Summary
The proteolytic conversion of prothrombin to thrombin catalysed by prothrombinase is one of the more extensively studied reactions of blood coagulation. Sophisticated biophysical and biochemical insights into the players of this reaction were developed in the early days of the field. Yet, many basic enzymological questions remained unanswered. I summarise new developments that uncover mechanisms by which high substrate specificity is achieved, and the impact of these strategies on enzymic function. Two principles emerge that deviate from conventional wisdom that has otherwise dominated thinking in the field. 1) Enzymic specificity is dominated by the contribution of exosite binding interactions between substrate and enzyme rather than by specific recognition of sequences flanking the scissile bond. Coupled with the regulation of substrate conformation as a result of the zymogen to proteinase transition, novel mechanistic insights result for numerous aspects of enzyme function. 2) The transition of zymogen to proteinase following cleavage is not absolute and instead, thrombin can reversibly interconvert between zymogen-like and proteinase-like forms depending on the complement of ligands bound to it. This establishes new paradigms for considering proteinase allostery and how enzyme function may be modulated by ligand binding. These insights into the action of prothrombinase on prothrombin have wide-ranging implications for the understanding of function in blood coagulation.
Keywords: Prothrombin, thrombin, prothrombinase, substrate specificity, serine proteinase, proteinase allostery, enzyme regulation
Introduction
Thrombin is produced by proteolytic activation of the precursor, prothrombin, in the first committed step of the pathway to thrombus formation. Thrombin catalyses thrombus formation and also a series of reactions that both positively and negatively regulate flux through the coagulation cascade. Thus, the transition of prothrombin to thrombin represents the only non-redundant step in thrombus formation and yields a critical regulatory end product of coagulation. In addition to its fundamental role in coagulation, prothrombin activation embodies key structural and functional features encountered in the other proteolytic activation steps of the cascade. Consequently, the wealth of biochemical and biophysical information that has accumulated in this system justifies its consideration as an archetypal reaction of coagulation.
The Catalyst
Factor Xa is the trypsin-like proteinase of coagulation that catalyses prothrombin activation. However, despite the fact that Xa readily cleaves peptidyl substrates, its ability to activate prothrombin is markedly low. Efficient thrombin formation requires the membrane-dependent interaction of Xa with the cofactor protein, factor Va, to assemble the prothrombinase complex (Fig. 1). The profound functional consequences of the reversible protein-membrane and protein-protein interactions that stabilise factor Xa within prothrombinase are evident from kinetic constants for prothrombin cleavage measured with Xa in binary and ternary complexes with membranes and Va (Table I). Saturation of Xa with membranes yields a large decrease in Km (Table I). As both factor Xa and prothrombin are vitamin K-dependent proteins that bind reversibly to phosphatidylserine-containing membranes in a Ca2+-dependent manner (1-3), approximation of the proteinase and substrate on the membrane surface is considered the source of the membrane-dependent increase in the apparent affinity for the substrate (4). Further addition of saturating Va to form membrane-assembled prothrombinase yields a large increase in the kcat for the reaction (Table I), previously attributed to the modulation of the active-site of Xa within prothrombinase by the cofactor (5,6). These improvements in substrate affinity and catalytic power produce the proverbial 105-fold increase in the rate of thrombin formation at the physiological concentration of prothrombin (1,7). It is for this reason that prothrombinase rather than Xa is considered the physiological catalyst for rapid thrombin formation following vascular damage. Analogous functional consequences in the cleavage of the cognate biological substrate are observed following assembly of the other membrane-dependent enzyme complexes of coagulation (1,2,8).
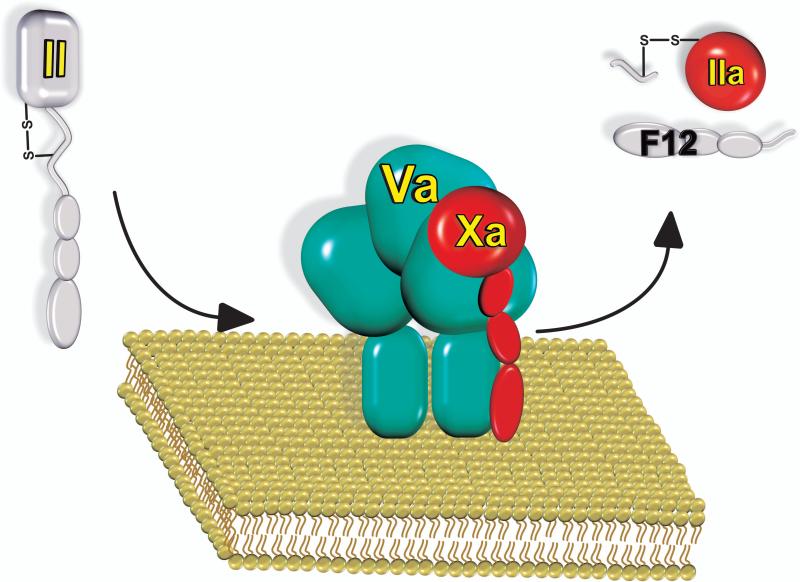
Figure 1. Prothrombin is activated by a membrane-bound enzyme complex.
Prothrombinase assembles through reversible interactions between the serine proteinase Xa and the protein cofactor Va on membranes containing phosphatidylserine. The enzyme complex cleaves the zymogen, prothrombin (II) at two sites to produce thrombin (IIa), which is composed of two chains in disulphide linkage and the release of the N-terminal propiece fragment 1.2 (F12).
Table 1.
Steady State Kinetic Constants for Xa and prothrombinase
| Prothrombin1 | Peptidyl Substrate2 | ||||
|---|---|---|---|---|---|
| Enzyme Species3 | Km (μM) | Vmax/ET (s-1) | Relative Rate4 | Km (μM) | Vmax/ET (s-1) |
| Xa | 84 | 0.01 | 1 | 98 | 172 |
| Xa/PCPS | 0.7 | 0.05 | 203 | 98 | 181 |
| Xa/Va/PCPS | 0.4 | 108 | 512,000 | 205 | 179 |
Kinetic constants for bovine prothrombin cleavage by bovine Xa alone (7), or for the activation of human prothrombin by human enzyme constituents in the presence of membranes (53).
Steady state constants for the hydrolysis of Spectrozyme Xa (methoxycarbonyl-D-cyclohexylglycyl-glycyl-L-arginine-p-nitroanilide ) measured with human Xa and Va (10).
The enzyme species correspond to Xa alone, Xa saturably bound to membranes (Xa/PCPS) and Xa assembled into prothrombinase (Xa/Va/PCPS) using saturating concentrations of membranes and Va.
Relative rates at 1.4 μM prothrombin calculated from the steady state kinetic constants.
However, the incorporation of Xa into prothrombinase is not associated with large improvements in the catalytic activity of factor Xa towards peptidyl substrates (Table I) (9-11). It follows that the mechanisms underlying Va function within prothrombinase cannot readily be explained by improvements in active-site function of Xa. Furthermore, peptidyl substrate studies have consistently failed to provide an explanation for the selective cleavage following the Glu/Asp-Gly-Arg sequence in prothrombin by prothrombinase (or Xa) (12,13). Thus, there are fundamental differences in the way that prothrombinase achieves specificity for its protein substrate not approximated by peptidyl substrates that interact in a limited way with the active-site of Xa within the enzyme complex. It also follows that questions related to how the cofactor may function within prothrombinase and how narrow enzymic specificity towards the protein substrate is achieved represent two sides of the same coin.
The Substrate
A complicating feature arises from the fact that prothrombin needs to be cleaved at two sites to produce thrombin. Consequently, prothrombin is converted to thrombin as a result of two sequential enzyme catalysed reactions. Thus the kinetic constants used to phrase the functional consequences of prothrombinase assembly are based on steady state kinetic constants determined by varying the substrate for the first reaction and measuring the product of the second enzyme catalysed reaction. As such kinetic constants cannot be interpreted in a meaningful way; basic features of the problem therefore need rephrasing and reconsideration.
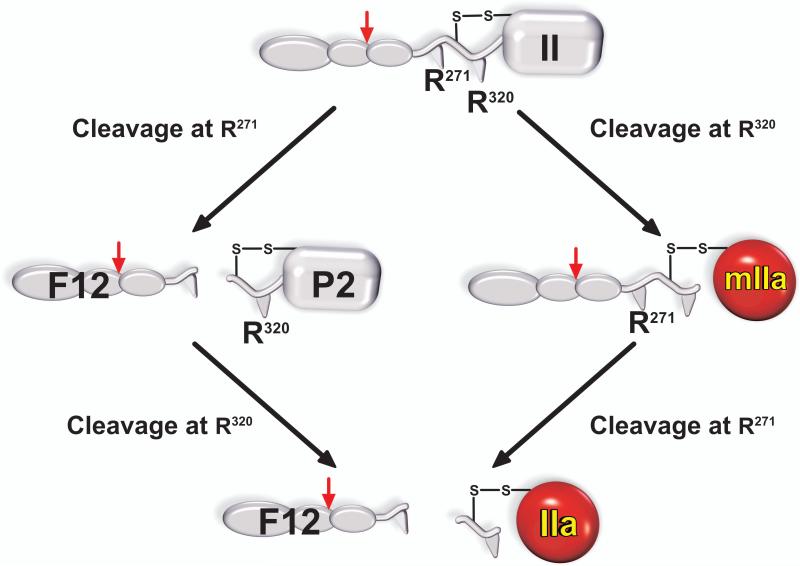
As cleavage at two sites is necessary, there are two possible pathways for the formation of thrombin (Fig. 2). Initial cleavage of prothrombin following Arg271 yields the N-terminal pro-piece fragment 1.2 (F12) and the zymogen prethrombin 2 (P2) which is then further cleaved following Arg320 to yield the disulphide-linked 2 chain form of thrombin. Conversely cleavage in the opposite order yields the proteinase meizothrombin (mIIa) as an intermediate, which is then processed at Arg271 to yield thrombin and F12. Early studies performed with Xa in the absence of Va established cleavage at Arg271 followed by cleavage at Arg320 as the prominent, or perhaps only, pathway by which prothrombin was activated (14). Subsequent studies with verifiably assembled prothrombinase revealed abundant levels of mIIa produced as an intermediate with little evidence for the accumulation of P2 (15,16). A series of subsequent studies have since shown that the reaction catalysed by prothrombinase assembled on synthetic phospholipid vesicles almost exclusively proceeds via mIIa formation with flux through the alternative cleavage pathway being experimentally indistinguishable from zero (17).
Figure 2. Pathways for the proteolytic conversion of prothrombin to thrombin.
The conversion of prothrombin to thrombin results from cleavages following Arg271 and Arg320. Initial cleavage following Arg271 yields the pathway on the left and produces the zymogen, prethrombin 2 (P2) and the propiece, fragment 1.2 (F12) as intermediates. P2 requires further processing at Arg320 to yield thrombin. The pathway on the right arises from initial cleavage following Arg320, which produces the proteinase meizothrombin (mIIa) as an intermediate. Further cleavage following Arg271 is required to yield IIa and the propiece, F12. The Arg155 site (Red Arrow) is susceptible to thrombin cleavage and separates the fragment 1 region from fragment 2 within F12.
The Question of Substrate Specificity
While both cleavage sites are accessible within the substrate to externally added proteinase, prothrombinase essentially exclusively acts at the Arg320 site when presented with prothrombin. Cleavage at Arg271 only occurs at an appreciable rate once the Arg320 site has been cleaved (17). Enzymic specificity at multiple levels comes to the fore in considering the specific action of prothrombinase on prothrombin. The first deals with the basis for substrate specificity at the level of the protein substrate: Why does prothrombinase selectively act to cleave prothrombin and not the other structurally related vitamin K-dependent zymogens of coagulation? The next relates to the mechanisms underlying specific cleavage at the two sites in prothrombin preceded by Glu/Asp-Gly-Arg, despite the fact that a kinetic preference for cleavage after this sequence is not evident from peptidyl substrate studies (12,13). The third reflects the ability of prothrombinase to discriminate between the two cleavage sites within prothrombin itself, yielding essentially ordered cleavage at Arg320 followed by cleavage at Arg271. Finally, considering the ambiguity in inferring the basis for sequence preference for cleavage by prothrombinase, it seems appropriate to question why the action of the enzyme is restricted to only two sites at the expense of 37 other arginines in the substrate. These are, after all, elementary questions regarding proteinase specificity, yet they remain major stumbling blocks in coagulation enzymology.
Exosite Binding
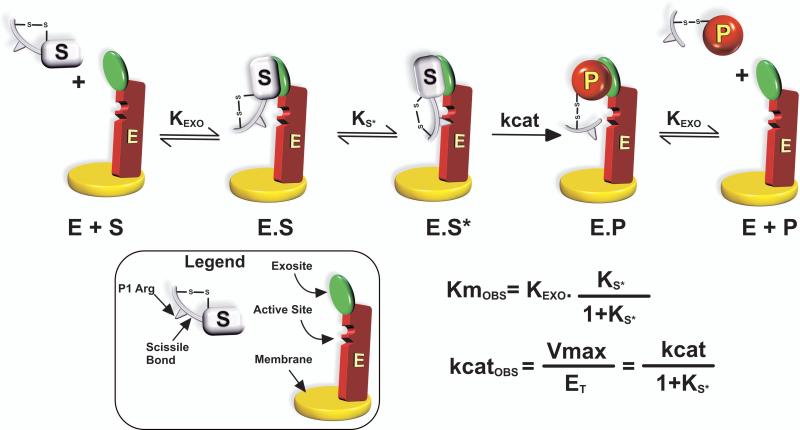
Efforts to derive mechanistic insights into the action of prothrombinase on its protein substrate were initially broached by studies of the conversion of P2 to thrombin (Fig. 2). P2 represents one of two simplest possible protein substrates containing a single cleavage site. In the absence of reversibly bound F12, P2 neither binds membranes (mediated by the fragment 1 region of F12) nor is it expected to participate in interactions with factor Va previously proposed to occur through the fragment 2 region (18,19). Despite the lack of these additional component interactions that are expected to complicate the pathway for substrate recognition, kinetic studies revealed a multi-step pathway for substrate recognition by prothrombinase (Fig. 3) (20,21). The initial capture of the substrate occurs through exosite interactions that are independent of the active-site of Xa within the complex. Active-site docking follows in a unimolecular binding step before catalysis. The cleaved product is tethered to the enzyme through similar exosite interactions and must dissociate before the next round of substrate binding and catalysis. Kinetic signatures of this mechanism are that inhibitors and alternate peptidyl substrates that bind to the active-site of Xa within prothrombinase act as classical non-competitive inhibitors of P2 activation, i.e. ligand binding to the active-site of the catalyst has no impact on protein substrate affinity. In addition, the product behaves as a classical competitive inhibitor of protein substrate cleavage but without affecting ligand or peptidyl substrate binding to the active-site of Xa within prothrombinase. The new mechanistic insights are that exosite binding dominates substrate affinity and that the inferred rate constant for catalysis is heavily influenced by the equilibrium constant for active-site docking (Fig. 3). These ideas deviate drastically from the “standard” interpretation of steady state kinetic constants drawn from the action of serine proteinases on peptidyl substrates (20).
Figure 3. Multi-step Pathway for Protein Substrate Recognition by Prothrombinase.
Kinetic scheme resolved for the action of prothrombinase on P2. The initial binding interaction between substrate (S) and prothrombinase (E) to form ES results from exosite-dependent interactions between S and E. Exosite binding is followed by a unimolecular binding step in which structures flanking the cleavage site engage the active-site of the enzyme before catalysis can occur. The product (P) is also bound to E by exosite interactions before it is released. The graphical legend highlights the important features of S and E. The composite nature of the steady state kinetic constants is illustrated by derivation employing the rapid equilibrium assumption. Ks* is defined as [E.S]/[E.S*].
Enforcement of Specificity
Exosite-dependent enforcement of binding specificity has been documented using a series of recombinant P2 species bearing altered P3-P1 sequences preceding the cleavage site (22). All variant substrates bound prothrombinase with comparable affinity as wild type P2 (22). Provided the P1 residue was Lys or Arg, the variant substrates were cleaved but with varyingly reduced Vmax. This was also surprisingly true for a variant bearing the P3-P1 sequence preceding the scissile bond in factor X (22). Thus, sub-optimal active-site engagement contributes in a minor way to affinity and instead affects Vmax. Indeed, replacement of the P1 Arg with Gln yielded an uncleavable substrate, but one that bound to prothrombinase with the same affinity as wild type P2. The uncleavable nature of this variant substrate arises from a Vmax defect associated with its inability to engage the active-site. These findings fly in the face of long-entrenched concepts implicating the engagement of structures flanking the scissile bond with the active-site in determining substrate-binding specificity. Instead, the catalytic site of Xa within prothrombinase is accepting of a variety of peptidyl sequences including those found at the activation sites of other coagulation zymogens. The action of prothrombinase on its protein substrate is restricted to prothrombin derivatives that can bind to the enzymic exosite thereby excluding cleavage of the other related coagulation zymogens that presumably cannot bind the enzyme in this way.
Exosite Surfaces on the Substrate and Enzyme
Concepts associated with exosite-dependent substrate recognition predict that affinity for the protein substrate is determined by substrate regions removed from substrate structures flanking the scissile bond and by enzymic structures distant from the catalytic site of Xa within prothrombinase. Fragmentation of P2 revealed that the COOH-terminal half of the substrate, physically separable from the scissile bond, could bind to the exosite within prothrombinase and act as a competitive inhibitor of P2 cleavage (21). Conversely, enzymic exosite structures have been probed using a monoclonal antibody and nematode anticoagulant peptide c2, both of which bind the proteinase domain of Xa within prothrombinase and selectively block protein substrate binding without occluding the active-site of the catalyst (10,23). Both sets of studies implicate a region in the proteinase domain, removed from the active-site of Xa, as contributing to exosite-dependent substrate binding (10,23). Related studies from the laboratory of Paul Bock have established an important role for the (pro)anion binding exosite 1 region - (pro)ABE1 - in prothrombin and its derivatives in binding Va (24-26). Since peptides that block (pro)ABE1 also inhibit the action of prothrombinase on these substrate species, it follows that the enzymic exosite is most likely constituted by extended surfaces on both Xa and Va within prothrombinase (27). Interference with either component interaction is sufficient to greatly reduce the affinity for exosite binding (23,26). Consequently, the mode of exosite-mediated substrate recognition delineated for prothrombinase is unlikely to prevail in the action of Xa alone on the substrate. If so, then large improvements in the kinetics of protein-substrate cleavage that accompany the incorporation of Xa into prothrombinase (Table 1) also derive from a change in the mechanism by which the substrate is recognised by the catalyst.
Second Half-Reaction of Prothrombin Activation
mIIa is the next more complex substrate with a single scissile bond at Arg271 (Fig. 2). Mechanistic details of the action of prothrombinase at Arg271 were obtained using a mIIa variant lacking the fragment 1 domain to reduce interpretation problems associated with membrane binding (28). Conclusions comparable to those established for cleavage at Arg320 in P2 were also found to apply for cleavage at Arg271 in this mIIa derivative (28). Thus, the dock and lock strategy, arising from the initial exosite-dependent tethering of the substrate followed by active-site engagement, applies to cleavage at both sites within the protein substrate. Both P2 and the mIIa derivative could bind to prothrombinase assembled using Xa containing a covalently occluded active-site with the same affinity as that inferred from functional studies (29). The two substrate forms as well as thrombin, bound in a mutually exclusive manner to prothrombinase (29). Thus, equivalent exosite binding interactions tether either substrate to prothrombinase and allow the presentation of the individual cleavage sites for active-site engagement and catalysis. Considering that the two cleavage sites are proposed to be distant from each other (30,31), it follows that there must be fundamental differences in the configuration in the bound forms of the two substrates to permit the presentation of the two scissile bonds to the catalytic site of prothrombinase.
Cleavage of Membrane-Binding Substrate Forms
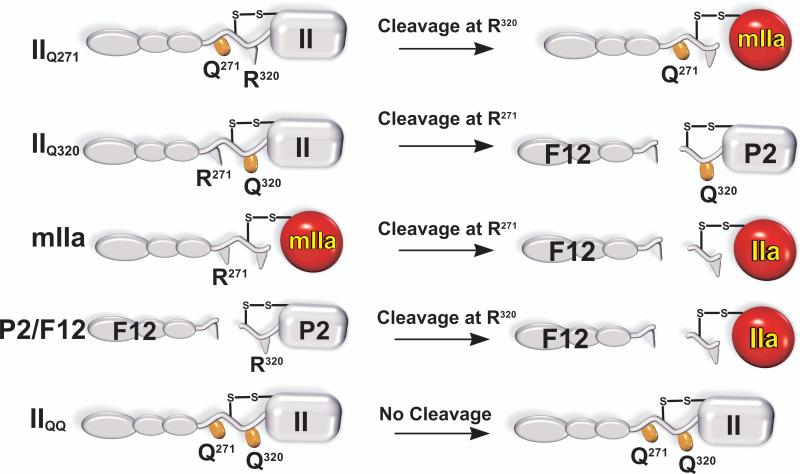
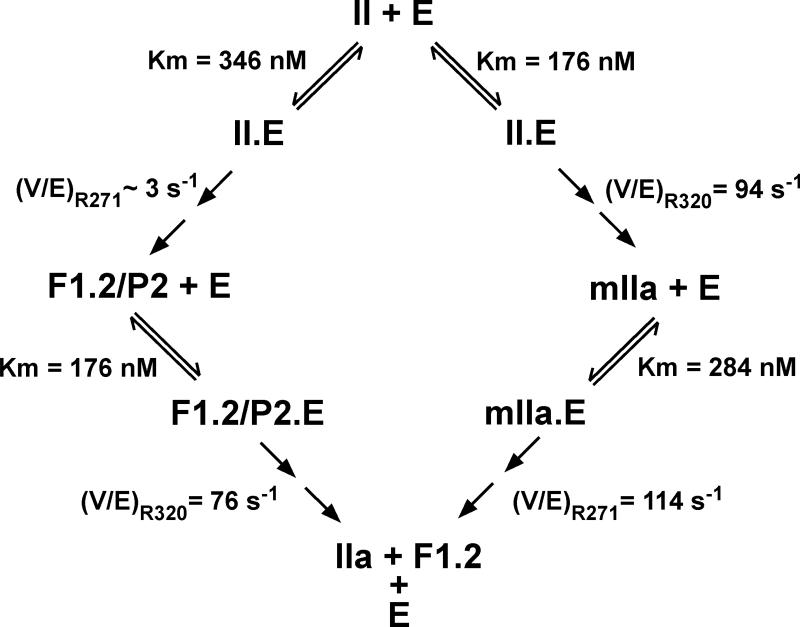
Initial work with intermediates lacking membrane binding was extended to full-length substrate species using prothrombin derivatives in which the individual cleavage sites were rendered uncleavable either singly or in combination, by substitution of the P1 Arg with Gln (Fig. 4). Along with full-length mIIa and P2 saturated with F12, these derivatives provided interpretable steady state kinetic constants for each of the four possible cleavage reactions (Fig. 5). The apparent affinity of each of the four substrate species were equivalent and ~10-30 fold higher than those observed for the non-membrane binding variants used in the initial work (17). Since the substrate-membrane interaction involves regions removed from the site of cleavage, it provides additional exosite interactions that add to the energetics of the binding of the substrate to the enzymic exosite thereby likely accounting for improved affinity. Accordingly, substrates for each of the four possible half reactions bound to prothrombinase in a mutually exclusive manner and the uncleavable IIQQ variant (Fig. 4) could compete with each of the four substrates with the same binding constant (17). While V/E was equivalent for 3 of the 4 half reactions, it was ~30 fold lower for cleavage at Arg271 in intact prothrombin (Fig. 5). Thus, the essentially ordered action of prothrombinase on the full-length zymogen, arises from the fact that Arg271 is cleaved at a ~30-fold lower rate than at Arg320 regardless of the substrate concentration. Defective cleavage at Arg271 in intact prothrombin is rectified following prior cleavage at Arg320 to produce mIIa (Fig. 5). Given the relationship between the equilibrium constant for the unimolecular active-site docking step and the perceived kcat (Fig. 3), the findings raise the possibility that the Arg271 site within prothrombin cannot efficiently engage the active-site of prothrombinase and this engagement is then facilitated once prior cleavage at Arg320 converts it to mIIa (17).
Figure 4. Substrate Derivatives for Kinetic Studies of all Possible Half-Reactions of Prothrombin Activation.
Cleavage of the individual sites in intact prothrombin was assessed using prothrombin variants in which the two arginines were individually rendered uncleavable by mutation to Gln. The intermediates mIIa and P2/F12 were used to assess cleavage at the individual sites following cleavage at the first site. IIQQ denotes a prothrombin variant in which both Arg side chains were mutated to Gln to yield an uncleavable derivative. The products formed upon the limiting action of prothrombinase on these substrate variants are illustrated.
Figure 5. Steady State Kinetic Constants for the Half-Reactions of Prothrombin Activation.
Kinetic constants measured for the individual half-reactions are listed. V/E denotes Vmax/E for the indicated cleavage reactions and is more appropriate than referring to this term as kcat. Taken from Orcutt & Krishnaswamy (17).
Role of the Zymogen to Proteinase Transition in Regulating Cleavage
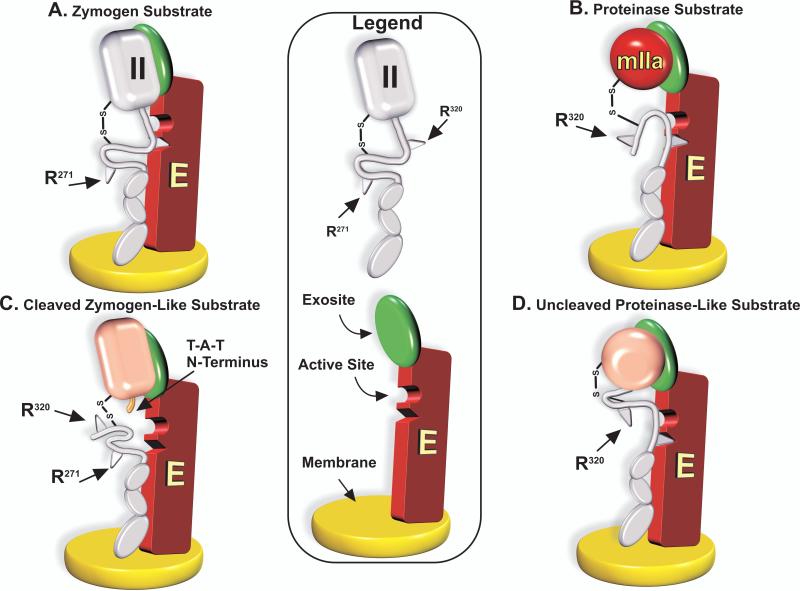
Cleavage at Arg320 corresponds to cleavage at the Arg15-Ile16 bond in trypsinogen that converts zymogen to proteinase (32). Proteinase formation results from the sequence-specific insertion of the newly generated N-terminal sequence into an N-terminal binding cleft and the formation of a salt bridge that triggers changes in the activation domains (32). The known structural transitions associated with the conversion of zymogen to proteinase could explain how prothrombin tethered by exosite binding allows for optimal placement of the Arg320 site for active-site docking and cleavage (Fig. 6, Panel A). In contrast, mIIa bound analogously to prothrombinase facilitates placement of the Arg271 site for active-site docking and cleavage (Fig. 6, Panel B). These ideas were tested using a recombinant prothrombin variant in which the Ile-Val-Glu sequence following Arg320 was replaced with Thr-Ala-Thr to yield a prothrombin derivative with a fully cleavable Arg320 site but with impaired ability to drive the conformation transitions to proteinase (33). Accordingly, this prothrombin variant could be converted to thrombin but failed to yield an active proteinase. While cleavage following Arg320 was normal, subsequent cleavage at Arg271 was impaired ~30-fold, yielding greatly reduced amounts of thrombin with large and persistent amounts of mIIa as an intermediate (33). Thus, a cleaved but zymogen-like substrate does not permit optimal presentation of Arg271 for cleavage (Fig. 6, Panel C). Alternatively, we employed the bacterial protein staphylocoagulase (SC), which employs its own N-terminus to affect conformational activation of prothrombin but without cleavage (34). Prothrombin was conformationally activated by SC, trapped in a proteinase-like configuration by covalent modification with d-Phe-l-Pro-l-Arg-chloromethylketone (FPRck) before dissociation of SC and re-purification. The use of uncleaved prothrombin trapped as proteinase as a substrate for prothrombinase increased the rate of cleavage at Arg271 by ~20-fold with a commensurate decrease in cleavage at Arg320 (33). Thus, favouring a proteinase-like configuration of prothrombin enhances cleavage at Arg271 at the expense of cleavage at Arg320 (Fig. 6, Panel D). These ideas, interpreted according to the scheme in Fig. 6, imply that “Ratcheting” of the substrate between the zymogen and proteinase configurations presents the two bonds in turn for active-site docking and cleavage, thereby accounting for the ordered action of prothrombinase on prothrombin.
Figure 6. Presentation of the Cleavage Sites for Active-site Docking is Driven by the Zymogen or Proteinase-Like Character of Exosite-Bound Substrate.
Exosite-binding constrains substrate presentation such that when the substrate is the zymogen, Arg320 preferentially engages the active-site and is cleaved (Panel A). Conversely, the Arg271 site is readily cleaved when the substrate is the proteinase (Panel B). Prior cleavage at Arg320 yields impaired subsequent cleavage at Arg271 in a variant that remains zymogen-like and is defective in making the transition to proteinase (Panel C). Conversely conformational activation and stabilisation of uncleaved prothrombin in a proteinase-like state yields increased cleavage at Arg271 at the expense of cleavage at Arg320 (Panel D).
Active-site Docking and Catalysis
Binding measurements of substrate with prothrombinase provided physical verification of the assumed relationship between active-site docking and the Vmax for cleavage at the two sites (35). This was achieved using fluorescence measurements of 4-aminobenzamidine binding to the active-site of prothrombinase assembled with a catalytically inactive variant of Xa bearing a substitution of the catalytic Ser195 with Ala. Competition studies arising from the displacement of the probe by various prothrombin variants (Fig. 4) established the equilibrium constant for active-site docking by the substrate but without subsequent catalysis. While titration with wild type prothrombin robustly displaced the fluorescent probe, the uncleavable IIQQ variant (Fig. 4) could not (35). The findings with wild type prothrombin could be replicated with IIQ271 (Ks* <0.02) while the IIQ320 variant yielded very poor probe displacement (Ks* ~3) (35). Thus, as schematically illustrated (Fig. 6), active-site binding by the Arg320 site is highly favoured in intact prothrombin while the Arg271 site engages the active-site ~100-fold weaker (35). This large difference in Ks* accounts for the ~30-fold difference in the Vmax for cleavage at the two sites (Fig. 3). In agreement with other findings, mIIa and IIQ320 stabilised in a proteinase-like configuration could now displace the probe by efficient docking of Arg271 to the active-site (35). Thus, active-site docking by Arg271 requires substrate in the proteinase-like configuration. These binding approaches verify the physical relationship between active-site docking and the Vmax for substrate cleavage and confirm the features schematically illustrated in Fig. 6. Susceptibility for cleavage at any one site within the substrate is determined by the degree of favourability with which it engages the active-site of prothrombinase.
Substrate Geometry and Cleavage Site Discrimination
Discrimination between the two distant cleavage sites by prothrombinase implies that exosite tethering leads to the precise geometric positioning of the scissile bonds to facilitate or restrict active-site docking (36). However, geometric constraints must not be absolute as the Arg271 site in uncleaved prothrombin is indeed capable of engaging the active-site weakly leading to its slow cleavage (33,35). This implies a flexible compound substrate wherein despite exosite-tethering, distant sites can also be captured by the catalytic site of prothrombinase with finite probability. The role of geometry in substrate recognition was assessed by creating a family of substrates in which additional arginines were introduced systematically shifted in one residue increments N-terminal to the Arg320 site (36). Provided the authentic Arg320 site was intact, no evidence was observed for cleavage at the additional introduced sites. However, substitution of Arg320 with Gln to render this site uncleavable revealed the ready ability of prothrombinase to cleave at newly introduced Arg sites shifted by as many as 3 residues. Shifts greater than 3 abruptly yielded a loss in cleavage at the shifted site and instead yielded slow cleavage at Arg271. Measurements of Ks* for active-site docking revealed that despite large changes in this binding constant associated with shifts in the P1 Arg, cleavage at these shifted sites was preferred provided Ks* remained more favourable than for the Arg271 site. Shifts beyond 3 residues increased Ks* sufficiently to allow an abrupt change to cleavage at Arg271. These findings reveal how cleavage at multiple sites is regulated in a potentially flexible compound substrate bearing two or more cleavage sites. Geometry plays an important role in favouring cleavage at a specific site but cleavage is prevented at other Arg residues also susceptible to cleavage by internal competition for active-site docking. The sites that engage the active-site with the smallest Ks* are preferentially cleaved at the expense of other sites that may engage the active-site less favourably. These findings provide a cogent explanation for how the action of prothrombinase is restricted to only two sites despite the presence of other potential cleavage sites.
Fate of Newly Formed Thrombin
Early studies established a nM affinity for the reversible interaction between thrombin and F12 at low ionic strength and pH 8 (37). Because of the membrane binding properties of F12, it was proposed that newly produced thrombin would remain membrane-bound and its release would require thrombin-mediated proteolysis between the fragment 1 and fragment 2 domains (38). More recent studies at physiological pH and ionic strength have revealed a much weaker affinity (μM) for this interaction indicating that only a modest fraction of thrombin may be bound to F12 in the best case (39,40). Thus, thrombin rapidly dissociates from F12 and the membrane surface once it is produced allowing it to participate in the remaining reactions of coagulation essentially unencumbered by the propiece or its fragment 2 derivative known to affect enzymic function (40).
Is Thrombin Always a Proteinase?
Substantial differences in affinity and energetics were noted for F12 binding to thrombin relative to the zymogen, P2, in isothermal titration calorimetry (40). The highly thermodynamically favoured binding of F12 to zymogen relative to proteinase provided an unforeseen avenue to assess the distribution of thrombin between zymogen- and proteinase-like forms (41). Profound differences in ΔH and ΔS with modest changes in ΔG were observed for the binding of F12 to P2, a cleaved but zymogen-like form of thrombin, the catalytically inactive variant of thrombin containing Ser195 substituted with Ala and thrombin covalently stabilised in the most proteinase-like form by covalent modification with FPRck (41). Large compensating changes in ΔH and ΔS for F12 binding were observed as the species increasingly became proteinase-like and were consistent with the binding of the propiece to structurally related zymogen- and proteinase-like forms expected to be in reversible equilibrium with each other. Accordingly, high concentrations of an active-site-directed ligand could transition the zymogen-like form of thrombin along this thermodynamic trajectory to resemble FPR-thrombin. Conversely, removal of Na+ at constant ionic strength could transition the thermodynamic constants along the same trajectory until binding resembled that seen with the cleaved by zymogen-like form of thrombin (41). Finally ligands directed towards anion binding exosite 1 (ABE1) could favour proteinase-like forms (42). In the extreme, uncleaved P2 could be imbued with catalytic activity in the absence of cleavage by strong ligand binding. These points illustrate that thrombin occupies a continuum of zymogen-like and proteinase-like forms following initial cleavage. The continuum and associated thermodynamic compensation in F12 binding derives from the fact that the series of changes associated with proteinase maturation following cleavage are additive. These ideas are surprising when taken in the context of the irreversibility of peptide bond hydrolysis but are in line with the findings with factor VIIa (43).
How Do These Transitions in Thrombin Work?
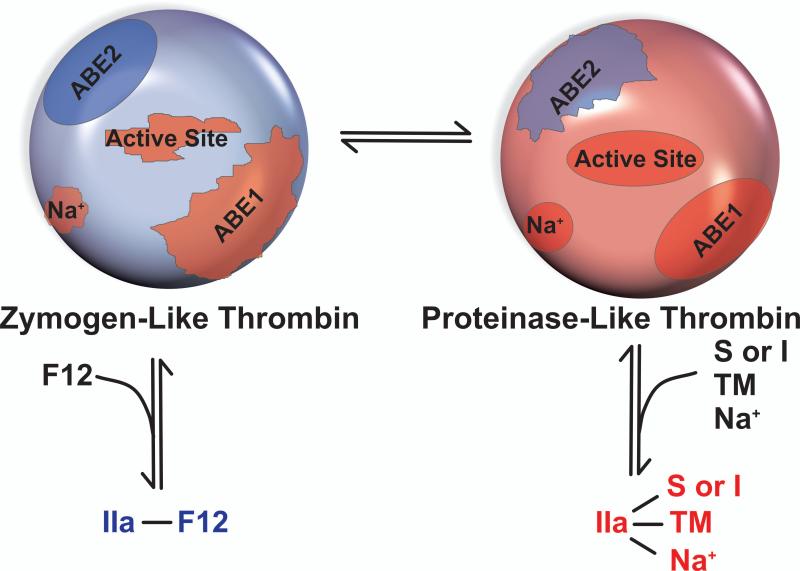
Ligand-dependent interconversions of cleaved thrombin between zymogen- and proteinase-like forms can formally be considered by a two-state model although multiple sub-states of thrombin are likely involved (Fig. 7). The reasoning behind this model lies in the fact that anion binding exosite 2 (ABE2), to which F12 binds, is properly configured in the zymogen-like form but distorted in the proteinase-like form (Fig. 7). Thus, high affinity and thermodynamically favoured binding of F12 stabilises the zymogen-like form. Ligation at the active-site, Na+ site and ABE1 is optimal in the proteinase-like form. Thus, ligands targeting these sites oppose the effects of F12 binding to ABE2 (41). The distribution between the two forms is determined by the complement of ligands available to bind to thrombin.
Figure 7. Ligand-Dependent Interconversions of Thrombin Between Zymogen- and Proteinase-Like Forms.
In the zymogen-like configuration, ABE1, the Na+ site and the active-site are not optimally configured while F12 binding to ABE2 is thermodynamically favoured. The reverse is true for the proteinase-like state in which ligands targeting ABE1, Na+ and the active-site bind more favourably while ABE2 is not optimally configured. Consequently, binding of F12 to ABE2 favours the zymogen-like form while substrates or inhibitors (S or I), Na+ or thrombomodulin (TM) favour the proteinase-like form. It remains to be established as to whether other ligands for ABE1 (i.e. proteinase activated receptor 1 (70), fibrinogen (71)) or ABE2 (i.e. heparin (72), glycoprotein 1bα (73,74)) replicate the effects seen with F12 and TM at biologically relevant concentrations.
Any mutation that destabilises the proteinase-like state will affect this equilibrium and favour the zymogen-like form. In the absence of ligands, the destabilised species is expected to exhibit reduced activity. In contrast, thrombomodulin binding to ABE1 will variably drive the equilibrium to the proteinase-like form yielding protein C activation. This likely represents the thermodynamic basis for how one can engineer anticoagulant-specific thrombin variants by a myriad of different mutations (44-46). Indeed, these special anticoagulant or slow thrombin forms appear variously zymogen-like in their thermodynamic binding signatures or in their distorted x-ray structures (47). These interpretations are also consistent with NMR studies showing the ability of ligands targeting the active-site, ABE1 and Na+ to order the various activation domains to yield the stabilised proteinase (48).
What about the voluminous literature on Na+ regulation of thrombin and the so-called E and E* forms? (47). A reasonable interpretation of our findings is that Na+ is part of the structure of the stabilised proteinase. The binding constants indicate that Na+ site will be near saturated at physiological Na+, ionic strength and pH (41). Thus, although it may be useful as a probe to uncover details of thrombin biochemistry, Na+-dependent regulation is unlikely to reflect a meaningful mechanism for modulating proteinase function in normal mammalian physiology (41). The equivalence between zymogen- and proteinase-like forms and the foregoing E and E* species is hotly denied for a series of technical reasons (49,50). Thus, we presently hesitate to equate our interpretations of zymogen- and proteinase-like forms with the E and E* forms that have been defined by studies done at varying Na+ (47).
Are Zymogen-Like Species Biologically Significant?
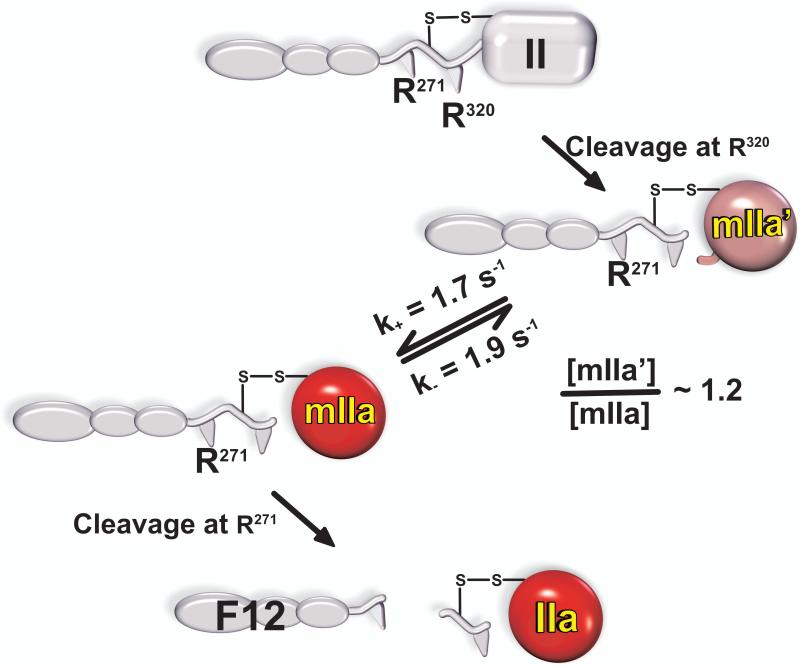
The ability of F12 to favour the zymogen-like state in thrombin lies at the heart of the ligand-dependent shuttling of thrombin between the two proposed states (Fig. 7). Given the relatively weak binding of F12 to thrombin, a significant fraction of the enzyme is unlikely to be regulated in this way (41). Fractional saturation of ABE2 by F12 is irrelevant in the case of mIIa in which the F12 region is covalently bound to the proteinase domain (Fig. 2). Our findings with thrombin imply that mIIa would be exceptionally zymogen-like. Indeed, rapid kinetic studies revealed the slow equilibration of mIIa between zymogen-like and proteinase-like forms only the latter of which could bind active-site ligands with high affinity (42). The two species were found to be approximately equally populated and the distribution between the two forms could be altered predictably by thrombomodulin or by decreasing Na+ (42). Single turnover rapid kinetic studies revealed that initial cleavage at Arg320 by prothrombinase yields a zymogen-like form of mIIa that only slowly equilibrates with the proteinase-like species (Fig. 8). Because of the essential role of the zymogen to proteinase transition in facilitating the second cleavage reaction by prothrombinase to yield thrombin, this slow reversible interconversion of mIIa between the two forms describes the ratcheting step proposed to separate the two half-reactions of prothrombin activation by prothrombinase. It represents a previously unexpected rate-limiting step in thrombin formation. The inclusion of this slow step now provides a comprehensive accounting of the fate of intermediates and products during the action of prothrombinase on prothrombin at a level not previously accomplished (42). The physiological significance of these findings could lie in the fact that zymogen-like mIIa, refractory to further cleavage and inhibition by antithrombin, may be susceptible to dispersal in flowing blood as a long-lived species. Its transit downstream from the site of its formation, as has been documented in capillary flow (51), followed by capture by TM and its participation in protein C activation may be important in limiting the size of the thrombus.
Figure 8. A New Rate-Limiting Step in the Conversion of Prothrombin to Thrombin.
Initial cleavage of prothrombin following Arg320 yields a zymogen-like form (mIIa’) that interconverts slowly and reversibly with the proteinase-like form (mIIa). Because the proteinase-like conformation is required for further cleavage at Arg271, it is only mIIa rather than mIIa’ that is processed to thrombin. The reversible conversion of mIIa’ to mIIa there represents a previously unanticipated rate-limiting step in thrombin formation.
Controversies and Challenges
Mechanistic details regarding prothrombin activation have become controversial in recent years (52-56). This is surprising considering that the area mostly lay fallow for almost 20 years since the numerous studies in the 1980s that established the essential features of prothrombin cleavage by prothrombinase assembled using synthetic phospholipids (15,16,57,58). One area of controversy is reflected in the proposal that two interconverting isomers of prothrombinase are responsible for the two cleavages (53). This proposal is inconsistent with the cleavage site independence of substrate binding to prothrombinase, the ability of the individual cleavage site mutants including IIQQ to bind prothrombinase in a mutually exclusive manner and the stoichiometries for substrate binding (17,35). Controversy also derives from experimental and computational measurements proposing that the two cleavages arise from “channelling” of sequential enzyme catalysed reactions or is even concerted (i.e. one or more enzymes cleave(s) the substrate at two sites without releasing intermediate). Accumulation of intermediate(s) in excess of the concentration of prothrombinase questions whether the idea of concerted cleavage is reasonable. Thus, while such ideas are within the realm of possibility, the available evidence suggests that the contribution of such concerted cleavage is likely very small (17). Compelling documentation of sequential cleavage of the substrate without intermediate release, will require resolution with far better experimental testing than the simple analysis of progress curves of intermediates produced during the activation of prothrombin (56). Another perennial area of controversy relates to whether studies with synthetic membranes properly recapitulate enzymic function on platelets where the physiologically relevant reaction occurs (59). It is interesting that there is increasing uncertainty as to whether the platelet plays the implied preeminent role in supporting coagulation enzyme function in vivo (60). Nevertheless, there is no arguing that platelets or other cells present a fundamentally more complex surface for enzyme binding than synthetic membranes. Evidence from several groups indicates that prothrombin cleavage with prothrombinase assembled on platelets yields substantial levels of P2 with no observed mIIa (56,59). Although these and other functional differences between synthetic membranes versus platelets are implicitly attributed to contributions from receptors, repeated claims of a protein receptor that facilitates prothrombinase binding and/or function have failed to yield tangible fruit for 30+ years (56,61). Instead, mechanistic explanations for some of the observations may lie in the membrane composition of activated cells and hindered diffusion on the crowded cell surface that is sufficient for prothrombinase assembly but sub-optimal for mediating delivery and constrained presentation of substrate to enzyme. Accordingly, prothrombin derivatives exhibiting impaired membrane binding yield significant amounts of P2/F12 even on synthetic phospholipids (62). Thus, while such rhetoric may rationalise the behaviour of prothrombinase on cell surfaces, the next major challenge will be to employ appropriate tools to extend the sophisticated enzymological concepts developed in purified systems to the behaviour of the enzyme complex on biologically relevant membranes.
Perspectives
Mechanistic studies of the action of prothrombinase on prothrombin have revealed details by which enzymic specificity and regulation is achieved. Many of these concepts likely apply to varying extents to the other proteolytic reactions of coagulation. The strategy and consequences of exosite-dependent substrate recognition probably likely apply widely to the coagulation proteinases and lie at the heart of the ability of these structurally related enzymes to act with narrow and defined specificity on their respective protein substrates. Thus, the secrets to the determinants of substrate specificity in this enzyme family, long sought from detailed investigation of residues surrounding the active-site, are likely to only reveal themselves when exosite-dependent substrate recognition is correctly accounted for. In this regard, many of the coagulation enzymes exhibit kinetic features signifying exosite-dependent recognition of the protein substrate. These include the VIIa-TF (extrinsic Xase) complex (63,64), the IXa-VIIIa complex (intrinsic Xase) (65), factor XIa (66,67) and thrombin (26,68). Furthermore, considering the details of the two cleavages in prothrombin may seem to focus on minutiae of a particularly complex activation reaction. However, zymogen or coagulation factor activation by cleavage at multiple sites and in a seemingly ordered fashion is the rule rather than the exception in coagulation biochemistry. Seemingly ordered cleavage is evident in fibrin formation, the activation of factors IX and XI, the reactions that lead to the activation of factors V and VIII as well as the proteolytic inactivation of Va and VIIIa. Thus, insights drawn from studies with prothrombin are likely to shed new light on a wide swathe of proteolytic reactions related to the blood coagulation response. The surprising ability of thrombin to interconvert between zymogen- and proteinase-like forms in a ligand-dependent manner has the potential to have the broadest impact on the field. This assertion is based on the fact that the proteinases of coagulation are structurally homologous and are likely to behave in analogous ways. Similar points have been made with VIIa (43) and even in early studies with chymotrypsin, the archetypal enzyme of this clan (69). The final perspective of note lies in the fact that it is F12 rather than the fragment 2 region in contact with ABE2 that can distinguish between zymogen- and proteinase-like forms of thrombin. This implies that the otherwise distant fragment 1 region within F12 regulates the zymogen-like character of the proteinase domain bound to the propiece. If similar features apply to the other coagulation enzymes, it is worth noting that all coagulation proteinase structures determined thus far, lack the membrane-binding Gla domain. Advances in proteinase structural biology that overcome this limitation may shed new light on our current understanding of coagulation enzymology.
Acknowledgement
Supported by grants HL-074124 and HL-108933 (to S.K.) from the National Institutes of Health.
Footnotes
Disclosure of Conflicts of Interest - The authors state that they have no conflict of interest.
References
- 1.Mann KG, Jenny RJ, Krishnaswamy S. Cofactor Proteins in the Assembly and Expression of Blood Clotting Enzyme Complexes. Annu. Rev. Biochem. 1988;57:915–956. doi: 10.1146/annurev.bi.57.070188.004411. [DOI] [PubMed] [Google Scholar]
- 2.Mann KG, Nesheim ME, Church WR, Haley P, Krishnaswamy S. Surface-dependent reactions of the vitamin K-dependent enzyme complexes. Blood. 1990;76:1–16. [PubMed] [Google Scholar]
- 3.Schenone M, Furie BC, Furie B. The blood coagulation cascade. Curr. Opin. Hematol. 2004;11:272–277. doi: 10.1097/01.moh.0000130308.37353.d4. [DOI] [PubMed] [Google Scholar]
- 4.Nesheim ME, Tracy RP, Mann KG. “Clotspeed,” a mathematical simulation of the functional properties of prothrombinase. J. Biol. Chem. 1984;259:1447–1453. [PubMed] [Google Scholar]
- 5.Higgins DL, Callahan PJ, Prendergast FG, Nesheim ME, Mann KG. Lipid mobility in the assembly and expression of the activity of the prothrombinase complex. J. Biol. Chem. 1985;260:3604–3612. [PubMed] [Google Scholar]
- 6.Husten EJ, Esmon CT, Johnson AE. The active site of blood coagulation factor Xa. Its distance from the phospholipid surface and its conformational sensitivity to components of the prothrombinase complex. J. Biol. Chem. 1987;262:12953–12961. [PubMed] [Google Scholar]
- 7.Rosing J, Tans G, Govers Riemslag JW, Zwaal RF, Hemker HC. The role of phospholipids and factor Va in the prothrombinase complex. J. Biol. Chem. 1980;255:274–283. [PubMed] [Google Scholar]
- 8.van Dieijen G, Tans G, Rosing J, Hemker HC. The role of phospholipid and factor VIIIa in the activation of bovine factor X. J. Biol. Chem. 1981;256:3433–3442. [PubMed] [Google Scholar]
- 9.Nesheim ME, Eid S, Mann KG. Assembly of the prothrombinase complex in the absence of prothrombin. J. Biol. Chem. 1981;256:9874–9882. [PubMed] [Google Scholar]
- 10.Buddai SK, Toulokhonova L, Bergum PW, Vlasuk GP, Krishnaswamy S. Nematode Anticoagulant Protein c2 Reveals a Site on Factor Xa That Is Important for Macromolecular Substrate Binding to Human Prothrombinase. J. Biol. Chem. 2002;277:26689–26698. doi: 10.1074/jbc.M202507200. [DOI] [PubMed] [Google Scholar]
- 11.Walker RK, Krishnaswamy S. The Influence of Factor Va on the Active Site of Factor Xa. J. Biol. Chem. 1993;268:13920–13929. [PubMed] [Google Scholar]
- 12.Bianchini EP, Louvain VB, Marque PE, Juliano MA, Juliano L, Le Bonniec BF. Mapping of the catalytic groove preferences of factor Xa reveals an inadequate selectivity for its macromolecule substrates. J. Biol. Chem. 2002;277:20527–20534. doi: 10.1074/jbc.M201139200. [DOI] [PubMed] [Google Scholar]
- 13.Hsu HJ, Tsai KC, Sun YK, Chang HJ, Huang YJ, Yu HM, Lin CH, Mao SS, Yang AS. Factor Xa Active Site Substrate Specificity with Substrate Phage Display and Computational Molecular Modeling. J. Biol. Chem. 2008;283:12343–12353. doi: 10.1074/jbc.M708843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esmon CT, Jackson CM. The conversion of prothrombin to thrombin. III. The factor Xa-catalysed activation of prothrombin. J. Biol. Chem. 1974;249:7782–7790. [PubMed] [Google Scholar]
- 15.Krishnaswamy S, Church WR, Nesheim ME, Mann KG. Activation of human prothrombin by human prothrombinase. Influence of factor Va on the reaction mechanism. J. Biol. Chem. 1987;262:3291–3299. [PubMed] [Google Scholar]
- 16.Krishnaswamy S, Mann KG, Nesheim ME. The prothrombinase-catalysed activation of prothrombin proceeds through the intermediate meizothrombin in an ordered, sequential reaction. J. Biol. Chem. 1986;261:8977–8984. [PubMed] [Google Scholar]
- 17.Orcutt SJ, Krishnaswamy S. Binding of Substrate in Two Conformations to Human Prothrombinase Drives Consecutive Cleavage at Two Sites in Prothrombin. J. Biol. Chem. 2004;279:54927–54936. doi: 10.1074/jbc.M410866200. [DOI] [PubMed] [Google Scholar]
- 18.Esmon CT, Jackson CM. The conversion of prothrombin to thrombin. IV. The function of the fragment 2 region during activation in the presence of factor V. J. Biol. Chem. 1974;249:7791–7797. [PubMed] [Google Scholar]
- 19.Luckow EA, Lyons DA, Ridgeway TM, Esmon CT, Laue TM. Interaction of clotting factor V heavy chain with prothrombin and prethrombin 1 and Role of activated protein C in regulating this interaction: Analysis by analytical ultracentrifugation. Biochemistry. 1989;28:2348–2354. doi: 10.1021/bi00431a055. [DOI] [PubMed] [Google Scholar]
- 20.Krishnaswamy S, Betz A. Exosites determine macromolecular substrate recognition by prothrombinase. Biochemistry. 1997;36:12080–12086. doi: 10.1021/bi970979+. [DOI] [PubMed] [Google Scholar]
- 21.Betz A, Krishnaswamy S. Regions remote from the site of cleavage determine macromolecular substrate recognition by the prothrombinase complex. J. Biol. Chem. 1998;273:10709–10718. doi: 10.1074/jbc.273.17.10709. [DOI] [PubMed] [Google Scholar]
- 22.Orcutt SJ, Pietropaolo C, Krishnaswamy S. Extended Interactions with Prothrombinase Enforce Affinity and Specificity for its Macromolecular Substrate. J. Biol. Chem. 2002;277:46191–46196. doi: 10.1074/jbc.M208677200. [DOI] [PubMed] [Google Scholar]
- 23.Wilkens M, Krishnaswamy S. The contribution of factor Xa to exosite-dependent substrate recognition by prothrombinase. J Biol Chem. 2002;277:9366–9374. doi: 10.1074/jbc.M110848200. [DOI] [PubMed] [Google Scholar]
- 24.Anderson PJ, Nesset A, Dharmawardana KR, Bock PE. Role of Proexosite I in Factor Va-dependent Substrate Interactions of Prothrombin Activation. J. Biol. Chem. 2000;275:16435–16442. doi: 10.1074/jbc.M001255200. [DOI] [PubMed] [Google Scholar]
- 25.Anderson PJ, Bock PE. Role of prothrombin fragment 1 in the pathway of regulatory exosite I formation during conversion of human prothrombin to thrombin. J. Biol. Chem. 2003;278:44489–44495. doi: 10.1074/jbc.M306916200. [DOI] [PubMed] [Google Scholar]
- 26.Bock PE, Panizzi P, Verhamme IM. Exosites in the substrate specificity of blood coagulation reactions. J. Thromb Haemost. 2007;5(Suppl 1):81–94. doi: 10.1111/j.1538-7836.2007.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishnaswamy S. Exosite-driven substrate specificity and function in coagulation. J. Thromb. Haemost. 2005;3:54–67. doi: 10.1111/j.1538-7836.2004.01021.x. [DOI] [PubMed] [Google Scholar]
- 28.Boskovic DS, Krishnaswamy S. Exosite Binding Tethers the Macromolecular Substrate to the Prothrombinase Complex and Directs Cleavage at Two Spatially Distinct Sites. J. Biol. Chem. 2000;275:38561–38570. doi: 10.1074/jbc.M006637200. [DOI] [PubMed] [Google Scholar]
- 29.Boskovic DS, Troxler T, Krishnaswamy S. Active Site-Independent Recognition of Substrates and Product by Prothrombinase. A Fluorescence Resonance Energy Transfer Study. J. Biol. Chem. 2004;279:20786–20793. doi: 10.1074/jbc.M400469200. [DOI] [PubMed] [Google Scholar]
- 30.Vijayalakshmi J, Padmanabhan KP, Mann KG, Tulinsky A. The isomorphous structures of prethrombin2, hirugen-, and PPACK- thrombin: changes accompanying activation and exosite binding to thrombin. Protein Sci. 1994;3:2254–2271. doi: 10.1002/pro.5560031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin PD, Malkowski MG, Box J, Esmon CT, Edwards BF. New insights into the regulation of the blood clotting cascade derived from the X-ray crystal structure of bovine meizothrombin des F1 in complex with PPACK. Structure. 1997;5:1681–1693. doi: 10.1016/s0969-2126(97)00314-6. [DOI] [PubMed] [Google Scholar]
- 32.Khan AR, James MNG. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 1998;7:815–836. doi: 10.1002/pro.5560070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bianchini EP, Orcutt SJ, Panizzi P, Bock PE, Krishnaswamy S. Ratcheting of the substrate from the zymogen to proteinase conformations directs the sequential cleavage of prothrombin by prothrombinase. Proc. Natl. Acad. Sci U. S. A. 2005;102:10099–10104. doi: 10.1073/pnas.0504704102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003;425:535–539. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- 35.Hacisalihoglu A, Panizzi P, Bock PE, Camire RM, Krishnaswamy S. Restricted Active Site Docking by Enzyme-bound Substrate Enforces the Ordered Cleavage of Prothrombin by Prothrombinase. J. Biol. Chem. 2007;282:32974–32982. doi: 10.1074/jbc.M706529200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradford HN, Micucci JA, Krishnaswamy S. Regulated cleavage of prothrombin by prothrombinase: repositioning a cleavage site reveals the unique kinetic behavior of the action of prothrombinase on its compound substrate. J Biol. Chem. 2010;285:328–338. doi: 10.1074/jbc.M109.070334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myrmel KH, Lundblad RL, Mann KG. Characteristics of the association between prothrombin fragment 2 and alpha-thrombin. Biochemistry. 1976;15:1767–1773. doi: 10.1021/bi00653a027. [DOI] [PubMed] [Google Scholar]
- 38.Nesheim ME, Abbott T, Jenny R, Mann KG. Evidence that the thrombin-catalysed feedback cleavage of fragment 1.2 at Arg154-Ser155 promotes the release of thrombin from the catalytic surface during the activation of bovine prothrombin. J. Biol. Chem. 1988;263:1037–1044. [PubMed] [Google Scholar]
- 39.Anderson PJ, Nesset A, Bock PE. Effects of activation peptide bond cleavage and fragment 2 interactions on the pathway of exosite I expression during activation of human prethrombin 1 to thrombin. J. Biol. Chem. 2003;278:44482–44488. doi: 10.1074/jbc.M306917200. [DOI] [PubMed] [Google Scholar]
- 40.Kamath P, Krishnaswamy S. Fate of membrane-bound reactants and products during the activation of human prothrombin by prothrombinase. J. Biol Chem. 2008;283:30164–30173. doi: 10.1074/jbc.M806158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamath P, Huntington JA, Krishnaswamy S. Ligand binding shuttles thrombin along a continuum of zymogen- and proteinase-like states. J. Biol Chem. 2010;285:28651–28658. doi: 10.1074/jbc.M110.154914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradford HN, Krishnaswamy S. Meizothrombin Is an Unexpectedly Zymogen-like Variant of Thrombin. J. Biol. Chem. 2012;287:30414–30425. doi: 10.1074/jbc.M112.394809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higashi S, Matsumoto N, Iwanaga S. Molecular mechanism of tissue factor-mediated acceleration of factor VIIa activity. J. Biol. Chem. 1996;271:26569–26574. doi: 10.1074/jbc.271.43.26569. [DOI] [PubMed] [Google Scholar]
- 44.Bah A, Carrell CJ, Chen Z, Gandhi PS, Di Cera E. Stabilization of the E* form turns thrombin into an anticoagulant. J. Biol Chem. 2009;284:20034–20040. doi: 10.1074/jbc.M109.012344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gandhi PS, Page MJ, Chen Z, Bush-Pelc L, Di Cera E. Mechanism of the anticoagulant activity of thrombin mutant W215A/E217A. J. Biol Chem. 2009;284:24098–24105. doi: 10.1074/jbc.M109.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bush-Pelc LA, Marino F, Chen Z, Pineda AO, Mathews FS, Di Cera E. Important role of the cys-191 cys-220 disulfide bond in thrombin function and allostery. J. Biol Chem. 2007;282:27165–27170. doi: 10.1074/jbc.M703202200. [DOI] [PubMed] [Google Scholar]
- 47.Di Cera E. Thrombin. Mol. Aspects Med. 2008;29:203–254. doi: 10.1016/j.mam.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lechtenberg BC, Johnson DJ, Freund SM, Huntington JA. NMR resonance assignments of thrombin reveal the conformational and dynamic effects of ligation. Proc. Natl. Acad. Sci U. S. A. 2010;107:14087–14092. doi: 10.1073/pnas.1005255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Page MJ, Di Cera E. Role of Na+ and K+ in enzyme function. Physiol Rev. 2006;86:1049–1092. doi: 10.1152/physrev.00008.2006. [DOI] [PubMed] [Google Scholar]
- 50.Di Cera E. Serine proteases. IUBMB. Life. 2009;61:510–515. doi: 10.1002/iub.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haynes LM, Dubief YC, Orfeo T, Mann KG. Dilutional Control of Prothrombin Activation at Physiologically Relevant Shear Rates. Biophys. J. 2011;100:765–773. doi: 10.1016/j.bpj.2010.12.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim PY, Nesheim ME. Further evidence for two functional forms of prothrombinase each specific for either of the two prothrombin activation cleavages. J. Biol. Chem. 2007;282:32568–32581. doi: 10.1074/jbc.M701781200. [DOI] [PubMed] [Google Scholar]
- 53.Brufatto N, Nesheim ME. Analysis of the kinetics of prothrombin activation and evidence that two equilibrating forms of prothrombinase are involved in the process. J. Biol. Chem. 2003;278:6755–6764. doi: 10.1074/jbc.M206413200. [DOI] [PubMed] [Google Scholar]
- 54.Lee C J, Wu S, Pedersen LG. A revisit to the one form kinetic model of prothrombinase. Biophys. Chem. 2010;149:28–33. doi: 10.1016/j.bpc.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim PY, Nesheim ME. A revisit of the two-form kinetic model of prothrombinase: A rebuttal. Biophys. Chem. 2012;160:75–76. doi: 10.1016/j.bpc.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 56.Haynes LM, Bouchard BA, Tracy PB, Mann KG. Prothrombin activation by platelet-associated prothrombinase proceeds through the prethrombin-2 pathway via a concerted mechanism. J. Biol. Chem. 2012;287:38647–38655. doi: 10.1074/jbc.M112.407791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tans G, Janssen-Claessen T, Hemker HC, Zwaal RFA, Rosing J. Meizothrombin Formation during Factor Xa-catalysed Prothrombin Activation. Formation in a purified system and in plasma. J. Biol. Chem. 1991;266:21864–21873. [PubMed] [Google Scholar]
- 58.Walker RK, Krishnaswamy S. The Activation of Prothrombin by the Prothrombinase Complex. The contribution of the substrate-membrane interaction to catalysis. J. Biol. Chem. 1994;269:27441–27450. [PubMed] [Google Scholar]
- 59.Wood JP, Silveira JR, Maille NM, Haynes LM, Tracy PB. Prothrombin activation on the activated platelet surface optimizes expression of procoagulant activity. Blood. 2011;117:1710–1718. doi: 10.1182/blood-2010-09-311035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vandendries ER, Hamilton JR, Coughlin SR, Furie B, Furie BC. Par4 is required for platelet thrombus propagation but not fibrin generation in a mouse model of thrombosis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:288–292. doi: 10.1073/pnas.0610188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouchard BA, Silveira JR, Tracy PB. On the role of EPR-1 or an EPR-1-like molecule in regulating factor Xa incorporation into platelet prothrombinase. Thromb Haemost. 2001;85:509–513. [PubMed] [Google Scholar]
- 62.Malhotra OP, Nesheim ME, Mann KG. The kinetics of activation of normal and gamma-carboxyglutamic acid-deficient prothrombins. J. Biol. Chem. 1985;260:279–287. [PubMed] [Google Scholar]
- 63.Baugh RJ, Dickinson CD, Ruf W, Krishnaswamy S. Exosite Interactions Determine the Affinity of Factor X for the Extrinsic Xase Complex. J. Biol. Chem. 2000;275:28826–28833. doi: 10.1074/jbc.M005266200. [DOI] [PubMed] [Google Scholar]
- 64.Dickinson CD, Shobe J, Ruf W. Influence of cofactor binding and active site occupancy on the conformation of the macromolecular substrate exosite of factor VIIa. J. Mol. Biol. 1998;277:959–971. doi: 10.1006/jmbi.1998.1639. [DOI] [PubMed] [Google Scholar]
- 65.Duffy EJ, Parker ET, Mutucumarana VP, Johnson AE, Lollar P. Binding of factor VIIIa and factor VIII to factor IXa on phospholipid vesicles. J. Biol. Chem. 1992;267:17006–17011. [PubMed] [Google Scholar]
- 66.Sun Y, Gailani D. Identification of a factor IX binding site on the third apple domain of activated factor XI. J Biol Chem. 1996;271:29023–29028. doi: 10.1074/jbc.271.46.29023. [DOI] [PubMed] [Google Scholar]
- 67.Gailani D. Activation of factor IX by factor XIa. Trends Cardiovasc. Med. 2000;10:198–204. doi: 10.1016/s1050-1738(00)00070-0. [DOI] [PubMed] [Google Scholar]
- 68.Stubbs MT, Bode W. The clot thickens: clues provided by thrombin structure. Trends Biochem. Sci. 1995;20:23–28. doi: 10.1016/s0968-0004(00)88945-8. [DOI] [PubMed] [Google Scholar]
- 69.Fersht A, Requena Y. Equilibrium and rate constants for the interconversion of two conformations of greek small letter alpha-chymotrypsin The existence of a catalytically inactive conformation at neutral pH. J. Mol. Biol. 1971;60:279–290. doi: 10.1016/0022-2836(71)90294-4. [DOI] [PubMed] [Google Scholar]
- 70.Gandhi PS, Chen Z, Di Cera E. Crystal structure of thrombin bound to the uncleaved extracellular fragment of PAR1. J. Biol Chem. 2010;285:15393–15398. doi: 10.1074/jbc.M110.115337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pechik I, Madrazo J, Mosesson MW, Hernandez I, Gilliland GL, Medved L. Crystal structure of the complex between thrombin and the central “E” region of fibrin. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2718–2723. doi: 10.1073/pnas.0303440101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carter WJ, Cama E, Huntington JA. Crystal structure of thrombin bound to heparin. J. Biol Chem. 2005;280:2745–2749. doi: 10.1074/jbc.M411606200. [DOI] [PubMed] [Google Scholar]
- 73.Dumas JJ, Kumar R, Seehra J, Somers WS, Mosyak L. Crystal structure of the GpIbalpha-thrombin complex essential for platelet aggregation. Science. 2003;301:222–226. doi: 10.1126/science.1083917. [DOI] [PubMed] [Google Scholar]
- 74.Celikel R, McClintock RA, Roberts JR, Mendolicchio GL, Ware J, Varughese KI, Ruggeri ZM. Modulation of alpha-thrombin function by distinct interactions with platelet glycoprotein Ibalpha. Science. 2003;301:218–221. doi: 10.1126/science.1084183. [DOI] [PubMed] [Google Scholar]