SUMMARY
The DNA damage response (DDR) protein 53BP1 protects DNA ends from excessive resection in G1, and thereby favors repair by non-homologous end joining (NHEJ) as opposed to homologous recombination (HR). During S phase, BRCA1 antagonizes 53BP1 to promote HR. The pro-NHEJ and anti-recombinase functions of 53BP1 are mediated in part by RIF1, the only known factor that requires 53BP1 phosphorylation for its recruitment to double strand breaks (DSBs). Here we show that a 53BP1 phospho-mutant 53BP18A, comprising alanine substitutions of the 8 most N-terminal S/TQ phosphorylation sites, mimics 53BP1 deficiency by restoring genome stability in BRCA1 deficient cells yet behaves like wild-type 53BP1 with respect to immunoglobulin class switch recombination (CSR). 53BP18A recruits RIF1 but fails to recruit the DDR protein PTIP to DSBs, and disruption of PTIP phenocopies 53BP18A. We conclude that 53BP1 promotes productive CSR and suppresses mutagenic DNA repair through distinct phospho-dependent interactions with RIF1 and PTIP.
INTRODUCTION
CSR is initiated by activation induced cytidine deaminase (AID), which generates multiple DSBs at highly repetitive immunoglobulin (Ig) switch regions. Paired distal DSBs are then rejoined by NHEJ thereby replacing Igμ by a downstream constant region (Igγ, Igε or Igα). Alternatively, if DSBs persist, a homology-driven pathway that involves resection of repetitive switch regions can repair DSBs locally. Such abortive “intra-switch” recombination events are increased at the expense of CSR in the absence of 53BP1(Reina-San-Martin et al., 2007), a key suppressor of end resection (Bothmer et al., 2010; Bouwman et al., 2010; Bunting et al., 2010; Cao et al., 2009; Difilippantonio et al., 2008).
In addition to its productive effect on CSR, 53BP1 blocks DNA ends from resection in BRCA1-deficient cells, leading to toxic radial chromosomes that arise from NHEJ (Bouwman et al., 2010; Bunting et al., 2012; Bunting et al., 2010; Cao et al., 2009). Deletion of 53BP1 leads to deposition of HR factors RPA and RAD51 on single strand DNA, which, in the case of recombining switch regions, promotes intra-switch recombination (Yamane et al., 2013), and in the setting of BRCA1-deficiency, restores HR (Bouwman et al., 2010; Bunting et al., 2010; Cao et al., 2009). Thus, DNA end protection by 53BP1 is critical for CSR in G1 but can unleash genome instability in S phase.
In addition to DNA end-blocking activities that disfavor HR and thereby promote NHEJ, 53BP1 has been suggested to directly mediate long-range chromosomal interactions and DSB mobility that facilitates the juxtaposition of distal DNA ends. These activities are believed to be responsible for 53BP1’s ability to support recombination of DSB ends that are far apart during V(D)J recombination and class switch recombination (Callen et al., 2007b; Difilippantonio et al., 2008), and to fuse uncapped telomeric DNA ends (Dimitrova et al., 2008). Both pro-NHEJ and anti-HR functions require the direct physical association of 53BP1 with DNA ends, but also necessitate the DSB induced phosphorylation of its N-terminal ATM/ATR kinase sites (Bothmer et al., 2011; Ward et al., 2006).
The DDR protein RIF1 was recently identified as an essential factor recruited by phosphorylated 53BP1 to promote NHEJ and block HR (Chapman et al., 2013; Di Virgilio et al., 2013; Escribano-Diaz et al., 2013; Feng et al., 2013; Zimmermann et al., 2013). Like 53BP1, RIF1 is required for CSR (Chapman et al., 2013; Di Virgilio et al., 2013; Escribano-Diaz et al., 2013). While the NHEJ of dysfunctional telomeres is abrogated in cells lacking 53BP1 or in cells expressing 53BP128A(Lottersberger et al., 2013), an allele harboring alanine substitutions at all 28 N-terminal ATM/ATR kinase phosphorylation targets sites, loss of RIF1 has considerably milder defect (Zimmermann et al., 2013). Moreover, while the generation of toxic radial chromosomes in BRCA1-deficient cells is prevented in 53BP1−/− or in 53BP128A-mutant cells (Bothmer et al., 2011; Bouwman et al., 2010; Bunting et al., 2012; Bunting et al., 2010), the loss of RIF1 only partially rescues HR in BRCA1- deficient cells (Escribano-Diaz et al., 2013; Feng et al., 2013; Zimmermann et al., 2013). This suggests that additional phosphorylation-dependent but RIF1-independent activities of 53BP1 might regulate the balance between HR and NHEJ.
PTIP is a ubiquitously expressed nuclear protein that associates constitutively with two of the known histone methyltransferases that catalyze tri-methylation of histone H3 at lysine 4 (H3K4me3), MLL3 and MLL4 (Cho et al., 2007; Patel et al., 2007). In addition to its well-established role in transcription initiation, a separate pool of PTIP functions in an unknown capacity in the DDR (Gong et al., 2009). Indeed, PTIP has been implicated in both HR (Wang et al., 2010) and NHEJ (Callen et al., 2012). PTIP is recruited to DSBs by its tandem BRCT (BRCA1 carboxyl-terminal) domains (Manke et al., 2003; Yu et al., 2003), which associate with the serine 25 phosphorylation site within the N-terminus of 53BP1 (Munoz et al., 2007). In contrast to RIF1, PTIP recruitment to DSBs was reported to be 53BP1 and ATM independent (Gong et al., 2009; Jowsey et al., 2004; Munoz et al., 2007). Thus, the mechanism by which PTIP is recruited to DSBs, its role in DSB repair, and physiological significance of PTIP interaction with 53BP1 remain unclear. Here we show that PTIP is required for 53BP1-mediated inhibition of HR in BRCA1-deficient cells, but is dispensable for 53BP1-initiated DSB repair during productive CSR. Thus, RIF1 and PTIP separate 53BP1 functions in productive and pathologic DSB repair.
RESULTS
A separation of function mutation in 53BP1
To determine whether 53BP1’s activities in NHEJ and HR are distinct, we compared 53BP18A, which disrupts phosphorylation of the 8 N-terminal ATM/ATR target sites (Figure S2A), to the 53BP1DB allele, which is indistinguishable from WT 53BP1 in all functional aspects (Bothmer et al., 2011). To assay for CSR, BRCA1/53BP1-deficient B cells were transduced with wild-type and 53BP1 mutant proteins by retroviral infection after activation with lipopolysaccharide (LPS) and interleukin-4 (IL4). As expected, 53BP1DB fully complemented the CSR defects (Figure 1A), and produced high levels of genome instability in PARPi treated BRCA1/53BP1-deficient cells (Figure 1B) (Bothmer et al., 2011). Surprisingly, despite rescuing CSR, the 53BP18A allele failed to promote genome instability in PARPi treated-BRCA1/53BP1-deficient cells above the levels observed in controls (Figure 1B). This effect was not due to differences in the expression levels of 53BP1 (Figure 1C) or in the recruitment of 53BP1 and RIF1 to DSBs (Figure 1D). Similar to B cells, BRCA1/53BP1 deficient MEFs complemented with 53BP1DB were hypersensitive to PARPi, whereas 53BP18A transduced MEFs were not (Figure S1). Thus, the mechanism by which 53BP1 promotes CSR and blocks HR in BRCA1 deficient cells is distinct. Moreover, the recruitment of RIF1 is insufficient to induce genome instability in PARPi-treated BRCA1-deficient cells.
Figure 1. Characterization of a separation of function mutant 53BP1.
(A) Top: Representative flow cytometry plots measuring CSR after stimulation of WT and BRCA1−/−53BP1−/− B cells infected with retroviruses expressing 53BP1DB (amino acids 1-1710), the N-terminal mutant 53BP18A or empty vector (EV). Numbers represent the percentages of IgG1 switched cells. B220 is a B cell marker. Bottom: Dot plot indicating IgG1 CSR as a percentage of WT value in the same experiment. Three independent experiments are shown. **: p<0.001 (two-tailed unpaired t-test); BRCA1/53BP1+DB vs. BRCA1/53BP1+8A, p>0.1 which is not significant (ns). (B) BRCA1−/−53BP1−/− B cells were reconstituted with empty vector, 53BP1DB and 53BP18A retroviruses and treated with PARPi. The arrows indicate representative images of aberrant chromosomes. Dot plot indicates the total aberrations per cell in three independent experiments. At least 50 metaphases were analyzed for each genotype in each experiment. **: p<0.01 (two-tailed unpaired t-test);ns: not significant. (C) Western blot analysis of 53BP1 expression in WT B cells and BRCA1−/−53BP1−/− B cells stimulated and infected with empty vector, 53BP1DB or 53BP18A. (D) BRCA1−/−53BP1−/− B cells infected with EV, 53BP1DB or 53BP18A retroviruses were assayed for IRIF (10 Gy, 2 hour recovery) for RIF1 (red, top panel) and 53BP1 (red, lower panel). Cells were counterstained with DAPI (blue). Scale bar represents 10 μm. See also Figure S1.
Role of PTIP in the DNA damage response
Upon DNA damage, PTIP binds to the serine 25 residue within the N-terminus of 53BP1(Munoz et al., 2007), which is located within the 8 N-terminal sites mutated in 53BP18A. Consistent with this, immunoprecipitation analysis revealed that PTIP association with 53BP1 after irradiation was abrogated in cells expressing S25A-harboring mutants 53BP18A, 53BP115A or 53BP128A (Figures 2A and Figure S2). In contrast, the damage-induced 53BP1/PTIP interaction was maintained in the 53BP17A-mutant, comprising alanine substitutions of 7 S/TQ phosphorylation sites C-terminus of those mutated in 53BP18A (Figure S2B).
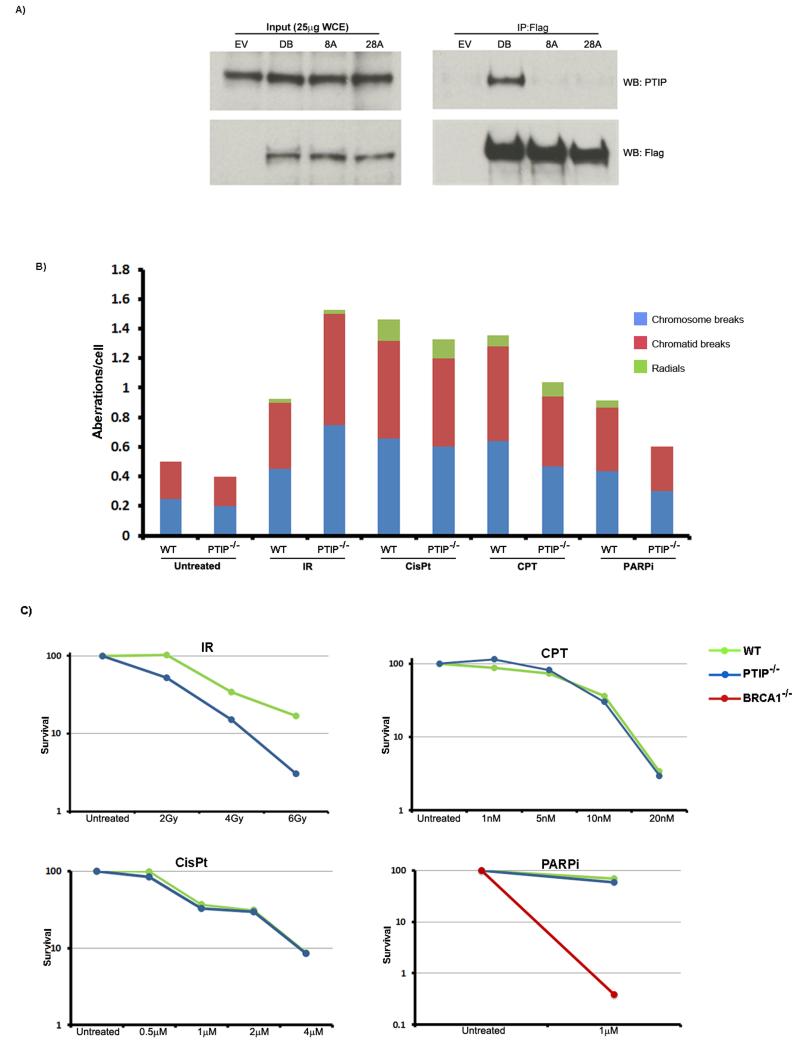
Figure 2. Response of PTIP to different DNA damaging agents.
(A) 53BP1−/− B cells were reconstituted with empty vector, 53BP1DB, 53BP18A, or 53BP128A retroviruses that were FLAG-tagged. Cells were irradiated (10 Gy, 45 minute recovery) and immunoprecipitation was performed with anti-FLAG antibodies. Western blot analysis of PTIP and FLAG are shown for input (left panel) and immunoprecipitated protein (right panel). (B) Isogenic immortalized WT and PTIP−/− MEFs were either untreated or treated with irradiation (IR, 2 Gy), cisplatin (CisPt, 0.5 μM), camptothecin (CPT, 10nM) or PARP inhibitor (PARPi, 1μM) and chromosomal aberrations (chromatid breaks, chromosome breaks and radials) were quantified in at least 50 metaphase spreads for each genotype and each treatment. Data from an independent experiment is shown in Figure S3. (C) WT (green lines) and PTIP−/− (blue line) MEFs were treated with different doses of the above drugs and colony formation was quantified relative to colonies formed in untreated cells from the same genotype. An experiment performed in parallel demonstrated that 1 μM PARPi treatment is toxic for BRCA1-mutant MEFs (red line). See also Figures S2-S5.
To explore the function of PTIP in the DDR, we asked whether PTIP deficient cells are sensitive to DNA damaging agents that are predominantly repaired by HR (Sonoda et al., 2006). WT and PTIP−/− MEFs were exposed to either cisplatin, camptothecin or PARPi, all of which sensitize HR deficient cells (Bryant et al., 2005; Farmer et al., 2005). Each of these agents induced a similar level of chromosomal aberrations and reduction in cell survival in WT and PTIP−/− MEFs (Figures 2B and 2C and Figure S3). In contrast, PTIP−/− MEFs were sensitive to irradiation (IR) (Figures 2B and 2C and Figure S3) (Gong et al., 2009; Jowsey et al., 2004; Munoz et al., 2007). Moreover, 53BP18A MEFs exhibited increased genome instability and reduced cell survival following IR (Figure S4). To examine the recruitment of HR proteins to DSBs, we evaluated BRCA1, RAD51 and γ-H2AX foci formation after IR in WT and PTIP−/− MEFs. All of these factors were normally recruited to DSBs in PTIP-deficient cells (Figure S5). Moreover 53BP1 also formed robust foci in the absence of PTIP (Figure S5). Thus, PTIP−/− MEFs are tolerant to agents that are highly toxic to HR deficient cells and the recruitment of several factors implicated in DSB repair is intact in the absence of PTIP. Nevertheless, both PTIP−/− and 53BP18A MEFs are sensitive to IR.
PTIP is dispensable for NHEJ during CSR but is required for NHEJ of dysfunctional telomeres
To explore the role of PTIP in NHEJ, we first assayed CSR. Deletion of PTIP in B cells leads to a defect in class switching to IgG3, IgG2b and IgG1 (Daniel et al., 2010; Schwab et al., 2011). By recruiting an MLL-like methyltransferase complex to the switch regions of these isotypes, PTIP promotes histone modifications and transcription initiation of IgG3/IgG2b/IgG1 germ-line switch regions, which is necessary for AID targeting (Daniel et al., 2010; Schwab et al., 2011). However, PTIP does not affect transcription at Igμ and Igε (Daniel et al., 2010), indicating that PTIP-associated methyltransferase complex promotes the accessibility of some but not all switch loci. To distinguish between PTIP’s effects on transcription vs. DSB repair we compared CSR to IgG1 and IgE on day 5 after stimulation with αCD40+IL4 as described (Wesemann et al., 2011). As expected PTIPf/fCD19CRE (PTIP−/−) B cells displayed a defect in switching to IgG1 (Figures 3A and 3B), which is consistent with decreased Igγ1 germ-line transcription (Daniel et al., 2010; Schwab et al., 2011). However there was no defect in IgE germline transcription (Daniel et al., 2010) or IgE CSR in PTIP-deficient cells (Figures 3A and 3B). Indeed, IgE CSR was consistently higher in the absence of PTIP, likely because Sγ1 is no longer a target for AID. In contrast to PTIP−/−, ablation of RIF1 in Rif1f/f CD19CRE (RIF1−/−) B cells impaired CSR to both IgG1 and IgE (Figures 3A and 3B). We conclude that loss of PTIP phenocopies the 53BP18A-mutant allele, in that neither has a significant impact on NHEJ during CSR.
Figure 3. PTIP is dispensable for CSR to IgE.
PTIPf/fCD19CRE (PTIP−/−), (Rif1f/fCD19CRE) RIF1−/− and littermate WT B cells were stimulated with αCD40 plus IL-4 and analyzed for IgG1 and IgE CSR on day 5. (A) Representative flow cytometry plots. The percentages of IgG1 switched cells (upper left quadrant) and IgE switched cells (lower right quadrant) is indicated. (B) Dot plot indicates IgG1 and IgE CSR in PTIP−/− and RIF1−/− as a percentage of the WT value in the same experiment. **:p<0.01 (two-tailed unpaired t-test);ns: not significant.
An alternative end-joining pathway can catalyze substantial CSR end joining to IgG1 and IgE even in the absence of classical NHEJ (Boboila et al., 2010). Loss of PTIP leads to IR sensitivity but tolerance to agents that are repaired by HR. We therefore speculated that PTIP might function in other reactions besides CSR that might rely on classical NHEJ, such as the fusion of dysfunctional telomeres. When the shelterin factor TRF2 is removed, deprotected telomeres trigger ATM-dependent phosphorylation of 53BP1, and the ends are processed by NHEJ to generate chromosome fusions (Celli et al., 2006; Rai et al., 2010; Zimmermann et al., 2013). Since ATM dependent phosphorylation of 53BP1 is also required for interaction between 53BP1 and PTIP (Figures 2A and Figure S2B)(Jowsey et al., 2004; Manke et al., 2003), we asked whether PTIP promotes NHEJ-mediated fusion of deprotected telomeres. To address this, we uncapped telomeres in SV40 immortalized WT and PTIP−/− MEFs by removing TRF2 with short hairpin RNA against TRF2 (Rai et al., 2010). Upon TRF2 depletion we observed a similar level of phosphorylation of the ATM target KAP-1 in WT and PTIP−/− MEFs, as measured by quantitative flow cytometry (Figure 4A). Consistent with this, there was an accumulation of cytologically discernable telomere-induced DNA damage foci (TIFs) containing γ-H2AX in WT and PTIP−/− cells (Figure 4B). Despite a robust DNA damage response and activation of ATM, the frequency of end-end chromosomal fusions was reduced by 2.8-fold in PTIP−/− MEFs relative to WT (Figures 4C and 4D). Whereas 42% of WT cells bearing fusions had more than 30% of their chromosome ends fused, only 13% of PTIP KO cells had greater than 30% of their ends fused (Figure 4E). Thus, PTIP deficiency results in a reduction in the number of long-chain telomere fusions when telomeres are de-protected. We conclude that PTIP contributes to the NHEJ of dysfunctional telomeres.
Figure 4. PTIP is required for NHEJ of dysfunctional telomeres.
(A) WT and PTIP−/− MEFs were infected with a retrovirus expressing either an empty vector or shRNA against TRF2 (shTRF2), and phosphorylated KAP1 (pKAP1) levels were measured by flow cytometry. (B) γ-H2AX (green) in telomere-dysfunction induced foci (TIF) generated in shTRF2-infected WT cells. PNA probe is shown in red, and images are merged on top of DAPI (blue). Scale bar represents 10 μm. (C) Representative images of a metaphase spread from WT and PTIP−/− MEFs infected with shTRF2. Telomere fusions are visualized by a telomeric PNA probe (red) and DAPI (blue). Arrows point to representative telomeric fusions. (D) Quantitation of telomeric fusion frequencies. At least 1800 chromosomes from each genotype were analyzed. Mean value derived from 3 independent experiments. **: p<0.01 (two-tailed unpaired t-test). (E) Distribution of telomeric fusions per metaphase in WT and PTIP−/− MEFs. At least 30 cells were examined in each of 3 independent experiments. p(chi-squared)<1×10−5.
PTIP promotes genome instability in BRCA1 deficient cells
In contrast to 53BP1, loss of RIF1 only partially reverses the chromosomal aberrations and hypersensitivity produced by PARPi treatment of BRCA1-deficient cells (Escribano-Diaz et al., 2013; Zimmermann et al., 2013). To determine whether PTIP could overcome the HR defects in BRCA1-deficient cells, we crossed PTIPf/f and BRCA1f(Δ11)/f(Δ11) mice with CD19 CRE transgenic mice to simultaneously delete PTIP and exon 11 of BRCA1 in primary B lymphocytes. When unchallenged, BRCA1+/+PTIP+/+ CD19CRE (WT), BRCA1f(Δ11)/f(Δ11) CD19CRE (BRCA1−/−), PTIPf/f CD19CRE (PTIP−/−) and BRCA1f(Δ11)/f(Δ11)PTIPf/f CD19CRE (BRCA1−/−PTIP−/−) doubly deficient B cells divided normally as determined by CFSE dye dilution (Figure 5A) and cell cycle distribution (Figure S6). Treatment with PARPi did not impair the proliferation of WT or PTIP−/− B cells (see also Figures 2B and 2C); however, BRCA1−/− cells underwent fewer divisions over the course of 72 hours (Figure 5A). In contrast, loss of PTIP completely reversed the BRCA1−/− growth defect (Figure 5A). Strikingly, while PARPi treatment generated chromatid breaks, chromosome breaks and radial chromosomes in BRCA1-deficient cells (Bunting et al., 2010), BRCA1−/−PTIP−/− B cells were insensitive to PARPi (Figure 5B). Thus, ablation of PTIP phenocopies both the 53BP18A mutation (Figure 1B) and 53BP1-deficiency (Bouwman et al., 2010; Bunting et al., 2010; Cao et al., 2009) in that it promotes genome stability and survival in BRCA1-mutant cells.
Figure 5. Ablation of PTIP rescues homologous recombination in BRCA1 deficient cells.
(A) WT, BRCA1−/−, PTIP−/− and BRCA1−/−PTIP−/− B cells were pulsed with CFSE and stimulated with (red) or without (green) PARPi. CFSE signal diminishes with increasing division. BRCA1−/− cells are sensitive to PARPi (arrow indicates sluggish cells) but loss of PTIP in BRCA1-deficient cells rescues the proliferation defect. (B) Analysis of genomic instability (radial chromosomes, chromatid breaks and chromosome breaks) in metaphases from B cells treated with 1 μM PARPi. At least 50 metaphases were analyzed for each genotype. (C) B cells were stimulated for 2 days, irradiated with 10 Gy and the percentage of cells with immunofluorescent RAD51 foci were quantified (at least 400 cells counted for each genotype). Data in (B) and (C) represent mean of three experiments +/− standard deviations. **: p<0.05 (two-tailed unpaired t-test), ns: not significant. (D) High-throughput microscopy quantification of RPA foci per cell in WT and PTIP−/− MEFs that were either untreated or treated with 30 Gy IR. Top: representative image of chromatin bound RPA in irradiated WT and PTIP−/− cells. Bottom: quantitation of RPA foci. Bar indicates the mean number of RPA foci per cell, and the blue box designates cells with more than 15 foci, whose percentage is indicated above each box. **: p<0.001 (E) BRCA1−/−PTIP−/− B cells were reconstituted with PTIPWT or PTIPW663R retroviruses (expressing a GFP marker driven by an internal ribosome entry site) and treated with PARPi. Cells were sorted (GFPpositive =infected and GFPnegative =uninfected) and metaphases were analyzed for radial chromosomes (n=50 metaphases analyzed in each case). See also Figures S6-S9.
Loss of PTIP increases HR in BRCA1 mutant cells by promoting DSB resection
BRCA1 and RAD51 function in a common HR pathway that promotes RAD51-mediated DNA strand exchange (Bhattacharyya et al., 2000; Moynahan et al., 1999; Scully et al., 1997). Loss of 53BP1 rescues RAD51 foci formation and HR in BRCA1-deficient cells (Bouwman et al., 2010; Bunting et al., 2010). To explore whether PTIP deficiency also promotes HR in BRCA1-deficient cells, we irradiated WT, BRCA1−/−, PTIP−/−, and BRCA1−/−PTIP−/−B cells and measured the frequency of immunofluorescent RAD51 foci. All mutant cells proliferated similarly to WT over the course of three days (Figure 5A), and as expected, RAD51 foci were reduced in IR-treated BRCA1−/− cells (Figure 5C). However in PTIP−/− cells, the frequency of RAD51 foci was greater than WT, and RAD51 foci were normalized to WT levels in BRCA1−/−PTIP−/ −B cells (Figure 5C). These results suggest that loss of PTIP reverses the HR defect in BRCA1 deficient cells, thereby explaining the insensitivity of BRCA1−/−PTIP−/ −B cells to PARPi.
Similar to 53BP1, loss of PTIP might promote RAD51 foci formation by allowing increased resection of DSBs (Bunting et al., 2010; Difilippantonio et al., 2008). Since 5′→3′ DSB end resection produces RPA-coated single strand DNA, we monitored RPA foci formation by high content microscopy. Irradiated PTIP−/− cells exhibited a significant increase in the mean number of RPA foci per cell relative to WT (Figure 5D); moreover, the fraction of PTIP−/− cells that had more than 15 RPA foci following IR was approximately 2-fold greater than WT (Figure 5D). Thus, PTIP limits the amount of chromatin bound RPA at IR-induced DSBs.
PTIP recruitment to DSBs promotes radial chromosomes in BRCA1 deficient cells
In the absence of DNA damage, PTIP is a subunit of the MLL3/4 methyltransferase complex and promotes histone H3 lysine 4 trimethylation and transcription initiation at specific promoters, such as the Sγ3/Sγ1 switch regions of the Igh locus (Daniel et al., 2010)(Figure S7A). To determine whether transcription of DDR genes is altered by PTIP ablation we profiled the transcriptome of WT and PTIP−/− B cells. Overall, there were 471 RefSeq annotated genes that were deregulated by more than 5-fold in PTIP−/− vs. WT (Figure S7B). However, HR and NHEJ DNA damage response genes were not among deregulated pathways (Figures S7B and S7C). This suggests that the functions of PTIP in suppressing HR might be unrelated to its role in transcriptional regulation.
To determine whether PTIP recruitment to DSBs is essential for its effects on HR, we made use of a point mutation in the BRCT domain 3 (W663R) of PTIP that selectively blocks its interaction with 53BP1 (Gong et al., 2009; Munoz et al., 2007) and is unable to form foci (Figure S8)(Daniel et al., 2010), but retains PTIP association with the MLL3/4 complex, which is dependent on BRCT (domains 5 and 6) (Patel et al., 2007). BRCA1−/− PTIP−/− B cells were infected with PTIPWT and PTIPW663R encoding retroviruses, treated with PARPi, and monitored for chromosomal damage (Figure 5E and Figure S9). Whereas PTIPWT expression in BRCA1−/−PTIP−/− cells led to an increase in the number of chromosomal radials relative to uninfected cells, BRCA1−/−PTIP−/− cells transduced with PTIPW663R remained insensitive (Figure 5E and Figure S9). Thus, PTIP recruitment to DSBs is necessary to block HR in BRCA1-deficient cells.
Recruitment of PTIP to DSBs is dependent on the 8 most N-terminal S/TQ phosphorylation sites of 53BP1
To explore the mechanism of PTIP recruitment to DSBs, we expressed FLAG-tagged PTIP in WT, 53BP1−/− and ATM−/− MEFs and irradiated them with 10 Gy (Figure 6A). Although PTIP ionizing irradiation induced foci (IRIF) were detectable in nearly all WT cells, PTIP IRIF formation was impaired in the absence of 53BP1 or ATM (Figure 6A). Measurements of colocalization coefficients of γ-H2AX (a marker of the DNA breaks) with PTIP in irradiated WT, 53BP1−/− and ATM−/− MEFs revealed that 80% of γ-H2AX foci in WT cells contained PTIP whereas less than 15% and 10% of γ-H2AX foci in the 53BP1−/− and ATM−/− cells, respectively, contained PTIP. Consistent with these findings, PTIP IRIF was highly sensitive to pharmacological inhibition of ATM (ATMi), less sensitive to ATRi treatment and insensitive to DNA-PKi. (Figure S10). These findings contrast with previous reports suggesting that PTIP, 53BP1, and ATM are independently recruited to DSBs (Gong et al., 2009; Jowsey et al., 2004; Munoz et al., 2007). Since available PTIP antibodies are unable to detect endogenous PTIP foci, we used laser microirradiation to generate DSBs in WT, 53BP1−/−, and ATM−/− MEFs. In WT cells, PTIP was recruited to laser scissors-induced DSBs, which colocalized with γ-H2AX. Consistent with our analysis of IRIF, PTIP recruitment to DNA damage sites was 53BP1 and ATM dependent (Figure 6B). Moreover, PTIP failed to be recruited to DSBs in 53BP1−/− MEFs reconstituted with a mutant protein lacking all 28 N-terminal S/TQ phosphorylation sites of 53BP1, 53BP128A (Figure 6B). We conclude that ATM-dependent phosphorylation of 53BP1 is necessary for PTIP recruitment to DSBs.
Figure 6. Recruitment of PTIP to DSBs is ATM and phospho-53BP1 dependent but RIF1-independent.
(A) WT, 53BP1−/− and ATM−/− MEFs were infected with a FLAG-tagged WT PTIP retrovirus. Cells were irradiated with 10 Gy, and FLAG (red) IRIF together with γ-H2AX (green) were assessed four hours post-IR. DAPI is indicated in blue. (B) WT, 53BP1−/−, ATM−/− and 53BP1−/− MEFs reconstituted with 53BP128A were treated with Hoecsht 33342 and then irradiated with a 364 nm laser line. Cells were allowed to recover for 15 minutes before processing for immunfluorescence analysis of PTIP and γ-H2AX. Hoechst counterstain is indicated in blue. (C) Cells expressing GFP-PTIP were irradiated with 10 Gy, and PTIPGFP (green) and RIF1 (red) IRIF were assessed four hours later. Representative image is shown. 82% of PTIP IRIF colocalized with RIF1 foci and 78% of RIF1 colocalized with PTIP foci (n≥800 foci examined. Cells had on average 28 foci). (D) RIF1 IRIF (red) in irradiated WT and PTIP−/− MEFs. (E) RIF1 (red) and PTIP (red) recruitment to laser scissors damage in WT and RIF1−/− MEFs. Damaged cells are indicated by γ-H2AX tracks (green). Scale bars represents 10 μm. See also Figure S10.
Given that RIF1 is also recruited to DSBs in a 53BP1 and ATM dependent manner (Chapman et al., 2013; Di Virgilio et al., 2013; Escribano-Diaz et al., 2013; Feng et al., 2013; Silverman et al., 2004; Zimmermann et al., 2013), we next monitored the co-dependency of PTIP and RIF1 for localization to DNA damage foci (Figure 6C). We found that 82% of PTIP IRIF colocalized with RIF1 foci and 78% of RIF1 colocalized with PTIP foci (Figure 6C, n>800 foci), However, RIF1 was recruited to DNA damage sites in PTIP−/− MEFs (Figure 6D) and visa-versa (Figure 6E). Thus, RIF1 and PTIP are independently recruited to IRIF in a phospho-53BP1 dependent manner.
To further define the residues required for recruitment to phospho-53BP1, we examined PTIP and RIF1 recruitment in 53BP1DB, 53BP18A and 53BP17A-mutant MEFs (Figures 2A and Figure S2B). Whereas expression of 53BP1DB in 53BP1−/− MEFs reconstituted PTIP IRIF (Figures 7A and Figure S11), PTIP recruitment to DSBs was abrogated in 53BP18A MEFs (Figure 7A and Figure S11). By contrast, RIF1 recruitment was independent of these 8 most N-terminus phosphorylation sites on 53BP1, partially dependent on the 7 S/TQ phosphorylation sites C-terminus to 53BP18A, and abrogated in 53BP115A mutant cells that lack all 8S/TQ and 7S/TQ phosphorylation sites (Figures 7A-C and Figure S11). Thus, PTIP and RIF1 exhibit distinct phosphorylation-dependent interactions with 53BP1 that guide them to DSBs. The association of PTIP with the 8S/TQ sites on 53BP1 upon DNA damage (Figures 2A and 7; Figure S11) likely explains why loss of PTIP phenocopies 53BP18A with respect to CSR, irradiation sensitivity and reversal of genome instability in BRCA1-deficient cells.
Figure 7. PTIP and RIF1 association with DSBs is dependent on distinct phosphorylation sites on 53BP1.
(A) 53BP1−/− MEFs (reconstituted with 53BP1DB, 53BP18A, 53BP17A, or 53BP115A were co-stained with RIF1 (red) and PTIP (green). Scale bar represents 10 μm. (B) Quantitation of percent 53BP1DB, 53BP18A, 53BP17A or 53BP115A cells with greater than ten 53BP1, PTIP or RIF1 foci. At least 100 cells were analyzed for each genotype. (C) Integrated intensity of individual IRIF in 53BP1−/− MEFs reconstituted with DB or 7A. Average RIF1 foci intensity (red line) is 1.6 fold greater in DB vs. 7A (** p<0.001, one-tailed unpaired t-test), and a greater percentage of very intense foci (z-score>3) are generated in 53BP1DB compared to 53BP17A (blue box). (D) Model for regulation of 53BP1 pro-NHEJ and anti-HR activities by distinct phospho-interactions with RIF1 and PTIP respectively. PTIP binds to the 8S/TQ sites. RIF1 recruitment is largely dependent on C-terminal 7S/TQ sites, but RIF1 may also be stabilized by interactions with 8S/TQ. An unknown factor (X) may bind directly to phosphorylated 53BP1 and mediate RIF1 recruitment, whereas PTIP interaction with 53BP1 is direct (Munoz et al., 2007). See also Figures S11-S13.
DISCUSSION
Regulation of DSB repair choice
53BP1 and BRCA1 play a critical role in channeling DSBs into either NHEJ or HR. 53BP1 promotes NHEJ in G1 by tethering DSBs together and by protecting these ends from exonuclease processing (Bothmer et al., 2010; Difilippantonio et al., 2008). In S phase, the inhibitory effect of 53BP1 on resection is antagonized by BRCA1 (Bouwman et al., 2010; Bunting et al., 2010). Loss of BRCA1 results in a shift towards a mutagenic NHEJ pathway that results in chromosomal abnormalities, tumorigenesis, and embryonic lethality, but all of these phenotypes are relieved by 53BP1 deletion (Bouwman et al., 2010; Bunting et al., 2010; Cao et al., 2009). In contrast, loss of classical NHEJ proteins (eg. Ku, Ligase IV, DNA-PKcs) does not overcome the HR defects associated with BRCA1 deficiency (Bunting et al., 2012; Bunting et al., 2010), perhaps because these factors play a more limited role in repressing 5′-3′ resection (Bunting et al., 2012; Sfeir and de Lange, 2012). Despite the striking rescue of BRCA1 deficiency, disrupting 53BP1 does not reverse the DNA repair defects associated with downstream mediators of the HR reaction (eg. XRCC2, BRCA2, or PALB2) (Bouwman et al., 2010; Bowman-Colin et al., 2013; Bunting et al., 2010). Thus, 53BP1 and BRCA1 oppose each other during critical initial stages of DSB repair, before commitment to repair the ends by NHEJ or HR.
Mechanism of PTIP and RIF1 association with 53BP1
The molecular events that are required for 53BP1 to promote the ligation of DNA ends during CSR and the aberrant chromosomal rearrangements in BRCA1-mutant cells were previously thought to be identical. Surprisingly our data suggest that the pro-NHEJ and anti-HR functions of 53BP1 are in fact distinct and separable activities that nevertheless require 53BP1 phosphorylation. These complementary aspects of 53BP1’s activities are mediated by the independent recruitment of RIF1 and PTIP respectively to phosphorylated 53BP1.
PTIP contains BRCT domains that interact directly with phosphorylated 53BP1 (Manke et al., 2003; Munoz et al., 2007). In contrast, RIF1 does not contain a known phospho-recognition motif, and it remains unclear how ATM dependent phosphorylation facilitates RIF1 association with 53BP1. RIF1 may associate with 53BP1 directly or through interactions with effector molecules that contain BRCT phospho-binding modules (Figure 7D). Based on the observation that there is no detectable defect in RIF1 foci in 53BP18A cells (Figures 7A and Figure S11), we suspected that a major RIF1-interaction motif would reside C-terminus of the 8S/TQ PTIP interaction sites. Consistent with this, the 53BP17A C-terminal mutant exhibits a reduction in RIF1 IRIF (Figure 7A-C and Figure S11) and CSR (Bothmer et al., 2011). RIF1 IRIF and CSR are further reduced in 53BP115A-mutant cells that lack 8S/TQ and 7S/TQ sites (Figures 7A and 7B) (Bothmer et al., 2011), suggesting that both regions contribute to RIF1 interactions with 53BP1 (Figure 7D). If so, we would predict some degree of competition between PTIP and RIF1 binding to 53BP1. Consistent with this, we have found an increased association between PTIP and 53BP1 in response to DNA damage in RIF1 deficient cells (Figure S12). Thus distinct from PTIP, RIF1 association with 53BP1 occurs via multi-domain interactions (Figure 7D).
Role of PTIP and RIF1 in DSB resection
Deletion of either PTIP or RIF1 leads to increased resection (Figure 5D)(Chapman et al., 2013; Di Virgilio et al., 2013; Escribano-Diaz et al., 2013; Feng et al., 2013; Zimmermann et al., 2013). However, whereas PTIP ablation rescues HR in BRCA1 deficient cells and is largely dispensable for NHEJ during CSR, RIF1 is essential for CSR and only partially contributes to the HR defects in BRCA1-deficient cells (Escribano-Diaz et al., 2013; Feng et al., 2013; Zimmermann et al., 2013). How can these observations be reconciled? One possibility is that distinct S/TQ kinase target sites in 53BP1 are phosphorylated during CSR in G1 and during replication fork collapse in S, resulting in independent recruitment of the two factors to DNA ends in distinct phases of the cell cycle. Consistent with this idea, it was reported that the localization of RIF1 to DSBs is mainly restricted to G1 and is suppressed by BRCA1 in S/G2 (Chapman et al., 2013; Escribano-Diaz et al., 2013; Feng et al., 2013). However, our finding that PTIP and RIF1 co-localize in the majority of irradiated cells, and that both proteins form IRIF during G1 and S/G2 (Figure S13) indicates that PTIP and RIF1 are not recruited to DSBs in distinct cell cycle phases.
Another possibility is that PTIP and RIF1 sites on 53BP1 are equally phosphorylated during the cell cycle but that these proteins might make the DSB-proximal chromatin refractory to a distinct set of nucleases. For example, initial DNA end resection is mediated by MRE11/RAD50/NBS1 and CTIP, whereas DNA2, EXO1 and BLM carry out more extensive resection (Symington and Gautier, 2011), and RIF1 appears to be involved in protection against initial but not sustained resection (Feng et al., 2013). In this model the level of resection supported by loss of RIF1 would be insufficient for complete rescue of HR in BRCA1-deficient cells, which might require more extensive 3′ single-strand tails. In contrast, ablation of PTIP supports the sustained resection required for the rescue of HR in BRCA1-deficient cells. Thus, RIF1 and PTIP may block different steps in resection or distinct nucleases that mediate HR.
Role of PTIP and RIF1 in telomeric end-joining
Depending on the nature of the break, RIF1 and PTIP might cooperate to promote NHEJ. For example, PTIP and RIF1 deficiency both result in IR sensitivity (Figures 2B and C)(Feng et al., 2013), and defective NHEJ of dysfunctional telomeres (Figure 4) (Chapman et al., 2013; Zimmermann et al., 2013). It has been demonstrated that 53BP1 has RIF1-independent roles in promoting telomeric end-joining, evidenced by the considerably higher frequency of telomeric fusions in RIF1−/−TRF2−/− vs. 53BP1−/−TRF2−/− or 53BP128ATRF2−/− MEFs (Lottersberger et al., 2013; Zimmermann et al., 2013). This RIF1-independent but phospho-53BP1 dependent function at telomeres has been linked to the induction of chromosome mobility (Zimmermann et al., 2013), which increases the probability that DNA ends fuse. Since PTIP binds to DSBs in a 53BP1-dependent but RIF1-independent manner, it is possible that this 53BP1-dependent/RIF1-independent increase in telomere mobility is mediated by PTIP.
Implications for cancer therapy
The identification of separation of function mutations that selectively disrupt anti-recombination functions of 53BP1 during replication fork collapse and CSR may open up new therapeutic opportunities. Breast cancers arising in BRCA1 mutation carriers frequently show low levels of 53BP1 expression (Bouwman et al., 2010), which might result in resistance to PARPi therapy, a promising strategy for treating HR deficient tumors (Bryant et al., 2005; Farmer et al., 2005). Consistent with this, 53BP1 was lost in a fraction of BRCA1 deficient mouse mammary tumors that acquired PARPi resistance in vivo (Jaspers et al., 2013). Interestingly, a fraction of PARPi-resistant tumors restored HR yet did not lose 53BP1. We speculate that PTIP mutation might emerge as a novel causal factor in PARPi resistance of BRCA1-deficient mammary tumors that restore HR. With respect to intervention, our study also suggests that it might be possible to increase HR in BRCA1 heterozygous carriers without compromising B cell immunoglobulin class switching by inhibiting the recruitment of PTIP to DSBs.
EXPERIMENTAL PROCEDURES
Mice, MEFs, B cell culture and infections
53BP1−/− (Ward et al., 2004), BRCA1f(Δ11)/f(Δ11)(NCI mouse repository), RIF1f/f (Buonomo et al., 2009; Di Virgilio et al., 2013) and PTIPf/f (Daniel et al., 2010) mice have been described. Resting splenic B cells were isolated from 8- to 12-week-old WT or mutant spleen with anti-CD43 Microbeads (anti-Ly48; Miltenyi Biotech) and were cultured with LPS (25 μg/ml; Sigma) and IL-4 (5 ng/ml; Sigma) or αCD40 (1μg/ml; eBiosciences) and IL4 as described (Barlow et al., 2013; Wesemann et al., 2011). WT, 53BP1−/−, and ATM−/− MEFs were immortalized by SV40. SV40T immortalized PTIPf/f (Cho et al., 2009) and RIF1f/f MEFs were infected with CRE viruses to delete PTIP and RIF1 respectively. PMX-PIE-based retroviruses encoding 53BP1DB and 53BP18A were previously described (Bothmer et al., 2011). Coding sequences for mouse PTIPWT/PTIPW663R and PTIP-GFP were cloned into the PMX-IRES-GFP and MIG-IRES-mCherry retro viral vectors respectively. PARP (KU58948), ATM (Ku55933) and DNA-PK (NU7026) inhibitors were obtained from Astra Zeneca and ATRi has recently been described(Toledo et al., 2011).
Flow Cytometry, metaphase analysis and telomere FISH
For FACs analysis, splenic B cells were stained with fluorochrome-conjugated anti-B220, anti-IgG1 and anti-IgE antibodies (PharMingen) as described (Wesemann et al., 2011). Carboxyfluorescein succinimidyl esther (CFSE) labeling was performed to track cell division. Samples were acquired on a FACSCalibur (Becton Dickinson) and cell sorting was preformed on a FACsAria (Becton Dickinson). Cells were harvested for metaphase analysis as described (Callen et al., 2007a). The murine TRF2 shRNA-targeting construct and MEF retroviral infection has been described (Rai et al., 2010). Telomere-induced foci were visualized by hybrization with anti-mouse γ-H2AX antibody (Upstate Biotechnology) together with PNA probe (Applied Biosystems). Phosphorylated Kap-1 was detected by flow cytometry after intracellular staining using the BD Cytofix/Cytoperm kit (BD Biosciences).
DNA damage, Laser microirradiation, immunoprecipitation and RNA-sequencing
Cells were treated with different DNA damaging agents (IR, CPT, CisPt, and PARPi), and colony survival was assessed after 14 days, or metaphase analysis was performed 24 hours after treatment. For immunofluorescent staining, cells were irradiated with indicated doses of ionizing radiation, allowed to recover, and then fixed and processed as described (Celeste et al., 2003). For microirradiation, cells were pre-sensitized in DMEM media containing 0.1ug/ml of Hoechst 33342 for 60 minutes before replacing with phenol red free media containing 5mM HEPES, and then irradiated with the 364nm laser line on a LSM510 confocal microscope (Zeiss) equipped with a heated stage. Cells were allowed to recover for 15 minutes prior to processing for immunofluorescence. Analysis of RPA foci was performed using an Opera High-Content Screening system as described (Lopez-Contreras et al., 2012). Primary antibodies for immunofluorescence were rabbit anti-53BP1 (Novus), mouse anti-γ-H2AX (Upstate Biotechnology), mouse or rabbit anti-FLAG-M2 (Sigma), mouse anti-AIM1 (Becton Dickinson), mouse anti-GFP (Roche), rabbit anti-RAD51 (Santa Cruz), rat anti-RPA (Cell Signaling), rabbit-anti-PTIP (Cho et al., 2009) and rabbit-anti-RIF1(Di Virgilio et al., 2013). DNA was counterstained with DAPI. For immunoprecipitation, primary 53BP1−/− B cells were infected with retroviral constructs. 96 hours post-activation, cells were irradiated (10 Gy), left to recover for 45 min, and collected by centrifugation. Cells were lysed, sonicated and cell lysates were incubated with magnetic beads (M-270 epoxy beads, Invitrogen) conjugated with anti-Flag M2 antibody (Di Virgilio et al., 2013). 53BP1-associated proteins were eluted by incubation in NuPAGE LDS sample buffer (Invitrogen) supplemented with 45 mM DTT for 10 min at 72°C. For RNA sequencing, reads from each cDNA library were mapped onto the Build 37 assembly of the National Center for Biotechnology Information mouse genome data (July 2007; NCBI37/mm9) using TopHat. Bioconductor (Gentleman et al., 2004) was used to calculate the RPKM (reads per kilobase exon model per million mapped reads) of the RefSeq annotated genes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the A. Nussenzweig lab and Davide Robbiani for discussions; Titia de Lange for RIF1f/f mice, Susan Sharrow for flow cytometry and Sandy Chang for the TRF2 shRNA construct. M.N., F.W.A and O.F.C. are HHMI investigators. D.R.W. was supported by NIH grants AI89972, the American Association of Allergy Asthma and Immunology and CSL Behring, and a Career Award for Medical Scientists (Burroughs Wellcome Fund). This work was supported by the Intramural Research Program of the NIH, the National Cancer Institute, and the Center for Cancer Research, and by a Department of Defense grant to A.N. (BC102335).
REFERENCES
- Barlow JH, Faryabi RB, Callen E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun HW, McKinnon P, Wright G, et al. Identification of Early Replicating Fragile Sites that Contribute to Genome Instability. Cell. 2013;152:620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. The Journal of biological chemistry. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, Fazeli A, Feldman L, Nussenzweig A, Nussenzweig M, et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Molecular cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. The Journal of experimental medicine. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature structural & molecular biology. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman-Colin C, Xia B, Bunting S, Klijn C, Drost R, Bouwman P, Fineman L, Chen X, Culhane AC, Bronson RT, et al. Palb2 synergizes with Trp53 to suppress mammary tumor formation in a model of inherited breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1305362110. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Molecular cell. 2012;46:125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SB, Wu Y, Ferguson D, de Lange T. Mammalian Rif1 contributes to replication stress survival and homology-directed repair. The Journal of cell biology. 2009;187:385–398. doi: 10.1083/jcb.200902039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Faryabi RB, Luckey M, Hao B, Daniel JA, Yang W, Sun HW, Dressler G, Peng W, Chi H, et al. The DNA damage- and transcription-associated protein paxip1 controls thymocyte development and emigration. Immunity. 2012;37:971–985. doi: 10.1016/j.immuni.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007a;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Callen E, Nussenzweig MC, Nussenzweig A. Breaking down cell cycle checkpoints and DNA repair during antigen receptor gene assembly. Oncogene. 2007b;26:7759–7764. doi: 10.1038/sj.onc.1210873. [DOI] [PubMed] [Google Scholar]
- Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Molecular cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature cell biology. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nature cell biology. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ. RIF1 Is Essential for 53BP1-Dependent Nonhomologous End Joining and Suppression of DNA Double-Strand Break Resection. Molecular cell. 2013 doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong S, Jin Q, Wang L, Lee JE, Gavrilova O, Ge K. Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell metabolism. 2009;10:27–39. doi: 10.1016/j.cmet.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. The Journal of biological chemistry. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, Jankovic M, Filsuf D, Chen HT, Gazumyan A, Yamane A, et al. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science. 2010;329:917–923. doi: 10.1126/science.1187942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, et al. A Cell Cycle-Dependent Regulatory Circuit Composed of 53BP1-RIF1 and BRCA1-CtIP Controls DNA Repair Pathway Choice. Molecular cell. 2013 doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Feng L, Fong KW, Wang J, Wang W, Chen J. RIF1 counteracts BRCA1-mediated end resection during DNA repair. The Journal of biological chemistry. 2013 doi: 10.1074/jbc.M113.457440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Cho YW, Kim JE, Ge K, Chen J. Accumulation of Pax2 transactivation domain interaction protein (PTIP) at sites of DNA breaks via RNF8-dependent pathway is required for cell survival after DNA damage. The Journal of biological chemistry. 2009;284:7284–7293. doi: 10.1074/jbc.M809158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers JE, Kersbergen A, Boon U, Sol W, van Deemter L, Zander SA, Drost R, Wientjens E, Ji J, Aly A, et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer discovery. 2013;3:68–81. doi: 10.1158/2159-8290.CD-12-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowsey PA, Doherty AJ, Rouse J. Human PTIP facilitates ATM-mediated activation of p53 and promotes cellular resistance to ionizing radiation. The Journal of biological chemistry. 2004;279:55562–55569. doi: 10.1074/jbc.M411021200. [DOI] [PubMed] [Google Scholar]
- Lopez-Contreras AJ, Gutierrez-Martinez P, Specks J, Rodrigo-Perez S, Fernandez-Capetillo O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. The Journal of experimental medicine. 2012;209:455–461. doi: 10.1084/jem.20112147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottersberger F, Bothmer A, Robbiani DF, Nussenzweig MC, de Lange T. Role of 53BP1 oligomerization in regulating double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1222617110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Molecular cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- Munoz IM, Jowsey PA, Toth R, Rouse J. Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic acids research. 2007;35:5312–5322. doi: 10.1093/nar/gkm493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Developmental cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Zheng H, He H, Luo Y, Multani A, Carpenter PB, Chang S. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. The EMBO journal. 2010;29:2598–2610. doi: 10.1038/emboj.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1−/− B cells. European journal of immunology. 2007;37:235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- Schwab KR, Patel SR, Dressler GR. Role of PTIP in class switch recombination and long-range chromatin interactions at the immunoglobulin heavy chain locus. Molecular and cellular biology. 2011;31:1503–1511. doi: 10.1128/MCB.00990-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T. Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes & development. 2004;18:2108–2119. doi: 10.1101/gad.1216004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA repair. 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annual review of genetics. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, Fernandez-Capetillo O. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nature structural & molecular biology. 2011;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Takenaka K, Takeda S. PTIP promotes DNA double-strand break repair through homologous recombination. Genes to cells: devoted to molecular & cellular mechanisms. 2010 doi: 10.1111/j.1365-2443.2009.01379.x. [DOI] [PubMed] [Google Scholar]
- Ward I, Kim JE, Minn K, Chini CC, Mer G, Chen J. The tandem BRCT domain of 53BP1 is not required for its repair function. The Journal of biological chemistry. 2006;281:38472–38477. doi: 10.1074/jbc.M607577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, et al. 53BP1 is required for class switch recombination. The Journal of cell biology. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesemann DR, Magee JM, Boboila C, Calado DP, Gallagher MP, Portuguese AJ, Manis JP, Zhou X, Recher M, Rajewsky K, et al. Immature B cells preferentially switch to IgE with increased direct Smu to Sepsilon recombination. The Journal of experimental medicine. 2011;208:2733–2746. doi: 10.1084/jem.20111155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Robbiani DF, Resch W, Bothmer A, Nakahashi H, Oliveira T, Rommel PC, Brown EJ, Nussenzweig A, Nussenzweig MC, et al. RPA Accumulation during Class Switch Recombination Represents 5′-3′ DNA-End Resection during the S-G2/M Phase of the Cell Cycle. Cell Rep. 2013;3:138–147. doi: 10.1016/j.celrep.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phosphoprotein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.