Abstract
Previous studies in monocytes and other cell types have provided evidence of a role for the NF-κB pathway in IL-6 induction. The purpose of the present study was to examine the involvement of NF-κB in the induction of the IL-6 promoter in skeletal muscle cells by endotoxin (LPS), TNFα or IL-1α. Transfection of C2C12 mouse myocytes with a luciferase reporter under the control of the IL-6 promoter indicated each immunomodulator enhanced IL-6 promoter activity. Mutation and inhibitor studies indicate this response was dependent on the IL-6 NF-κB binding site, but independent of NF-IL6, AP-1, CREB or C/EBP. Cotransfection with an expression vector which constitutively activates the RelA pathway increased IL-6 promoter activity, and activity could not be further enhanced by cytokines or LPS. However, cotransfecting various dominant negative upstream NF-κB kinase expression vectors which inhibited RelA or RelB pathways either individually or in combination had no effect on LPS-induced activation of the IL-6 promoter, but abolished induction from a NF-κB-based promoter. This lack of effect was not due to a lack of NF-κB pathway activation in C2C12 myocytes because both Western analysis and EMSA supershifting showed an LPS-induced increase in nuclear RelA and RelA phosphorylation. However, another protein was observed bound to the IL-6 NF-κB site that does not bind to a consensus NF-κB site. The present findings provide novel insights regarding inflammation-induced stimulation of IL-6 promoter activity in skeletal muscle which is an important but non-traditional component of the innate immune system.
Keywords: Lipopolysaccharide, TNFα, IL-1, myocytes, promoter activity, inflammation
Introduction
Muscle wasting is a major consequence of infection (eg sepsis, AIDS) or injury (eg trauma, burn, alcohol).1 During such conditions, tumor necrosis factor-α (TNFα), interleukin 1 (IL-1), and IL-6 are released by the damaged tissue and activated leukocytes as a result of stimulating the innate immune system.2 These cytokines are responsible for at least part of the observed metabolic responses. 3–6 Gram-negative lipopolysaccharide (LPS) also induces the expression of these cytokines. The presence of LPS, as well as TNFα and IL-1, stimulates the transcription of IL-6 in different cell types, such as endothelial cells, fibroblasts and monocytes/macrophages located at the site of injury as well as from fixed macrophages within the liver and spleen. Skeletal muscle also participates in the innate immune response by promoting the synthesis and secretion of IL-6 in response to infection and injury.1,2
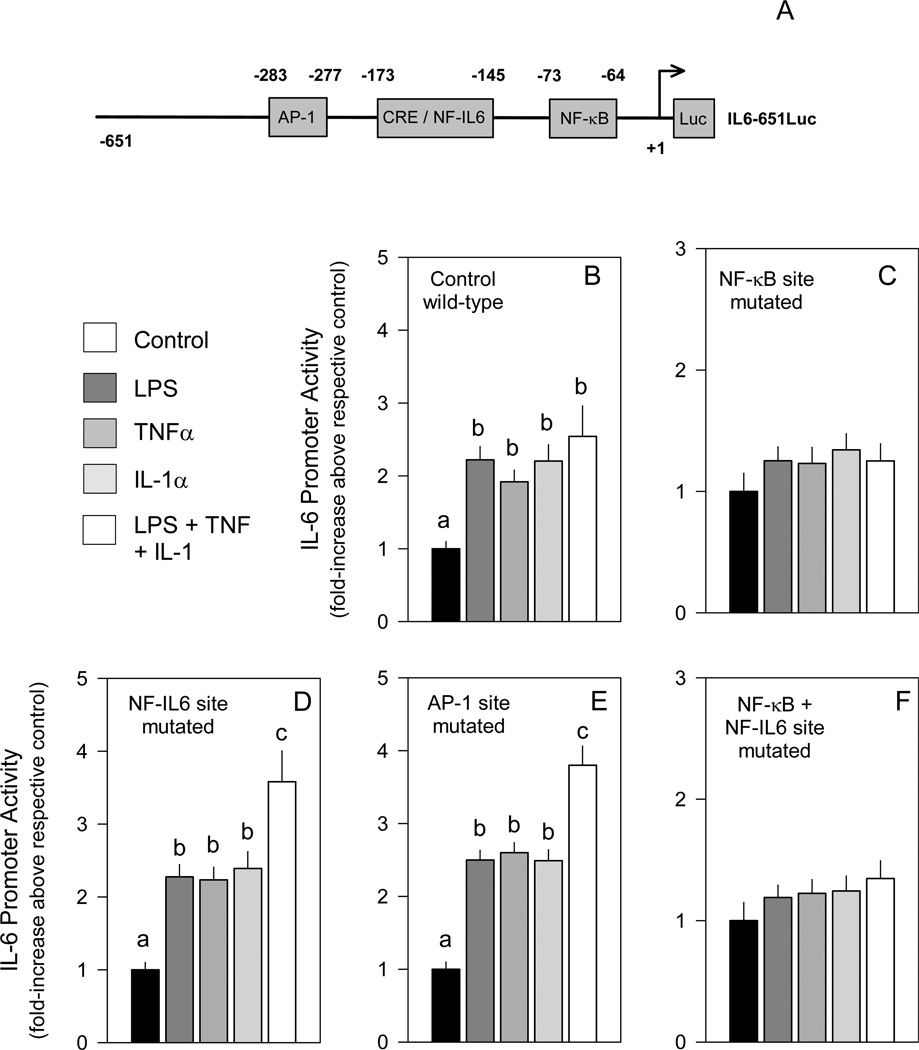
The IL-6 promoter contains multiple regulatory elements (Figure 1A)7,8 There is an AP-1 site located between −283 to −277 and two partially overlapping multiple regulatory element (MRE) regions between −225 and −113. MRE I, −173 to −151, contains a typical CRE/TRE responsive motif (eg cAMP, phorbol esters), while MRE II, −158 to −145, contains a NF-IL6 (eg C/EBPβ) binding site, but is also responsive to cAMP, TNFα and IL-1. There is also a NF-κB site, −73 to −64, in the IL-6 promoter which is necessary for induction by IL-1 and LPS. Interestingly, the transcription factors binding to the IL-6 promoter which increase IL-6 expression appear to be dependent on the inducer as well as the cell type. For example, IL-6 is induced by cAMP in fibroblasts and epithelial cells,9,10 but not in monocytes and macrophages where cAMP agonists inhibit LPS induction. TNFα can also induce IL-6 expression in numerous cell types including fibroblasts, endothelial cells and keratinocytes, and the NF-κB site is necessary for induction by IL-1 in Hela cells and for LPS stimulation of monocytes.
Figure 1.
Involvement of known cis elements in IL-6 promoter activity. A, Map of the IL-6 promoter indicating the elements present. Panels B-F, C2C12 myocytes were transfected with different luciferase-containing IL-6 promoters in which selected regulatory sequences were mutated and subsequently cultured with LPS, TNFα, IL-1α or the combination as described in “Experimental Procedures.” Myocytes transfected with IL6-651Luc, a luciferase reporter under the control of a wild-type IL-6 promoter, were used as controls. In general, under basal conditions (eg no LPS or cytokines), there was no difference in promoter activity between the wild-type control and the promoter with the NF-kB mutation (data not shown). However, the three other IL-6 promoters with mutated sequences had a 15–20% lower basal activity than that detected in myocytes transfected with the wild-type (WT) IL-6 promoter (data not shown) but such differences did not influence data interpretation.
Notes: The results shown represent the mean ± SEM of 4 to 7 independent experiments. Promoter activity is presented as fold increase over the respective control transfection for each mutated promoter. One-way ANOVA was performed followed by Stundent-Neuman-Keuls post-hoc analysis as appropriate. Values within the same treatment group with different letters are statistically different (P < 0.05).
Injection of mice with LPS increases TNFα and IL-6 mRNA and protein content in skeletal muscle and this increase precedes the rise in the plasma IL-6 concentration.11–13 An elevation in IL-6 mRNA and secretion is also observed in C2C12 mouse myocytes incubated with LPS, TNFα, and IL-1. However, culturing myocytes with an inhibitor of either TNFα (eg TNF binding protein) or IL-1 (eg IL-1 receptor antagonist) does not prevent the LPS-induced increase in IL-6 transcription suggesting LPS directly stimulates IL-6 expression.13 As enhanced IL-6 synthesis after LPS stimulation was inhibited by an Ikappa kinase (IKK)-2 inhibitor (eg TPCA-1) and a general proteasome inhibitor (eg MG-132), such data implicate the need for a functional NF-κB pathway in IL-6 induction in myocytes. In contrast, other studies report that IL-6 mRNA content is induced by MG-132 and suggest that NF-κB-independent pathways may be important for IL-6 transcription as well.
The present studies address the hypothesis that LPS-stimulated IL-6 promoter activity in skeletal muscle cells is mediated by both NF-kB-dependent and –independent mechanisms. Various known cis-acting elements within the IL-6 promoter and the trans-acting factors which bind them were investigated. We have found many of the known sites are not involved in the induction of the IL-6 promoter by LPS, TNFα and IL-1α in muscle, and only the NF-κB site was required for induction. EMSA and Western analysis show that the nuclear levels of RelA, but not RelB, are increased by LPS, consistent with earlier reports in other cell types.
Methods
Cells and constructs
C2C12 cells were obtained frozen from American Type Culture Collection (Manassas, VA) and plated directly in one 100 mm plate. The cells were maintained in minimal essential media (Mediatech, Inc, cat #: 10-010-CM) supplemented with 5% fetal bovine serum (FBS; Mediatech, cat #: 35-010-CV) and 5% newborn calf serum (NCS; Atlanta Biologicals: cat #: S11250) in addition to 100 IU/ml penicillin and 100 µg/ml streptomycin (Mediatech, cat #: 30-004-Cl)). Cells were used between the 4th and the 20th passage, and observed changes were qualitatively similar regardless of passage number.
IL-6 promoter luciferase reporter vectors were generously provided by Dr. Oliver Eickelberg.14,15 All the vectors contained the IL-6 promoter from −651 to +1 and were either wild-type (IL6-651luc) or contained mutations at the NF-IL6 (mutNF-IL6luc), NF-κB (mutNF-κBluc), AP-1 (mutNF-AP1luc), or NF-IL6/NF-κB sites (mutNF-IL6/NF-κBluc). The NF-κB-based reporter vector, NF-κB-Luc, was purchased as pNF-κB-TA-Luc from BD Biosciences (San Diego, CA). The constitutively active (CA) and dominant negative (DN) NF-κB pathway effector expression vectors, IKKB-S/E (CA), IKKβ-K44A (DN), IκBα-37-317 (DN), IκBα-S/A (DN), NIK-K/M (DN), p100-SSS/AAA (DN) were kindly provided by Dr. Shao-Cong Sun.16,17 The dominant negative A-ZIP inhibitor expression vectors, A-CREB, A-C/EBP, A-VBP, and A-Fos were originally obtained from Dr. Patrick Quinn (PSU College Medicine, Hershey, PA) with the permission from Dr. Charles Vinson, NCI, NIH.18 All vectors were verified by restriction digest analysis. The transcribed regions of the NF-κB pathway effector expression vectors were also sequenced to verify expression of the correct effector.
Transfection analyses
C2C12 cells were grown in 100 mm dishes until ~80–90% confluent, harvested with trypsin/EDTA (Mediatech, cat #: 25-053-Cl), reseeded into 6-well plates and used for study after acclimating for ~2–4 h. During this time a 100 ng/µl lipofectamine solution was prepared in serum free media. The firefly luciferase reporters (3 µg) and expression plasmids (6 µg) were added, as well as a pRL-SV-Luc (Renilla luciferase; Promega Corp, Madison, WI) reporter (3 ng) to correct for transfection efficiency, and the DNA/lipofectamine mixture was incubated at room temperature for 20 minutes. In all studies, an equivalent amount of empty vector plasmid was included in the control myocytes so that each experiment included cells which contained the same amount of DNA. Myocytes were washed once with serum free media and then covered with serum free media containing the DNA/lipofectamine mixture. After incubation of cells (2 h at 37 °C with 5% CO2), myocytes were again washed with serum free media and covered with media containing 5% FBS and 5% NCS, and the cells incubated overnight (22–24 h). In most experiments, treatment (100 ng/ml LPS [Escherichia coli O26:B6], 40 ng/ml TNFα and/or 40 ng/ml IL-1α) was added immediately before the overnight incubation, while in the experiments using MG-132, treatment was initiated the next morning and the cells allowed to incubate for another 22–26 hours. Preliminary studies of IL-6 promoter activity demonstrated a comparable effect of IL-1α and IL-1β, and thus all studies were subsequently performed with only IL-1α (data not shown). All drug doses were based on previous results showing effective stimulation of IL-6 synthesis and secretion in C2C12 myocytes.11,12,19,20 Cells were harvested with trypsin/EDTA and ruptured at −80° C using Passive Lysis Buffer (Promega) and stored there until analyzed. For analysis the samples were thawed in a room temperature water bath and clarified for 10 minutes (13,000 rpm). The activities of the firefly and Renilla luciferase reporters were measured with a dual luciferase kit (Promega, cat #: E1960) using either a Turner Biosystems 20/20 luminometer or an ALL Monolight 3010 dual injector luminometer. Each experiment was repeated at least three times with different preparations of plasmids, and data normalization described in each respective figure legend.
Western blotting analyses
C2C12 myoblasts were treated with LPS as described above for 30 min and nuclear extracts were prepared by a modification of the method of Hurst et al, 21 some of which was described previously.18,22 Plates were incubated on ice for 20 minutes after the addition of media to inhibit the trypsin during harvesting. Aliquots were loaded onto two gels and the proteins separated by 7% SDS-PAGE in parallel. The separated proteins were transferred from the gels onto polyvinylidene difluoride (PVDF) membranes. The membranes were washed and one probed with anti-RelA antibody [NF-κB p65 (C-20), sc-372] and the other with anti-RelB antibody [RelB (C-19), sc-226] obtained from Santa Cruz Biotechnology, Inc (Santa Cruz, CA). The proteins were detected using ECL Western Blotting Detection Reagents (Amersham Biosciences) using the recommended protocol.
EMSA and supershift analyses
Nuclear extracts were prepared from C2C12 myocytes 30 min after LPS, as described above. Double-stranded DNA probes for the NF-κB site on the IL-6 promoter (IL-6 NF-κB) TGATTTATCAAATGTGGGATTTTCCCATGAGTCTCAAT, the mutated NF-κB site (IL-6 mutNF-κB) TGATTTATCAAATGTCTCATTTTCCCATGAGTCTCAAT, a single NF-κB consensus site (1XNF-κB) TGATTTATCAAATGCGGGAATTTCCCATGAGTCTCAAT, three tandem NF-κB consensus sites like those present on NF-κB-Luc (3XNF-κB) TGTAGCGGGAATTTCCGGGAATTTCCGGGAATTTCCAT, and the NF-κB site on the immunoglobulin promoter (IgNF-κB) TGATTTATCAAATGGGGGACTTTCCCATGAGTCTCAAT were first prepared by annealing single-stranded oligonucleotides obtained from Integrated DNA Technologies (IDT; San Diego, CA). The probes were labeled by filling in the overhang with Klenow in the presence of [α-32P]dCTP. 10 fmol of radiolabeled DNA probe was incubated with 5 µg of nuclear protein for 15 minutes at room temperature, as previously described.18 The reactions were electrophoresed in a 6% polyacrylamide gel (25 mM Tris, 190 mM glycine, 1 mM EDTA; 150V; 3 h), which was dried and analyzed by autoradiography. For competition studies, 50-fold molar excess of unlabeled double-stranded oligonucleotide was mixed with the radiolabeled probe prior to the addition of nuclear protein. For the supershift experiments, anti-RelA [NF-κB p65 (C-20), sc-372], anti-RelB [RelB (C-19), sc-226], anti-CREB [CREB-1 (24H4B), sc-271], anti-C/EBPα [C/EBPα (14AA), sc-61], and anti-C/EBPβ [C/EBPβ (C-19), sc-150] antibodies were obtained from Santa Cruz Biotechnology and mixed with radiolabeled probe prior to the addition of nuclear protein.
Statistical analysis
Each experimental condition was tested in triplicate as described above and each experiment was repeated at least three times on separate days. The values are presented as the mean ± SEM. Data were analyzed by either a one-way or two-way ANOVA (using immunomodulator and transfection agent as factors) followed by the Student-Newman-Keuls post-hoc test when appropriate (GraphPad Prism, version 5; La Jolla,CA). In studies where two groups were compared, data were analyzed by unpaired Student’s t-test to determine treatment effect. When necessary, data were logarithmically transformed to normalize the distribution of residuals and to obtain homogeneity as assessed using the Kolmogorov-Smirnov test (InStat). Differences were considered significant when P<0.05.
Results
NF-κB site mutation inhibits induction by LPS, TNFα and IL-1
NF-κB, NF-IL6 and AP-1 are all involved in cytokine-induced IL-6 transcription in different cell types, and LPS, TNFα and IL-1 can increase IL-6 mRNA and protein expression in skeletal muscle and C2C12 mouse myocytes.11,12,19,20 Therefore, we transfected C2C12 cells with various IL-6 promoters in which these regulatory sequences were mutated and myocytes subsequently incubated with different inducers. Transfection of myocytes with a luciferase reporter under the control of a wild-type IL-6 promoter (IL6-651Luc), showed an approximately 2-fold induction by LPS, TNFα or IL-1α at the doses used (Figure B). Both IL-1α and IL-1β stimulated the wild-type IL-6 promoter to the same extent (data not shown) and therefore all subsequent studies were performed with only IL-1α. Simultaneous addition of LPS, TNFα and IL-1α did not significantly increase luciferase activity, compared to values seen with each agent alone. Mutation of the putative NF-κB site abolished induction by all the stimuli examined (Figure 1C), while mutation of the NF-IL6 site or the AP-1 site show no such inhibitory effect (Figure 1D and 1E). Myocytes transfected with the IL-6 promoter containing either the mutated NF-IL6 or AP-1 site and then incubated with the combination of LPS + TNFα + IL-1α demonstrated a greater increase in activity than cells incubated with each stimuli alone. The mechanism for this exaggerated response was not obvious and was not further investigated. Concurrent mutation of both the NF-κB and NF-IL6 sites abolished LPS-, TNFα-, or IL-1α-induced IL-6 transcription (Figure 1F). Cotransfection of the wild-type control IL-6 promoter with A-ZIP inhibitors of CREB, C/EBP, VBP, or Fos did not blunt the LPS, TNFα or IL-1α stimulation of promoter activity (data not shown).
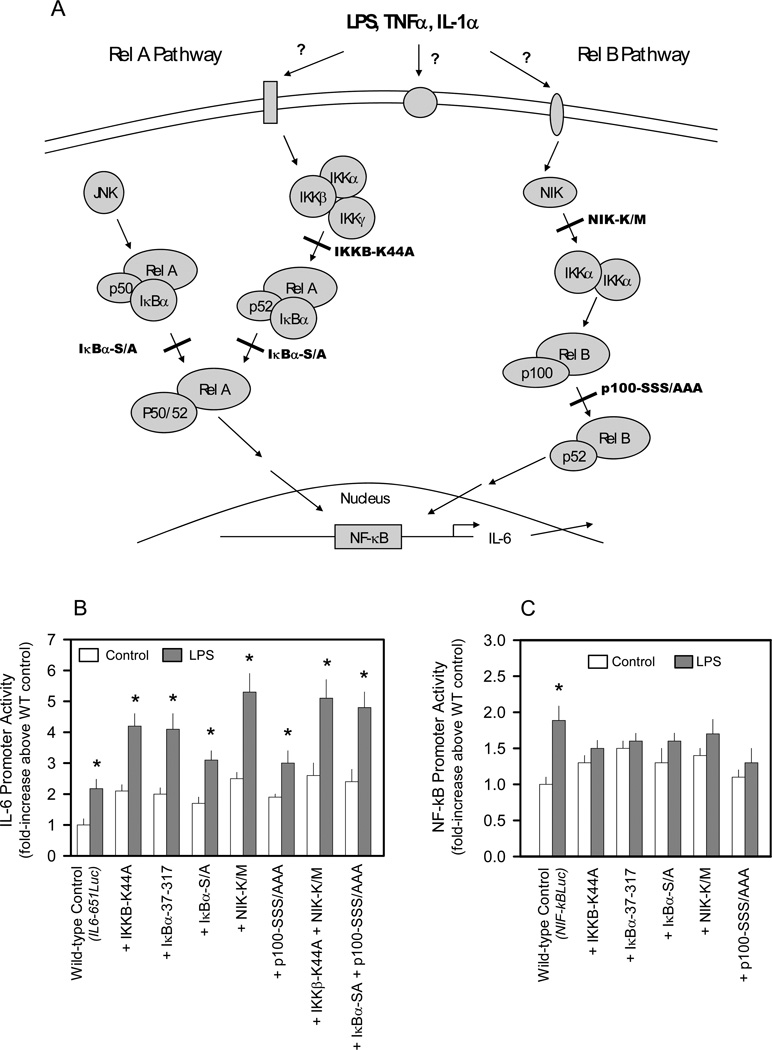
LPS induces IL-6 transcription in the presence of NF-κB pathway inhibitors
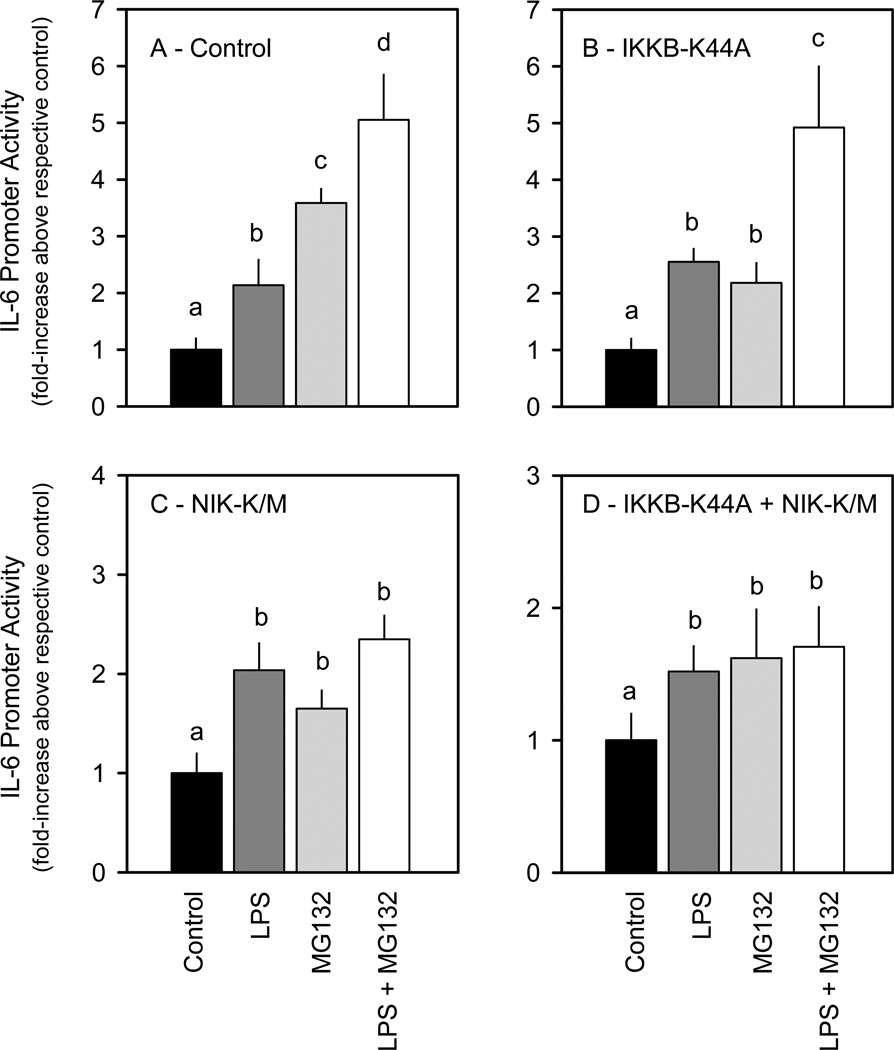
We next cotransfected various dominant negative effectors which function at different select points along the RelA or RelB pathways to determine which branch of the NF-κB pathway was involved in regulating IL-6 transcription (Figure 2A). Although the dominant negative effectors generally increased basal IL-6 promoter activity by 1.5– 2.2-fold, it is clear that the ability of LPS to increase IL-6 promoter activity was not impaired by any single dominant negative effector examined (Figure 2B). The dominant negative constructs IKKB-K44A, IKBα-37–317, and IKBα-S/A, which inhibit the RelA pathway in other cells, 16,17,23–30 did not prevent an LPS-induced increase in IL-6 promoter activity. Dominant negative NIK-K/M and p100-SSS/AAA, which inhibit the RelB pathway, also did not impair LPS stimulation. Furthermore, simultaneous inhibition of both the RelA and RelB pathways by cotransfection with either IKKβ-K44A + NIK-K/M or IκBα-S/A + p100-SSS/AAA also did not prevent the LPS-induced increase in IL-6 promoter activity (Figure 2B). In contrast, the presence of either a single RelA pathway inhibitor (IKKB-K44A, IKBα-37-317 or IKBα-S/A) or a RelB pathway inhibitor (NIK-K/M or p100-SSS/AAA) reduced NF-κB luciferase activity to basal levels (Figure 2C).
Figure 2.
Effect of cotransfecting dominant negative NF-κB pathway effectors on LPS induction of IL-6 or NF-κB-based promoters. A, Diagram of the known NF-κB pathways indicating the point of inhibition by the dominant negative effectors. B, C2C12 myocytes were cotransfected with the indicated dominant negative expression vector(s) and a luciferase reporter under the control of the wild-type (WT) IL-6 promoter (IL6-651Luc) in the presence or absence of LPS as described in “Experimental Procedures.” C, C2C12 myocytes were cotransfected with the indicated dominant negative expression vectors and a luciferase reporter under the control of a wild-type NF-κB-based promoter (NF-κBLuc) in the presence or absence of LPS as described in “Experimental Procedures.”
Notes: The results shown represent the mean ± SEM of 4 independent experiments. Promoter activity is presented as fold increase over the wild-type control transfection under basal conditions. Data from each treatment group were analyzed separately by unpaired Student’s t-test to determine treatment effect. *P < 0.5, compared to control value from the same treatment group.
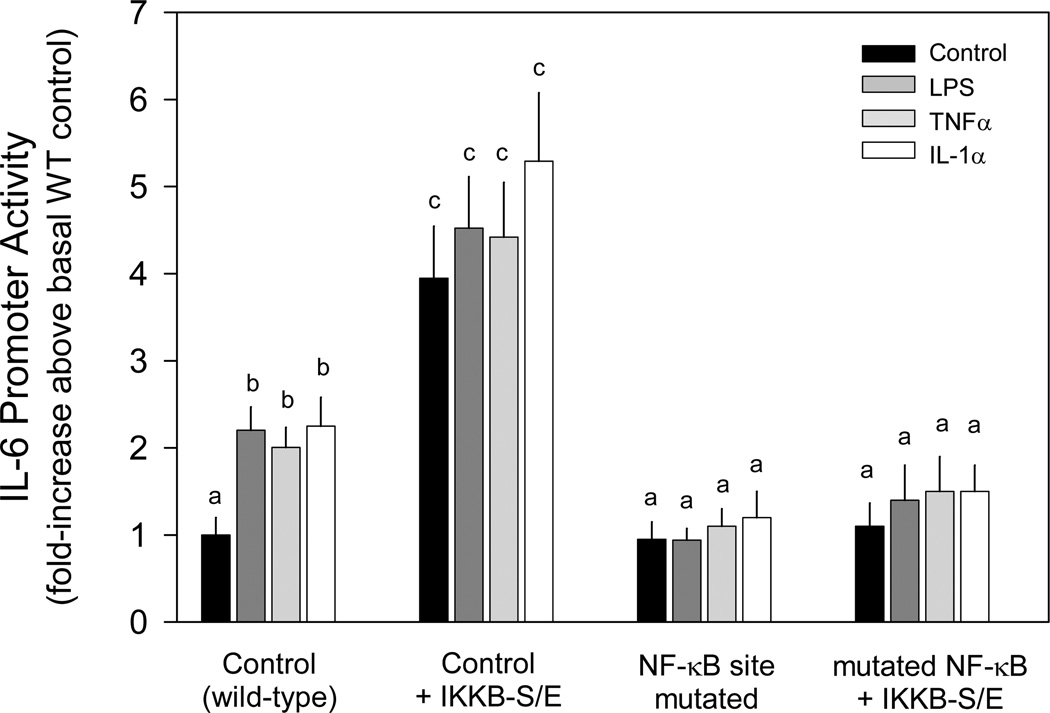
Constitutively active NF-κB pathway effectors increase IL-6 promoter activity
To investigate whether the putative NF-κB site in the IL-6 promoter is functionally involved in NF-κB stimulating IL-6 transcription, we cotransfected a constitutively active RelA pathway effector (IKKB-S/E) along with either the wild-type or the mutated NF-κB site IL-6 promoter. The presence of the constitutively active IKKB increased IL-6 transcription approximately 2-fold more than LPS, TNFα or IL-1α stimulation of the wild-type promoter (Figure 3). No further increase in activity was detected in IKKB-S/E transfected myocytes incubated with these immunomodulators. The stimulatory effect of IKKB-S/E and/or cytokines was not observed when the putative NF-κB site was mutated (Figure 3).
Figure 3.
Effect of constitutive activation of NF-κB pathway on basal and cytokine induction of IL-6 promoter activity. C2C12 myocytes were cotransfected with the indicated constitutively active expression vector (IKKBβ-S/E) and a luciferase reporter under the control of the IL-6 promoter [wild-type (WT) control; IL6-651Luc] or the same promoter with the NF-κB site mutated to inhibit binding (mutNF-κB) in the presence or absence of LPS, TNFα or IL-1α, as described in the “Experimental Procedures.”
Notes: The results shown represent the mean ± SEM of 3–5 independent experiments. Promoter activity is presented as fold increase over the wild-type control transfection under basal conditions. Two-way ANOVA was performed followed by Stundent-Neuman-Keuls post-hoc analysis as appropriate. Values with different letters are statistically different (P < 0.05).
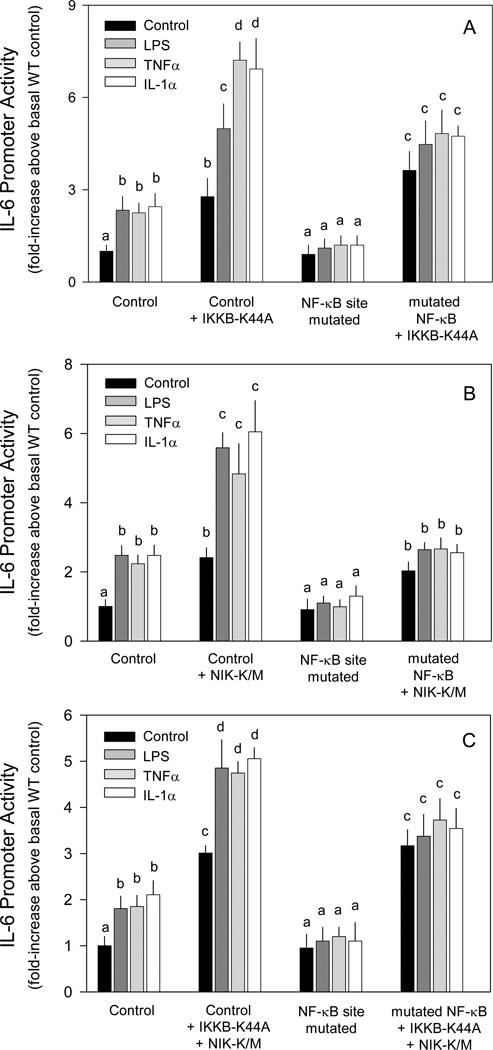
Dominant negative NF-κB pathway effectors increase IL-6 promoter activity
As activation of the RelA pathway increased IL-6 promoter activity, inhibition of the pathway was expected to reduce activity. However, the dominant negative IKKB (eg IKKB-K44A) also increased basal IL-6 promoter activity (Figure 4A). Furthermore, activity was increased approximately 2-fold more by addition of LPS, TNFα or IL-1α to cells transfected with IKKB-K44A. Mutation of the NF-κB site had no effect on the increase produced by IKKB-K44A under control conditions, but prevented the stimulatory effect of the immunomodulators.
Figure 4.
Effect of dominant negative NF-κB pathway effectors on basal and cytokine induction of IL-6 promoter activity. C2C12 myocytes were cotransfected with the indicated dominant negative expression vector (A, IKKβ-K44A; B, NIK-K/M; C, both) and a luciferase reporter under the control of the IL-6 promoter [wild-type (WT) control; IL6-651Luc] or the same promoter with the NF-κB site mutated to inhibit binding (mutNF-κB) in the presence or absence of LPS, TNFα or IL-1α as described in the “Experimental Procedures.”
Notes: Values in bar graphs represent the mean ± SEM of 3–5 independent experiments. Promoter activity for each panel is presented as fold increase over the respective wild-type control transfection under basal control conditions. Two-way ANOVA was performed followed by Stundent-Neuman-Keuls post-hoc analysis as appropriate. Values within the same treatment group with different letters are statistically different (P < 0.05).
To determine the potential role of RelB, myocytes were transfected with a dominant negative NIK construct (eg NIK-K/M). Similar to transfection of myocytes with the dominant negative IKKB, cotransfection with NIK-K/M increased IL-6 promoter activity (Figure 4B). This enhanced response was further increased approximately 2-fold by the presence of LPS, TNFα or IL-1α. Again, mutation of the putative NF-κB site had no effect on the basal increase observed by NIK-K/M, but prevented promoter stimulation by immunomodulators.
To examine the possibility that inhibition of either the RelA or RelB pathway was overcome by compensatory activation through the alternate pathway, cells were cotransfected with both IKKB-K44A and NIK-K/M. The presence of both dominant negative effectors increased basal IL-6 transcription and this increase was not affected by mutation of the putative NF-κB site (Figure 4C). However, addition of LPS, TNFα or IL-1α, which further increased IL-6 expression in the presence of the NF-κB pathway effectors, had no additional effect when the putative NF-κB site was mutated.
Proteasome inhibition does not alter LPS-induced increase in IL-6 promoter activity
Myocytes were incubated with the general proteosome inhibitor MG-132 to inhibit all Rel pathway activity. Similar to our findings using the dominant negative NF-κB inhibitors, the presence of MG-132 increased basal IL-6 promoter activity and LPS further induced activity in the presence of MG-132 (Figure 5A). This effect of MG132 was also seen when cells were cotransfected with IKKB-K44A (Figure 5B). However, the near additive effect of MG-132 and LPS was not seen when myocytes were cotransfected with NIK-K/M, even though we observed an induction with each separately (Figure 5C). The lack of an additive effect was also evident in myocytes cotransfected with both IKKB-K44A and NIK-K/M (Figure 5D).
Figure 5.
Effect of MG-132 on LPS induction of IL-6 promoter activity in the presence or absence of dominant negative NF-κB pathway effectors. C2C12 myocytes were cotransfected with a control reporter plasmid (panel A) or the indicated dominant negative expression vector, IKKβ-K44A (panel B), NIK-K/M (panel C) or the combination of IKKβ-K44A + NIK-K/M (panel D) in the presence or absence of MG-132 and/or LPS as described in the “Experimental Procedures.”
Notes: The results shown represent the mean ± SEM of n = 4 independent experiments. Under control conditions, IL-6 promoter activity was 2- to 3-fold greater in the myocytes transfected with IKKB-K44A, NIK-K/M or the combination (data not shown), as previously illustrated in Figures 3 and 4. In this figure, promoter activity is presented as fold increase over the respective control transfection within each treatment group. One-way ANOVA was performed followed by Stundent-Neuman-Keuls post-hoc analysis. Values within the same treatment group with different letters are statistically different (P < 0.05).
LPS increases RelA in nuclear extracts
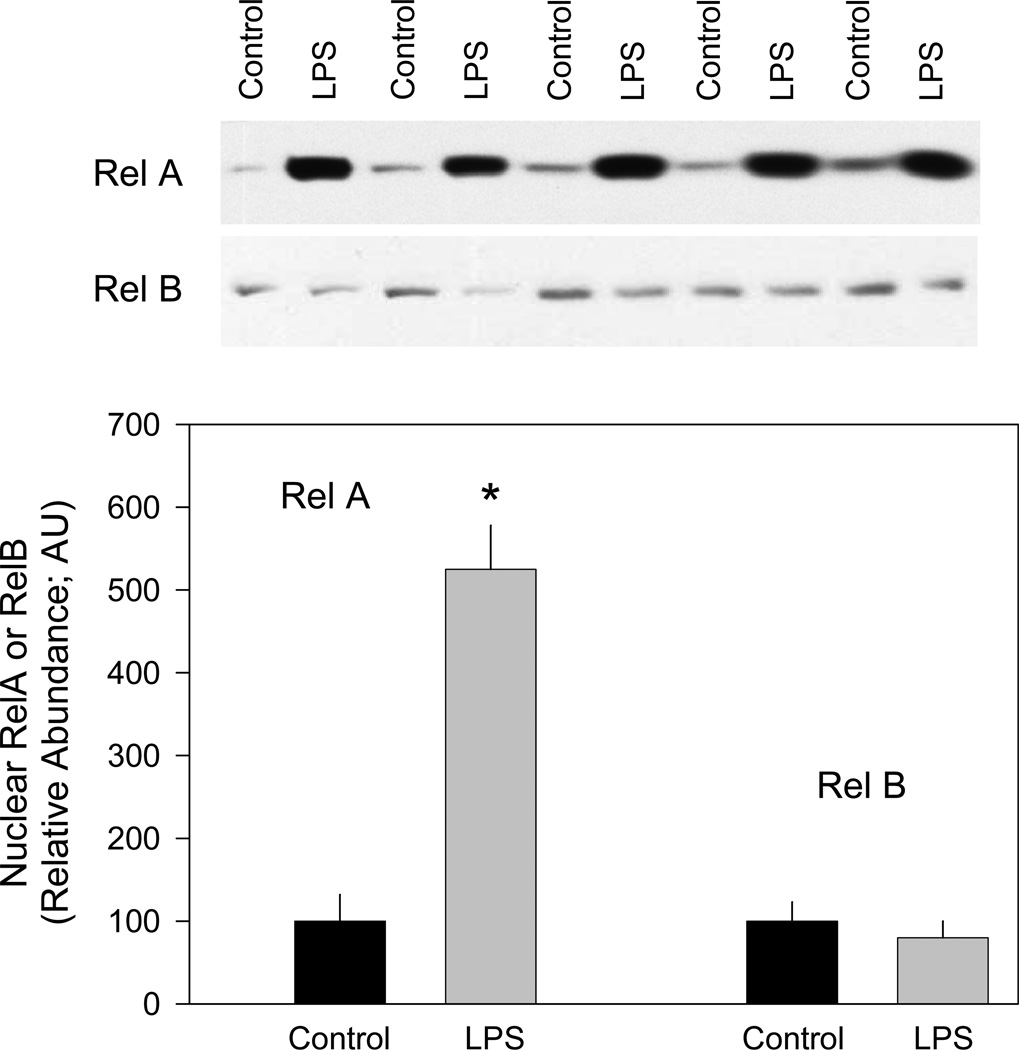
The aforementioned data suggest that although the canonical NF-κB pathways are not necessary, the NF-κB site is required for LPS or cytokine induction of the IL-6 promoter. Therefore, we assessed whether LPS increased nuclear translocation of either RelA or RelB which is necessary for generation of the biological response. Cells were treated with LPS for 30 minutes, after which time nuclear extracts were prepared and analyzed for RelA and RelB by Western analysis. LPS increased the nuclear content of RelA several-fold, while the levels of RelB were not increased above time-matched control values (Figure 6). Similar increases in nuclear translocation of RelA were seen in response to TNFα and IL-1α (data not shown).
Figure 6.
Effect of LPS on nuclear levels of NF-κB transcription factors. Nuclear extracts were prepared from C2C12 cells and proteins separated by SDS-PAGE and transferred to PVDF membrane as described in “Experimental Procedures”. Individual membranes were probed with RelA or RelB specific antibodies. Results of a typical experiment are shown. Similar results were obtained with independent preparations of nuclear extracts. The control values for RelA and RelB abundance were arbitrarily set at 1.0 AU.
Notes: The results shown represent the mean ± SEM of n = 4 independent experiments. *P < 0.05 compared to respective control value.
EMSA binding site competition and supershift analysis
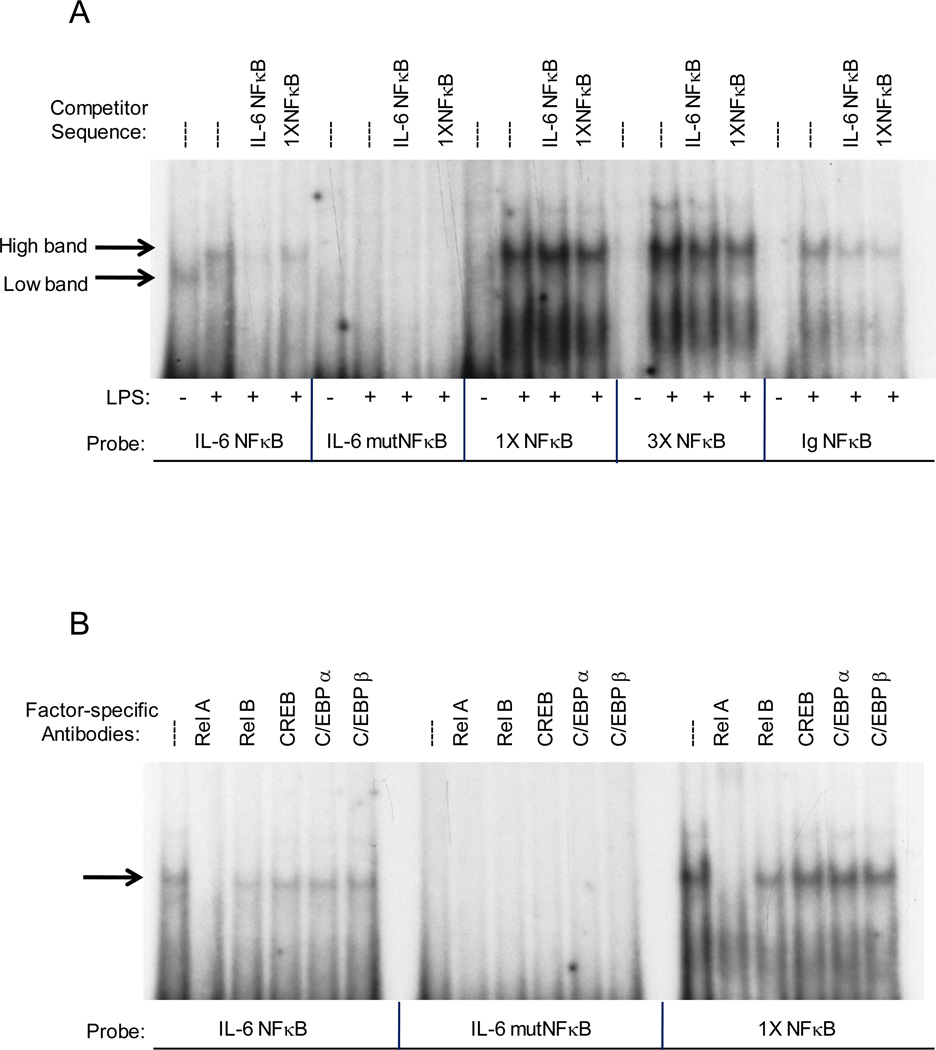
We next performed EMSA with nuclear extracts using probes containing either the IL-6 NF-κB site (IL-6 NF-κB), the mutated IL-6 NF-κB site used in the transfection experiments (IL-6 mutNF-κB), a consensus NF-κB site (1X NF-κB), three tandem NF-κB sites consistent with those in the NF-κB-Luc promoter used in the transfection experiments (3X NF-κB), and an immunoglobulin NF-κB site (Ig NF-κB). A single band is present using the IL-6 NF-κB probe when uninduced (eg no LPS) which is not observed when using any of the other probes (Figure 7A). However, a single higher migrating band was detected bound to the IL-6 NF-κB, 1X NF-κB, 3X NF-κB, and Ig NF-κB probes in LPS-treated myocytes (Figure 7A). The strongest binding was seen with the 1X NF-κB and 3X NF-κB consensus sequence probes, with a lesser amount bound to the IL-6 NF-κB and Ig NF-κB probes. No binding was observed using the IL-6 mutNF-κB probes. Binding to the other probes was similarly competed with NF-IL6 and 1XNF-κB competitor oligonucleotides suggesting a similar protein complex is likely to have been formed on all the probes.
Figure 7.
Effect of binding site competitors and factor specific antibodies on protein-DNA complex formation on various NF-κB site probes. Nuclear extracts were prepared from LPS-treated C2C12 myocytes and incubated with 10 fmol of radiolabeled NF-κB site probe, in the presence or absence of the indicated competitor oligonucleotides (panel A) or factor-specific antibody (panel B). The binding reactions were electrophoresed on a 6% native polyacrylamide gel to produce this autoradiograph. Results of a typical experiment are shown. Similar results were obtained with independent preparations of nuclear extracts.
To investigate the protein(s) binding to the different NF-κB binding sequences, antibodies to RelA, RelB, CREB, C/EBPα, and C/EBPβ were obtained and used in EMSA supershift experiments. The addition of RelA antibodies caused a loss of the higher migrating band formed on the IL-6 NF-κB and 1X NF-κB probes when induced with LPS. These data suggest this band is p65/RelA (Figure 7B) and are consistent with our results of increased nuclear RelA levels with LPS induction. A slight reduction in band intensity in the presence of RelB antibodies could indicate the presence of RelB in the band or possibly a slight cross-reactivity of the RelB antibody for the RelA protein. The addition of CREB, C/EBPα or C/EBPβ specific antibodies had little or no affect on band intensity, indicating these proteins are not involved in complex formation at the NF-κB site.
Discussion
Skeletal muscle is one of the largest organs in the body accounting for approximately 45% of total body weight. As such, it represents a major reservoir of circulating amino acids to support the acute phase response to infection and traumatic injury. Moreover, it is now well accepted that skeletal muscle has important immune functions and myocytes, like other more traditional immune cells, process both afferent and efferent limbs of the innate immune system. That is, because of the presence of various toll-like receptors and cytokine receptors these myocytes respond to a large number of endogenous and exogenous immune mediators as well as substances released in response to tissue injury.1,2 In turn, once stimulated, these myocytes synthesize and secrete a wide array of inflammatory mediators which are at least under partial control by NF-κB. These mediators can affect the immune and metabolic response of the host in both a classical endocrine manner as well as via an autocrine/paracrine mechanism. IL-6 is one of the predominant proinflammatory cytokines released both by skeletal muscle in vivo and myocytes in vitro after appropriate stimulation.11–13 Hence, a greater understanding of the transcriptional regulation of this cytokine in muscle per se is of importance.
In the current study we show the putative NF-κB binding site is necessary for IL-6 induction by LPS, TNFα and IL-1α in C2C12 myocytes thereby extending reports in other cell systems.7,13,31–35 Furthermore, the ability of LPS to activate the IL-6 promoter appears independent of the NF-IL6 and AP-1 sites. Our data also demonstrate that these immune modulators increased NF-κB activity by increasing the nuclear content of RelA, but not RelB. As expected, constitutive activation of the NF-κB pathway increased IL-6 transcription to the cytokine-induced level, consistent with the involvement of the NF-κB pathway. However, our data suggest that IL-6 transcription may be regulated through an alternative mechanism when the RelA pathway is inhibited or, alteratively, that the dominant negative plasmids induce IL-6 promoter activity from either the NF-IL6 or AP-1 sites as a result of the accumulation of some intercellular mediator. Contrary to expectations, inhibiting RelA and RelB pathways, either individually or in tandom, using dominant negative upstream kinases or the general proteosome inhibitor MG-132 had no effect on the ability of LPS to stimulate the IL-6 promoter. These data suggest that NF-κB per se is not involved in the induction of IL-6, at least at the time point studied. Therefore, we speculate that some other factor may be involved in regulating IL-6 transcription by binding to the putative NF-κB site within the IL-6 promoter and stimulating the formation of a transcription complex.36,37 Data from our EMSA supershift analysis revealed that RelA binds weakly to the putative NF-κB site in response to LPS. However, a different protein also appears to bind to that site, but not to a consensus NF-κB site. The binding of this unknown protein to the putative NF-κB site in the IL-6 promoter was only observed in the uninduced basal state. This finding may suggest the factor acts as an inhibitor preventing the binding of an NF-κB protein until its nuclear level increases upon induction. Alternatively, these data might suggest RelA becomes phosphorylated, translocates to the nucleus and has a greater ability to interact with CBP and RNA polymerase II to enhance transcription. Further investigations are necessary to identify the unknown protein and additional regulatory sequences, as well as to determine how this putative factor functions within myocytes to regulate IL-6 transcription.
It has been previously reported that both NF-κB and NF-IL6 are required for LPS and cytokine induction of the IL-6 gene in monocytes.7,38,39 However in myocytes, mutation of the NF-IL6 binding site had no effect on IL-6 promoter activity and only the loss of NF-κB binding abolished LPS and cytokine induction. Also the presence of an A-ZIP inhibitor for C/EBPβ had no effect on the induction of the IL-6 reporter. Interestingly, both NF-κB and NF-IL6 binding sites are required for hypoxia-induced IL-6 expression in cardiac myocytes,40 suggesting that different inducers regulate IL-6 transcription by different mechanisms in different cell types.
Overall, the current data show activation of the IL-6 promoter by inducers which are typically present during infection or injury does not use the canonical NF-κB pathways, even though the putative NF-κB binding site in the IL-6 promoter is required. We also show that a protein that is neither RelA nor RelB can bind to this site. Therefore, either an unrecognized NF-κB pathway exists in myocytes or a non-NF-κB factor binds to the site to induce complex formation. Additional studies are needed to resolve these or other possibilities.
Acknowledgment
This work was supported in part by a grant from the National Institutes of Health GM-38032 and AA11290.
References
- 1.Frost RA, Lang CH. Regulation of muscle growth by pathogen-associated molecules. J Anim Sci. 2008;86:E84–E93. doi: 10.2527/jas.2007-0483. [DOI] [PubMed] [Google Scholar]
- 2.Frost RA, Lang CH. Skeletal muscle cytokines: regulation by pathogen-associated molecules and catabolic hormones. Curr Opin Clin Nutr Metab Care. 2005;8:255–263. doi: 10.1097/01.mco.0000165003.16578.2d. [DOI] [PubMed] [Google Scholar]
- 3.Heinrich PC, Castell JV, Andus T. Interleukin-6 and the acute phase response. Biochem J. 1990;265:621–636. doi: 10.1042/bj2650621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ray A, Tatter SB, Santhanam U, et al. Regulation of expression of interleukin-6. Molecular and clinical studies. Ann N Y Acad Sci. 1989;557:353–361. [PubMed] [Google Scholar]
- 5.Kishimoto T, Hibi M, Murakami M, et al. The molecular biology of interleukin 6 and its receptor. Ciba Found Symp. 1992;167:5–16. doi: 10.1002/9780470514269.ch2. [DOI] [PubMed] [Google Scholar]
- 6.Sehgal PB. Regulation of IL6 gene expression. Res Immunol. 1992;143:724–734. doi: 10.1016/0923-2494(92)80011-9. [DOI] [PubMed] [Google Scholar]
- 7.Dendorfer U, Oettgen P, Libermann TA. Multiple regulatory elements in the interleukin-6 gene mediate induction by prostaglandins, cyclic AMP, lipopolysaccharide. Mol Cell Biol. 1994;14:4443–4454. doi: 10.1128/mcb.14.7.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akira S, Isshiki H, Nakajima T, et al. A nuclear factor for the IL-6 gene (NF-IL6) Chem Immunol. 1992;51:299–322. [PubMed] [Google Scholar]
- 9.Zhang Y, Lin JX, Vilcek J. Synthesis of interleukin 6 (interferon-beta 2/B cell stimulatory factor 2) in human fibroblasts is triggered by an increase in intracellular cyclic AMP. J Biol Chem. 1988;263:6177–6182. [PubMed] [Google Scholar]
- 10.Zhang YH, Lin JX, Yip YK, et al. Enhancement of cAMP levels and of protein kinase activity by tumor necrosis factor and interleukin 1 in human fibroblasts: role in the induction of interleukin 6. Proc Natl Acad Sci U S A. 1988;85:6802–6805. doi: 10.1073/pnas.85.18.6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frost RA, Nystrom GJ, Lang CH. Lipopolysaccharide regulates proinflammatory cytokine expression in mouse myoblasts and skeletal muscle. Am J Physiol Regul Integr Comp Physiol. 2002;283:R698–R709. doi: 10.1152/ajpregu.00039.2002. [DOI] [PubMed] [Google Scholar]
- 12.Frost RA, Nystrom GJ, Lang CH. Lipopolysaccharide and proinflammatory cytokines stimulate interleukin-6 expression in C2C12 myoblasts: role of the Jun NH2-terminal kinase. Am J Physiol Regul Integr Comp Physiol. 2003;285:R1153–R1164. doi: 10.1152/ajpregu.00164.2003. [DOI] [PubMed] [Google Scholar]
- 13.Frost RA, Nystrom GJ, Lang CH. Multiple toll-like receptor ligands induce an IL-6 transcriptional response in skeletal myocytes. Am J Physiol Regul Integr Comp Physiol. 2006;290:R773–R784. doi: 10.1152/ajpregu.00490.2005. 2006. [DOI] [PubMed] [Google Scholar]
- 14.Eickelberg O, Pansky A, Mussmann R, et al. Transforming growth factor-beta1 induces interleukin-6 expression via activating protein-1 consisting of JunD homodimers in primary human lung fibroblasts. J Biol Chem. 1999;274:12933–12938. doi: 10.1074/jbc.274.18.12933. [DOI] [PubMed] [Google Scholar]
- 15.Smart DE, Vincent KJ, Arthur MJ, et al. JunD regulates transcription of the tissue inhibitor of metalloproteinases-1 and interleukin-6 genes in activated hepatic stellate cells. J Biol Chem. 2001;276:24414–24421. doi: 10.1074/jbc.M101840200. [DOI] [PubMed] [Google Scholar]
- 16.Xiao G, Cvijic ME, Fong A, et al. Retroviral oncoprotein Tax induces processing of NF-kappaB2/p100 in T cells: evidence for the involvement of IKKalpha. EMBO J. 2001;20:6805–6815. doi: 10.1093/emboj/20.23.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao G, Harhaj EW, Sun SC. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol Cell. 2001;7:401–409. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 18.Yeagley D, Moll J, Vinson CA, et al. Characterization of elements mediating regulation of phosphoenolpyruvate carboxykinase gene transcription by protein kinase A and insulin. Identification of a distinct complex formed in cells that mediate insulin inhibition. J Biol Chem. 2000;275:17814–17820. doi: 10.1074/jbc.M909842199. [DOI] [PubMed] [Google Scholar]
- 19.Frost RA, Lang CH, Gelato MC. Transient exposure of human myoblasts to tumor necrosis factor-alpha inhibits serum and insulin-like growth factor-I stimulated protein synthesis. Endocrinology. 1997;138:4153–4159. doi: 10.1210/endo.138.10.5450. [DOI] [PubMed] [Google Scholar]
- 20.Frost RA, Nystrom GJ, Lang CH. Tumor necrosis factor-alpha decreases insulin-like growth factor-I messenger ribonucleic acid expression in C2C12 myoblasts via a Jun N-terminal kinase pathway. Endocrinology. 2003;144:1770–1779. doi: 10.1210/en.2002-220808. [DOI] [PubMed] [Google Scholar]
- 21.Hurst HC, Masson N, Jones NC, et al. The cellular transcription factor CREB corresponds to activating transcription factor 47 (ATF-47) and forms complexes with a group of polypeptides related to ATF-43. Mol Cell Biol. 1990;10:6192–6203. doi: 10.1128/mcb.10.12.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeagley D, Agati JM, Quinn PG. A tripartite array of transcription factor binding sites mediates cAMP induction of phosphoenolpyruvate carboxykinase gene transcription and its inhibition by insulin. J Biol Chem. 1998;273:18743–18750. doi: 10.1074/jbc.273.30.18743. [DOI] [PubMed] [Google Scholar]
- 23.Qu Z, Qing G, Rabson A, et al. Tax deregulation of NF-kappaB2 p100 processing involves both beta-TrCP-dependent and -independent mechanisms. J Biol Chem. 2004;279:44563–44572. doi: 10.1074/jbc.M403689200. [DOI] [PubMed] [Google Scholar]
- 24.Akiba H, Nakano H, Nishinaka S, et al. CD27, a member of the tumor necrosis factor receptor superfamily, activates NF-kappaB and stress-activated protein kinase/c-Jun N-terminal kinase via TRAF2, TRAF5, and NF-kappaB-inducing kinase. J Biol Chem. 1998;273:13353–13358. doi: 10.1074/jbc.273.21.13353. [DOI] [PubMed] [Google Scholar]
- 25.Holtmann H, Winzen R, Holland P, et al. Induction of interleukin-8 synthesis integrates effects on transcription and mRNA degradation from at least three different cytokine- or stress-activated signal transduction pathways. Mol Cell Biol. 1999;19:6742–6753. doi: 10.1128/mcb.19.10.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foehr ED, Lin X, O'Mahony A, et al. NF-kappa B signaling promotes both cell survival and neurite process formation in nerve growth factor-stimulated PC12 cells. J Neurosci. 2000;20:7556–7563. doi: 10.1523/JNEUROSCI.20-20-07556.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geleziunas R, Ferrell S, Lin X, et al. Human T-cell leukemia virus type 1 Tax induction of NF-kappaB involves activation of the IkappaB kinase alpha (IKKalpha) and IKKbeta cellular kinases. Mol Cell Biol. 1998;18:5157–5165. doi: 10.1128/mcb.18.9.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun S, Elwood J, Greene WC. Both amino- and carboxyl-terminal sequences within I kappa B alpha regulate its inducible degradation. Mol Cell Biol. 1996;16:1058–1065. doi: 10.1128/mcb.16.3.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uhlik M, Good L, Xiao G, et al. NF-kappaB-inducing kinase and IkappaB kinase participate in human T-cell leukemia virus I Tax-mediated NF-kappaB activation. J Biol Chem. 1998;273:21132–21136. doi: 10.1074/jbc.273.33.21132. [DOI] [PubMed] [Google Scholar]
- 30.Delhase M, Hayakawa M, Chen Y, et al. Positive and negative regulation of IkappaB kinase activity through IKKbeta subunit phosphorylation. Science. 1999;284:309–313. doi: 10.1126/science.284.5412.309. [DOI] [PubMed] [Google Scholar]
- 31.Zhang YH, Lin JX, Vilcek J. Interleukin-6 induction by tumor necrosis factor and interleukin-1 in human fibroblasts involves activation of a nuclear factor binding to a kappa B-like sequence. Mol Cell Biol. 1990;10:3818–3823. doi: 10.1128/mcb.10.7.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomita N, Morishita R, Tomita S, et al. Transcription factor decoy for nuclear factor-kappaB inhibits tumor necrosis factor-alpha-induced expression of interleukin-6 and intracellular adhesion molecule-1 in endothelial cells. J Hypertens. 1998;16:993–1000. doi: 10.1097/00004872-199816070-00013. [DOI] [PubMed] [Google Scholar]
- 33.Luo G, Hershko DD, Robb BW, et al. IL-1beta stimulates IL-6 production in cultured skeletal muscle cells through activation of MAP kinase signaling pathway and NF-kappa B. Am J Physiol Regul Integr Comp Physiol. 2003;284:R1249–R1254. doi: 10.1152/ajpregu.00490.2002. [DOI] [PubMed] [Google Scholar]
- 34.Sparacio SM, Zhang Y, Vilcek J, et al. Cytokine regulation of interleukin-6 gene expression in astrocytes involves activation of an NF-kappa B-like nuclear protein. J Neuroimmunol. 1992;39:231–242. doi: 10.1016/0165-5728(92)90257-l. [DOI] [PubMed] [Google Scholar]
- 35.Libermann TA, Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol Cell Biol. 1990;10:2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanden BW, De BK, Boone E, et al. The nuclear factor-kappaB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J Biol Chem. 1999;274:32091–32098. doi: 10.1074/jbc.274.45.32091. [DOI] [PubMed] [Google Scholar]
- 37.Vanden BW, Vermeulen L, De WG, et al. Signal transduction by tumor necrosis factor and gene regulation of the inflammatory cytokine interleukin-6. Biochem Pharmacol. 2000;60:1185–1195. doi: 10.1016/s0006-2952(00)00412-3. [DOI] [PubMed] [Google Scholar]
- 38.Matsusaka T, Fujikawa K, Nishio Y, et al. Transcription factors NF-IL6 and NF-kappa B synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci U S A. 1993;90:10193–10197. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akira S, Kishimoto T. NF-IL6 and NF-kappa B in cytokine gene regulation. Adv Immunol. 1997;65:1–46. [PubMed] [Google Scholar]
- 40.Matsui H, Ihara Y, Fujio Y, et al. Induction of interleukin (IL)-6 by hypoxia is mediated by nuclear factor (NF)-kappa B and NF-IL6 in cardiac myocytes. Cardiovasc Res. 1999;42:104–112. doi: 10.1016/s0008-6363(98)00285-5. [DOI] [PubMed] [Google Scholar]