncharacterized serine hydrolases acyl peptide hydrolase (APEH), platelet activating factor acetylhydrolase 2 (PAFAH2), and ABHD11, respectively [1]. Adibekian et al ascertained the selectivity of these inhibitors both in situ and in vivo using a quantitative isotopic labeling-based proteomic approach adapted for competitive ABPP--“quantitative stable isotope labeling of amino acids in culture/mice” (SILAC/SILAM)-based competitive ABPP-MudPIT (ABPP-SILAC/SILAM). This procedure consists of labeling or feeding cells or mice, respectively with “light” [12C6, 14N2]-L-lysine and [12C6, 14N2]-L-arginine (treated with inhibitor) or “heavy” [13C6, 15N2]-L-lysine and [13C6, 15N2]-L-arginine (treated with DMSO or vehicle). The cells or tissue lysates are then labeled with the biotin-labeled ABP and mixed together in a 1:1 ratio. Enzyme activities are enriched through avidin beads, subjected to on-bead tryptic digest, and analyzed by high-mass accuracy MS [1].
While gel-based and MS-based ABPP have yielded invaluable pharmacological inhibitors for investigating enzyme function, gel-based formats are limited in throughput to compound libraries of modest size (200–300 compounds). Advancements in robotics technologies coupled with the generation of structurally diverse small-molecule libraries have led to the expansion of high-throughput screening (HTS) programs in academia and industry. The screens range from traditional in vitro substrate assays for enzyme inhibitors to in situ screens that profile cellular phenotypes [38,4]. Requisite to these HTS screens for enzyme inhibitors is a reproducible, robust, and highly sensitive assay for measuring enzyme activity. While this is manageable for enzymes with known substrates, these assays are very challenging for uncharacterized enzymes. As such, the unannotated portion of the human proteome (30–50%) has been largely neglected for inhibitor discovery. To overcome these challenges, Bachovchin et al recently developed a highly versatile HTS-compatible ABPP platform that uses fluorescence polarization as a readout for competitive ABPP (fluopol-ABPP) (Figure 1) [5]. This method can be readily adapted to different classes of enzymes and ABPP probes and has been successfully used to identify inhibitors for a multitude of enzymes, including retinoblastoma binding protein 9 (RBBP9), glutathione-S-transferase omega (GSTO), protein methyl esterase 1 (PME1), protein arginine deiminase (PAD), and protein arginine methyltransferases (PRMT) [5,35,8].
While competitive ABPP with enzyme-class or reactivity-specific ABPs can be used to assess inhibitor selectivity across its enzyme or reactivity class, a caveat of this technology is its inability to assess inhibitor selectivity outside of its cognate class. However, the highly selective inhibitor can itself be turned into an ABP, if the inhibitor is irreversibly binding to its target, to comprehensively assess its selectivity by appending an alkyne for subsequent conjugation of a biotin or rhodamine handle for MS or in-gel fluorescent detection, respectively, of off-targets. This has been successfully achieved with the FAAH inhibitor PF-3845 and PME1 inhibitor ABL127, and their alkyne-appended analogs PF-3845yne and ABL112, showing that these inhibitors bind only to FAAH and PME1, respectively (Figure 2) [8,2].
Competitive ABPP (gel-based ABPP, ABPP-MudPIT, fluopol ABPP, or ABPP-SILAC/SILAM) is thus a powerful platform for developing pharmacological tools to inhibit metabolic enzymes and investigate the associated biochemical and (patho)physiological functions. Should the characterized metabolic enzymes or resultant inhibitors be found to be therapeutically useful, these approaches can provide lead compounds for further clinical development.
Functional Metabolomics for Mapping and Elucidating (Patho)Physiological Roles of Metabolic Pathways
We have thus far introduced chemical approaches that can be used to directly assess enzyme activities in complex biological samples for identification of pathologically relevant metabolic enzymes, as well as platforms for developing selective small-molecule inhibitors to inactivate these enzymes. These enzyme inhibitors can in-turn be used to investigate the role that these enzymes play in metabolism, physiology, and disease using functional metabolomic technologies. Functional metabolomics can yield information on the substrate/product relationship of an enzyme as well as metabolites that are up or downstream of the substrate and product in a specific biological system. This information can be used in-turn to place the enzyme into a biochemical pathway or larger metabolic network. Here, we introduce several metabolomic approaches and will highlight recent examples of how these technologies have yielded novel insights into the biochemical and (patho)physiological roles of enzymes (Figure 3).
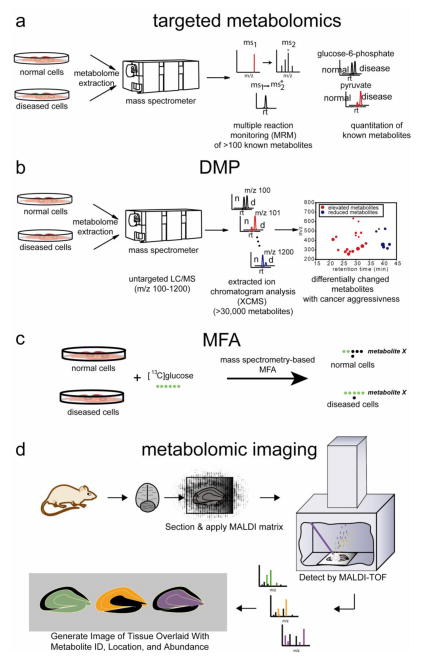
Figure 3. Metabolomic approaches for investigating metabolism.
a) For targeted metabolomics, specific known metabolites are targeted by commonly through multiple reaction monitoring (MRM) by LC-MS/MS, which measures the abundance of a metabolite-specific fragment (ms2) that arises from fragmentation of its corresponding parent mass (ms1). b) For untargeted or DMP-based metabolomics, the mass spectrometer scans a large mass range (m/z 100–1200) in an unbiased manner for both known and unknown metabolites. The resulting large datasets are then analyzed by bioinformatics programs (e.g. XCMS), which aligns, quantifies, and identifies metabolites that are significantly altered between two comparison groups. (n=normal, d=disease) c) For metabolic flux analysis, cells or mice can be treated with isotopic tracers such as [13C]glucose or [13C]glutamine and MRM-based LC-MS/MS or GC/MS can be used to quantify 13C-incorporation rates into cellular metabolites.
Traditional metabolomic approaches utilize a targeted approach, in which one comparatively quantitates the abundance of a known set of metabolites by targeting for their cognate mass over charge ratios (m/z), fragments of parent m/z ions, or magnetic resonance using mass spectrometry or NMR. Using these approaches, one can quantify the levels of several hundred to several thousand known metabolites [58,30,15]. While targeted metabolomics is a useful approach, it is limited to quantifying metabolites that are known. While a known set of nucleotides and amino acids constitute DNA, RNA, and proteins, the metabolome is significantly more physicochemically diverse in its content and complexity, and our knowledge of the metabolomic landscape is incomplete. Furthermore, the vast numbers of uncharacterized enzymes in the mammalian genome may catalyze reactions on metabolites that have not yet been identified. Thus, discovery-based metabolomics (DMP) has arisen as a powerful untargeted and unbiased approach for globally identifying metabolites that may be altered upon manipulation of enzyme activity or in disease states [59]. Instead of targeting for specific masses on MS, DMP consists of scanning a large m/z range. The large resulting datasets are then analyzed through bioinformatics platforms (e.g. XCMS) which identify, align, integrate, and compare all m/z ion intensities between different biological samples [71,70]. While both targeted and untargeted approaches measuring steady-state levels of metabolites have been instrumental in elucidating enzyme and pathway functions, these methods do not provide information on metabolic flux. Metabolic flux analysis (MFA) provides this information by measuring the amount and rates of isotopic incorporation of metabolites into metabolic intermediates [28]. Isotopic metabolites that have been used for MFA include metabolites such as [13C]glucose, [13C]glutamine, [13C]acetate, or 2H2O. Important considerations in MFA include the need for detailed information on the metabolic network being studied and appropriately selected isotopically labeled tracer substrate [18]. MFA has typically been used in a targeted fashion to trace the series of enzymatic reactions in which the selected tracer substrate is used to quantify the metabolic rate of utilization of that substrate [65]. Unique advantages of MFA include non-invasive measurements of metabolism and importantly, the ability to calculate kinetics i.e. the movement of a given metabolite through enzymatic networks [27]. We will also discuss recent advances in mass-spectrometry-based metabolomic imaging approaches that allow for morphological mapping of metabolic pathways. Each of these approaches has been used to provide valuable insights into enzyme function and mapping novel metabolic pathways in normal physiology and disease.
Cancer cells possess fundamentally altered metabolism that drives cancer pathogenicity and progression. Recent years have seen an explosion of interest in mapping the altered biochemistry that underlies cancer transformation and proliferation. Jain et al recently used multiple-reaction monitoring (MRM)-based liquid chromatography MS (LC-MS/MS) metabolomic profiling to systematically map the consumption and release (CORE) profiles of 219 metabolites spanning the major pathways of intermediary metabolism across the NCI-60 panel of human cancer cell lines [30]. CORE provides a systematic and quantitative assessment of cellular metabolic activity by relating metabolite concentrations in spent medium from cultured cell to metabolite concentrations in baseline medium. The authors identified 140 metabolites that were either present in fresh media or released by at least one cancer cell line. They found that glycine and mitochondrial glycine biosynthetic pathways were correlated with rates of proliferation across these cells, and mortality in breast cancer patients. Antagonizing glycine uptake or biosynthesis preferentially impaired rapidly proliferating cells, indicating that glycine metabolism may be a pathway that can be targeted for cancer therapy [30].
In another study, Cheng et al profiled 45 distinct metabolites in plasma by targeted LC-MS/MS to identify metabolic risk factors in individuals that were susceptible to diabetes mellitus or cardiovascular disease. The authors showed that metabolic risk factors were associated with multiple metabolites including branched-chain amino acids, other hydrophobic amino acids, tryptophan breakdown products, and nucleotides. They observed strong associations of metabolic risk factors with glutamate (a well documented effect), though the inverse effect was observed with glutamine and a glutamine-to-glutamate ratio, a heretofore unreported phenomenon. To confirm the epidemiological data, the authors found that glutamine administration in mice reduced blood pressure and improved glucose tolerance [15].
In another example of using targeted metabolomics to map altered metabolism in biology, the LIPID MAPS consortium performed targeted lipidomic and transcriptomic analyses of the response of the mouse macrophage RAW cell line to Kdo2-Lipid A2. They found that RAW cell stimulation elicits immediate responses in fatty acid metabolism represented by increases in eicosanoid synthesis, and delayed responses characterized by sphingolipid and sterol biosynthesis. They also showed lipid remodeling of glycerolipids, glycerophospholipids, and prenols, indicating that activation of the innate immune system by inflammatory mediators leads to alterations in a majority of mammalian lipid categories [20].
DMP-based metabolomics has been used numerous times to identify the role of enzymes in normal physiology and in disease. Saghatelian et al used DMP to investigate the function of FAAH, an enzyme that was known to degrade the endogenous cannabinoid signaling lipid N-arachidonoyl ethanolamine (anandamide). Previous studies had shown that FAAH is the primary hydrolase for N-acylethanolamines (NAEs) [24]. Untargeted metabolomic profiling of FAAH −/− and FAAH +/+ brains revealed substantial elevations in NAE levels in the −/− brain, consistent with the role of FAAH in degrading NAEs [62]. However, this profiling effort also revealed a distinct class of N-acyl taurines (NATs) that were also elevated greater than 10-fold, revealing a new class of substrates for this enzyme [62]. Subsequent studies have shown that NATs can activate TRP ion channels, indicating that NATs may act as signaling lipids in vivo [61]. FAAH inhibitors developed through screening and competitive ABPP efforts, such as PF-3845 and PF-04457845, have been shown to recapitulate elevations in brain NAE levels and exhibit cannabinoid-mediated anti-hyperalgesia in several inflammatory pain models, and FAAH inhibitors are now in clinical development for pain [2,3].
Untargeted metabolomics was also used to investigate the role of the uncharacterized serine hydrolase KIAA1363, identified by ABPP to be highly upregulated across multiple types of aggressive human cancer cells and primary tumors [16]. Competitive ABPP was employed to develop the highly selective, irreversible KIAA1363 inhibitor AS115, and pharmacological or genetic inactivation of KIAA1363 led to reductions in the ether lipid monoalkylglycerol ether (MAGE). Further analysis showed that KIAA1363 deacetylates 2-acetyl monoalkylglycerol ether, and that inhibition of KIAA1363 reduced MAGE and the levels of the downstream metabolite alkyl lysophosphatidic acid (alkyl LPA), a highly potent oncogenic signaling lipid. Furthermore, the authors showed that genetic knockdown of KIAA1363 reduced LPA signaling to thwart cancer cell migration and ovarian tumor xenograft growth [16]. Chang et al showed that pharmacological blockade of KIAA1363 with JW480 (developed through competitive ABPP platforms) impaired ether lipid metabolism and reduce prostate cancer xenograft growth in mice [46]. Recently, Chang et al developed an in vivo imaging probe for KIAA1363 in tumors, that KIAA1363 is largely localized to the endoplasmic reticulum of cancer cells [12].
ABPP was also used to identify MAGL as a highly upregulated serine hydrolase in aggressive human cancer cells and high-grade human tumors [54,53]. Both pharmacological inhibition of MAGL by the highly selective inhibitor JZL184 (developed using competitive ABPP) and genetic inactivation of MAGL in aggressive cancer cells led to elevations in substrate monoacylglycerols (MAG), but also reductions in global free fatty acid (FFA) levels. Additional downstream metabolites such as the protumorigenic signaling lipids prostaglandins and LPA were also reduced upon blockade of MAGL. These results were quite intriguing since MAGL does not control FFA levels under normal physiological conditions. The authors thus found that aggressive cancer cells rewire their metabolism to confer a novel pathophysiological function to MAGL in order to control a FFA network enriched in lipids that drive cancer progression [54]. Consistent with this premise, the authors showed that blocking MAGL led to impairments in cancer cell migration, invasion, and in vivo tumor growth, which could be rescued by adding back fatty acids, prostaglandins, or LPA in situ or in vivo [54]. Stable isotopic labeling of cancer cells with isotopic MAGs or FFAs were used to confirm that these pathways were driven through direct biochemical conversion of MAGs to FFAs by MAGL to form oncogenic lipids [54].
MAGL was previously known to hydrolyze MAGs as the penultimate step in triacylglycerol lipolysis [41,43]. One of these MAGs is the endocannabinoid signaling lipid 2-arachidonoylglycerol (2-AG) [42]. Several studies have shown that MAGL blockade with JZL184 has been shown to exert cannabinoid-mediated anti-hyperalgesia in several pain models and anxiolysis [42,34,33]. Interestingly, genetic ablation or chronic and complete pharmacological inactivation of MAGL causes functional desensitization of the cannabinoid system, negating the cannabinoid-mediated beneficial effects [66]. This finding underscores the utility of pharmacological tools in investigating the biological roles of enzymes before compensatory effects can occur. In a recent study, targeted and untargeted metabolomic profiling of MAGL −/− mouse brains revealed not only elevations in the endocannabinoid 2-AG, but also significant reductions in bulk arachidonic acid (AA) and downstream cyclooxygenase-mediated pro-inflammatory eicosanoids which include prostaglandins and thromboxanes [55]. This was surprising since historically, AA has been thought to derive from phospholipase-mediated hydrolysis of phospholipids. Instead, metabolomic profiling revealed that MAGL coordinately regulates endocannabinoid and eicosanoid levels in certain tissues such as the brain, liver, and lung, whereas cytosolic phospholipase A2 mediates AA release in the spleen and gut. The authors showed that pharmacological or genetic inactivation of MAGL with the selective inhibitor JZL184 produces potent anti-inflammatory effects and elicits neuroprotective effects in Parkinson’s disease and Alzheimer’s disease mouse models [55,57]. In a different study, Patti et al recently used untargeted metabolomics to identify novel metabolites that were linked to neuropathic pain. The authors showed that sphingomyelin-ceramide metabolism is altered in the dorsal horn of rats with neuropathic pain and that the upregulated, endogenous metabolite N,N-dimethylsphingosine induces mechanical hypersensitivity in vivo [56].
Another intriguing finding that has arisen out of DMP surrounds the neomorphic function of mutant isocitrate dehydrogenase 1 (IDH1), which had been previously shown to be mutated in a major subset of low-grade gliomas and secondary glioblastomas and leukemic cancers [51,77,29]. These mutations were found to occur at a single amino acid residue of IDH1, arginine 132, which is most commonly mutated to histidine. Dang et al found that stable transfection of this mutant IDH1 into glioblastoma cells resulted in a dramatic accumulation of a novel oncometabolite 2-hydroxyglutarate (2-HG), using untargeted metabolomic profiling [19]. Surprisingly, while the wild-type IDH1 uses isocitrate and NADP+ as a substrate to make -ketoglutarate and NADPH, Dang et al found that mutant IDH1 consumed NAPDH and reduced -ketoglutarate to 2-HG. These studies provided the first evidence for a mutated enzyme in cancer conferring not only a loss of endogenous function, but also a neomorphic function to yield an unforeseen metabolite, and underscored the utility of using untargeted metabolomic approaches towards revealing novel aspects of metabolism. Subsequent studies have shown that 2-HG acts as an inhibitor of histone demethylases and the TET family of 5-methylcytosine hydroxylases [23,17]. Recent studies have shown that mutations in IDH1 are sufficient to establish the glioma hypermethylator phenotype, in which introduction of mutant IDH1 into primary human astroyctes alters specific histone marks, induces extensive DNA hypermethylation, and reshapes the methylome in a similar manner to alterations observed in certain gliomas [45,72].
MFA has been used for decades to map biochemical pathways in living systems in situ and in vivo, but we will focus our examples on recent studies that have uncovered unique aspects of metabolism in disease. Locasale et al coupled isotopic labeling approaches in cancer cells with sensitivity-enhanced nuclear magnetic resonance (NMR)-based two dimensional heteronuclear single quantum correlation spectroscopy (HSQC) to quantify steady-state levels of glucose-derived carbons after 24 h labeling of [13C]glucose in human cancer cells [40]. Consistent with the “Warburg effect” of cancer cells, the highest intensity bins contained lactate peaks. Locasale et al interestingly also found that a bin containing [13C]glycine was nearly as abundant as those containing [13C]lactate. The authors showed that cancer cells divert a relatively large amount of glycolytic carbon into serine and glycine metabolism through phosphoglycerate dehydrogenase (PHGDH). They found that human cancers have a high frequency of PHGDH genomic amplification and that decreasing PHGDH expression impairs proliferation in amplified cancer cell lines. Additionally, Locasale et al reported that increasing PHGDH in mammary epithelial cells disrupts acinar morphogenesis and induces other phenotypic alterations that may predispose cells to transformation [40]. Several recent studies performing metabolic flux analysis on cancer cells using [13C]glucose and [13C]glutamine have shown that under hypoxia or mitochondrial dysfunction, cancer cells undergo a switch in which citrate, an important lipogenic precursor, is produced not from glucose carbons, but primarily from glutamine via reductive carboxylation of ketoglutarate to isocitrate via isocitrate (IDH)1 or IDH2 [76,49,47,67]. These studies showed differential incorporation of 13C-labeled carbons arising from [13C]glucose or [13C]glutamine labeling in cells by mass spectrometry or NMR. Wise et al showed that IDH2 knockdown impairs cell proliferation [76].
A relatively recent development in the field of metabolomics is imaging mass spectrometry, using matrix-assisted laser desorption ionization (MALDI IMS) (Figure 3). The MS-based metabolomics techniques described thus far have relied on extraction of metabolites from a cell or tissue, destroying metabolite location data [50]. MALDI IMS, on the other hand, is able to detect abundance and identify metabolites in a tissue while maintaining the location of those metabolites. This technology allows the generation of an image containing four dimensions of data – metabolite localization on x and y axes, selected metabolite m/z, and relative abundance of the metabolite as represented by pixel density [50]. A thin section of tissue is coated by one of various methods with the MALDI matrix, followed by a laser that generates ions at the site of the laser spot on the tissue. These ions are then analyzed by a time of flight (TOF) mass analyzer to give accurate mass to all ions generated. An image is then generated containing location and density information for a selected m/z. Murphy et al validated this method of MALDI IMS [50] using a direct sublimation approach of depositing MALDI matrix (2,5-dihydroxybenzoic acid) and imaging using MALDI-TOF on mouse kidney and brain sections. The authors were able to determine the locations of multiple lipid species based on the ions produced by each species, generating an ion map that represented the distribution of each lipid. A study by Hankin et al where traumatic-brain injury (TBI) and ischemia/reperfusion injury models in rat brain were characterized using MALDI MSI revealed unique, localized events related to lipid biochemistry following injury [26]. In the ischemia/reperfusion model, where the CA1 region of the hippocampus is particularly vulnerable, pits, holes, and a loss of substructure definition were observed when imaging with the lipid palmitoyl-oleoyl-phosphatidyl choline as the abundant ion. An additional ion, ceramide (d18:1/18:0) was detected in greater abundance in the ischemic brain compared to control, and this ion was the most abundant ion in the analyzed mass range of the injured brain. MALDI IMS showed a specific increase of ceramides in the CA1 region of the hippocampus, suggesting a role for sphingomyelin metabolism in hippocampal neuronal cell death [26]. These findings underscore the importance metabolomic imaging approaches and the ability to localize metabolite changes in a location-specific manner. As the sensitivity of detecting metabolites improves, this technology will likely become a much more widely used approach for metabolomics profiling.
Challenges in Mapping Metabolic Pathways
While the approaches described here have been useful in annotating the (patho)physiological roles of metabolic pathways, there are still several challenges moving forward. While ABPP and competitive ABPP are powerful approaches to assess enzyme activities and develop pharmacological tools to perturb these activities, we do not yet have activity-based or reactivity-based probes for every enzyme class, and therefore lack full coverage of metabolic activities in proteomes. While the metabolomic platforms described here collectively provide an ever-expanding view of the metabolomic landscape, we are likely still not able to detect metabolites that are low abundance or possess intransigent analytical properties (e.g. difficult to detect due to poor ionization). There are thus areas of metabolism that are still currently not easily amenable for characterization. These challenges will have to be addressed with the advent of new chemical tools coupled with advancements in metabolomic technologies. Furthermore, any enzyme that is annotated through these approaches will have to be incorporated into larger metabolic networks and will likely have to be coupled with more advanced computational approaches for metabolic flux modeling and will have to be integrated with currently known metabolic pathways.
Conclusion
In this review, we have provided an overview of specific chemical approaches towards investigating enzyme function and mapping metabolic pathways in normal physiology and disease. We have showed examples of how these individual technologies have been used to elucidate important biochemical features that drive physiological processes or disease progression. We have also provided a few examples (e.g. MAGL and KIAA1363) of how these chemical approaches can be integrated with metabolomic profiling to fully annotate enzyme function. These studies provide a general workflow for future investigations, wherein chemoproteomic approaches can identify enzyme activities of interest, pharmacological tools can be developed to perturb enzyme function, and in-turn be used in conjunction with targeted, untargeted DMP, MFA, and MS-based metabolomics imaging approaches to comprehensively annotate biochemical pathways. While the technologies and chemical tools employed in studying metabolism and metabolic enzymes need to be advanced and expanded upon, we currently now have the technology for annotating the vast landscape of uncharacterized or dysregulated metabolic pathways. Tapping into these uncharacterized metabolic pathways will undoubtedly open up novel targets for therapeutic intervention of diseases.
Acknowledgments
We thank the members of the Nomura Research Group for helpful discussion and critical reading of the manuscript. This work was supported by the University of California, Berkeley startup funds, Searle Scholars Program, and the National Institutes of Health (NIDA/NIH) (R00DA030908).
References
- 1.Adibekian A, Martin BR, Wang C, Hsu KL, Bachovchin DA, Niessen S, Hoover H, Cravatt BF. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nature chemical biology. 2011;7(7):469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn K, Johnson DS, Mileni M, Beidler D, Long JZ, McKinney MK, Weerapana E, Sadagopan N, Liimatta M, Smith SE, Lazerwith S, Stiff C, Kamtekar S, Bhattacharya K, Zhang Y, Swaney S, Van Becelaere K, Stevens RC, Cravatt BF. Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chemistry & biology. 2009;16(4):411–420. doi: 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn K, Smith SE, Liimatta MB, Beidler D, Sadagopan N, Dudley DT, Young T, Wren P, Zhang Y, Swaney S, Van Becelaere K, Blankman JL, Nomura DK, Bhattachar SN, Stiff C, Nomanbhoy TK, Weerapana E, Johnson DS, Cravatt BF. Mechanistic and pharmacological characterization of PF-04457845: a highly potent and selective fatty acid amide hydrolase inhibitor that reduces inflammatory and noninflammatory pain. The Journal of pharmacology and experimental therapeutics. 2011;338(1):114–124. doi: 10.1124/jpet.111.180257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An WF, Tolliday N. Cell-Based Assays for High-Throughput Screening. Mol Biotechnol. 2010;45(2):180–186. doi: 10.1007/s12033-010-9251-z. [DOI] [PubMed] [Google Scholar]
- 5.Bachovchin DA, Brown SJ, Rosen H, Cravatt BF. Identification of selective inhibitors of uncharacterized enzymes by high-throughput screening with fluorescent activity-based probes. Nature biotechnology. 2009;27(4):387–394. doi: 10.1038/nbt.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachovchin DA, Cravatt BF. The pharmacological landscape and therapeutic potential of serine hydrolases. Nature reviews Drug discovery. 2012;11(1):52–68. doi: 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(49):20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachovchin DA, Mohr JT, Speers AE, Wang C, Berlin JM, Spicer TP, Fernandez-Vega V, Chase P, Hodder PS, Schurer SC, Nomura DK, Rosen H, Fu GC, Cravatt BF. Academic cross-fertilization by public screening yields a remarkable class of protein phosphatase methylesterase-1 inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(17):6811–6816. doi: 10.1073/pnas.1015248108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barglow KT, Cravatt BF. Discovering disease-associated enzymes by proteome reactivity profiling. Chemistry & biology. 2004;11(11):1523–1531. doi: 10.1016/j.chembiol.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 10.Blum G, von Degenfeld G, Merchant MJ, Blau HM, Bogyo M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nature chemical biology. 2007;3 (10):668–677. doi: 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- 11.Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nature genetics. 1999;21(1 Suppl):33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 12.Chang JW, Moellering RE, Cravatt BF. An activity-based imaging probe for the integral membrane hydrolase KIAA1363. Angew Chem Int Ed Engl. 2012;51(4):966–970. doi: 10.1002/anie.201107236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang JW, Niphakis MJ, Lum KM, Cognetta AB, 3rd, Wang C, Matthews ML, Niessen S, Buczynski MW, Parsons LH, Cravatt BF. Highly Selective Inhibitors of Monoacylglycerol Lipase Bearing a Reactive Group that Is Bioisosteric with Endocannabinoid Substrates. Chemistry & biology. 2012;19(5):579–588. doi: 10.1016/j.chembiol.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang JW, Nomura DK, Cravatt BF. A potent and selective inhibitor of KIAA1363/AADACL1 that impairs prostate cancer pathogenesis. Chemistry & biology. 2011;18(4):476–484. doi: 10.1016/j.chembiol.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng S, Rhee EP, Larson MG, Lewis GD, McCabe EL, Shen D, Palma MJ, Roberts LD, Dejam A, Souza AL, Deik AA, Magnusson M, Fox CS, O’Donnell CJ, Vasan RS, Melander O, Clish CB, Gerszten RE, Wang TJ. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125(18):2222–2231. doi: 10.1161/CIRCULATIONAHA.111.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chemistry & biology. 2006;13(10):1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IKH, Li XS, Woon ECY, Yang M, McDonough MA, King ON, Clifton IJ, Klose RJ, Claridge TDW, Ratcliffe PJ, Schofield CJ, Kawamura A. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. Embo Rep. 2011;12(5):463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crown SB, Antoniewicz MR. Selection of tracers for C-13-Metabolic Flux Analysis using Elementary Metabolite Units (EMU) basis vector methodology. Metab Eng. 2012;14(2):150–161. doi: 10.1016/j.ymben.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Heiden MGV, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–U752. doi: 10.1038/Nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dennis EA, Deems RA, Harkewicz R, Quehenberger O, Brown HA, Milne SB, Myers DS, Glass CK, Hardiman G, Reichart D, Merrill AH, Jr, Sullards MC, Wang E, Murphy RC, Raetz CR, Garrett TA, Guan Z, Ryan AC, Russell DW, McDonald JG, Thompson BM, Shaw WA, Sud M, Zhao Y, Gupta S, Maurya MR, Fahy E, Subramaniam S. A mouse macrophage lipidome. The Journal of biological chemistry. 2010;285(51):39976–39985. doi: 10.1074/jbc.M110.182915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N, Bogyo M. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nature medicine. 2009;15(8):967–973. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chemical reviews. 2006;106(8):3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 23.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li YS, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun ZX, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD, Godley LA, Delwel R, Valk PJM, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 Mutations Result in a Hypermethylation Phenotype, Disrupt TET2 Function, and Impair Hematopoietic Differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giang DK, Cravatt BF. Molecular characterization of human and mouse fatty acid amide hydrolases. Proceedings of the National Academy of Sciences of the United States of America. 1997;94 (6):2238–2242. doi: 10.1073/pnas.94.6.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286 (5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 26.Hankin JA, Farias SE, Barkley R, Heidenreich K, Frey LC, Hamazaki K, Kim HY, Murphy RC. MALDI Mass Spectrometric Imaging of Lipids in Rat Brain Injury Models. J Am Soc Mass Spectr. 2011;22(6):1014–1021. doi: 10.1007/s13361-011-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hellerstein MK, Murphy E. Stable isotope-mass spectrometric measurements of molecular fluxes in vivo: emerging applications in drug development. Current opinion in molecular therapeutics. 2004;6 (3):249–264. [PubMed] [Google Scholar]
- 28.Hiller K, Metallo C, Stephanopoulos G. Elucidation of Cellular Metabolism Via Metabolomics and Stable-Isotope Assisted Metabolomics. Curr Pharm Biotechno. 2011;12 (7):1075–1086. doi: 10.2174/138920111795909096. [DOI] [PubMed] [Google Scholar]
- 29.Ho PA, Alonzo TA, Kopecky KJ, Miller KL, Kuhn J, Zeng R, Gerbing RB, Raimondi SC, Hirsch BA, Oehler V, Hurwitz CA, Franklin JL, Gamis AS, Petersdorf SH, Anderson JE, Reaman GH, Baker LH, Willman CL, Bernstein ID, Radich JP, Appelbaum FR, Stirewalt DL, Meshinchi S. Molecular alterations of the IDH1 gene in AML: a Children’s Oncology Group and Southwest Oncology Group study. Leukemia. 2010;24(5):909–913. doi: 10.1038/Leu.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336(6084):1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(16):10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd, Jeffrey SS, Cravatt BF. A streamlined platform for high-content functional proteomics of primary human specimens. Nature methods. 2005;2(9):691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 33.Kinsey SG, Long JZ, O’Neal ST, Abdullah RA, Poklis JL, Boger DL, Cravatt BF, Lichtman AH. Blockade of endocannabinoid-degrading enzymes attenuates neuropathic pain. The Journal of pharmacology and experimental therapeutics. 2009;330(3):902–910. doi: 10.1124/jpet.109.155465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinsey SG, O’Neal ST, Long JZ, Cravatt BF, Lichtman AH. Inhibition of endocannabinoid catabolic enzymes elicits anxiolytic-like effects in the marble burying assay. Pharmacology, biochemistry, and behavior. 2011;98(1):21–27. doi: 10.1016/j.pbb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knuckley B, Jones JE, Bachovchin DA, Slack J, Causey CP, Brown SJ, Rosen H, Cravatt BF, Thompson PR. A fluopol-ABPP HTS assay to identify PAD inhibitors. Chem Commun (Camb) 2010;46(38):7175–7177. doi: 10.1039/c0cc02634d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobe B, Kemp BE. Active site-directed protein regulation. Nature. 1999;402(6760):373–376. doi: 10.1038/46478. [DOI] [PubMed] [Google Scholar]
- 37.Kodadek T. Protein microarrays: prospects and problems. Chemistry & biology. 2001;8 (2):105–115. doi: 10.1016/s1074-5521(00)90067-x. [DOI] [PubMed] [Google Scholar]
- 38.Lea WA, Simeonov A. Fluorescence polarization assays in small molecule screening. Expert Opin Drug Dis. 2011;6(1):17–32. doi: 10.1517/17460441.2011.537322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, Blankman JL, Cravatt BF. A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. Journal of the American Chemical Society. 2007;129(31):9594–9595. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- 40.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature genetics. 2011;43(9):869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chemical reviews. 2011;111(10):6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavon FJ, Serrano AM, Selley DE, Parsons LH, Lichtman AH, Cravatt BF. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nature chemical biology. 2009;5(1):37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long JZ, Nomura DK, Cravatt BF. Characterization of monoacylglycerol lipase inhibition reveals differences in central and peripheral endocannabinoid metabolism. Chemistry & biology. 2009;16(7):744–753. doi: 10.1016/j.chembiol.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Long JZ, Nomura DK, Vann RE, Walentiny DM, Booker L, Jin X, Burston JJ, Sim-Selley LJ, Lichtman AH, Wiley JL, Cravatt BF. Dual blockade of FAAH and MAGL identifies behavioral processes regulated by endocannabinoid crosstalk in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(48):20270–20275. doi: 10.1073/pnas.0909411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O’Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–U130. doi: 10.1038/Nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo WB, Hu HX, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate Kinase M2 Is a PHD3-Stimulated Coactivator for Hypoxia-Inducible Factor 1. Cell. 2011;145(5):732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moellering RE, Cravatt BF. How chemoproteomics can enable drug discovery and development. Chemistry & biology. 2012;19(1):11–22. doi: 10.1016/j.chembiol.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy RC, Hankin JA, Barkley RM. Imaging of lipid species by MALDI mass spectrometry. J Lipid Res. 2009;50:S317–S322. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 Mutations as Molecular Signature and Predictive Factor of Secondary Glioblastomas. Clin Cancer Res. 2009;15(19):6002–6007. doi: 10.1158/1078-0432.Ccr-09-0715. [DOI] [PubMed] [Google Scholar]
- 52.Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nature reviews Cancer. 2010;10(9):630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nomura DK, Lombardi DP, Chang JW, Niessen S, Ward AM, Long JZ, Hoover HH, Cravatt BF. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chemistry & biology. 2011;18(7):846–856. doi: 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140(1):49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nomura DK, Morrison BE, Blankman JL, Long JZ, Kinsey SG, Marcondes MCG, Ward AM, Hahn YK, Lichtman AH, Conti B, Cravatt BF. Endocannabinoid Hydrolysis Generates Brain Prostaglandins That Promote Neuroinflammation. Science. 2011;334(6057):809–813. doi: 10.1126/science.1209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patti GJ, Yanes O, Shriver LP, Courade JP, Tautenhahn R, Manchester M, Siuzdak G. Metabolomics implicates altered sphingolipids in chronic pain of neuropathic origin. Nature chemical biology. 2012;8(3):232–234. doi: 10.1038/nchembio.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Piro Justin R, Benjamin Daniel I, Duerr James M, Pi Y, Gonzales C, Wood Kathleen M, Schwartz Joel W, Nomura Daniel K, Samad Tarek A. A Dysregulated Endocannabinoid-Eicosanoid Network Supports Pathogenesis in a Mouse Model of Alzheimer’s Disease. Cell Reports. 2012;1 (6):617–623. doi: 10.1016/j.celrep.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reaves ML, Rabinowitz JD. Metabolomics in systems microbiology. Curr Opin Biotech. 2011;22(1):17–25. doi: 10.1016/j.copbio.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saghatelian A, Cravatt BF. Discovery metabolite profiling--forging functional connections between the proteome and metabolome. Life sciences. 2005;77(14):1759–1766. doi: 10.1016/j.lfs.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 60.Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(27):10000–10005. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saghatelian A, McKinney MK, Bandell M, Patapoutian A, Cravatt BF. A FAAH-regulated class of N-acyl taurines that activates TRP ion channels. Biochemistry. 2006;45(30):9007–9015. doi: 10.1021/bi0608008. [DOI] [PubMed] [Google Scholar]
- 62.Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43(45):14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 63.Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(4):1171–1176. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis. 2000;21(6):1054–1070. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 65.Sauer U. Metabolic networks in motion: 13C-based flux analysis. Molecular systems biology. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schlosburg JE, Blankman JL, Long JZ, Nomura DK, Pan B, Kinsey SG, Nguyen PT, Ramesh D, Booker L, Burston JJ, Thomas EA, Selley DE, Sim-Selley LJ, Liu QS, Lichtman AH, Cravatt BF. Chronic monoacylglycerol lipase blockade causes functional antagonism of the endocannabinoid system. Nat Neurosci. 2010;13(9):1113–U1111. doi: 10.1038/Nn.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scott DA, Richardson AD, Filipp FV, Knutzen CA, Chiang GG, Ronai ZA, Osterman AL, Smith JW. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. The Journal of biological chemistry. 2011;286(49):42626–42634. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shields DJ, Niessen S, Murphy EA, Mielgo A, Desgrosellier JS, Lau SK, Barnes LA, Lesperance J, Bouvet M, Tarin D, Cravatt BF, Cheresh DA. RBBP9: a tumor-associated serine hydrolase activity required for pancreatic neoplasia. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(5):2189–2194. doi: 10.1073/pnas.0911646107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. The Journal of biological chemistry. 2010;285(15):11051–11055. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78(3):779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 71.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Anal Chem. 2012;84(11):5035–5039. doi: 10.1021/Ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AWM, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LGT, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–U137. doi: 10.1038/Nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 74.Weerapana E, Simon GM, Cravatt BF. Disparate proteome reactivity profiles of carbon electrophiles. Nature chemical biology. 2008;4(7):405–407. doi: 10.1038/nchembio.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468(7325):790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, Dematteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(49):19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan H, Parsons DW, Jin GL, McLendon R, Rasheed BA, Yuan WS, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 Mutations in Gliomas. New Engl J Med. 2009;360 (8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yates JR., 3rd Mass spectral analysis in proteomics. Annual review of biophysics and biomolecular structure. 2004;33:297–316. doi: 10.1146/annurev.biophys.33.111502.082538. [DOI] [PubMed] [Google Scholar]