Abstract
Memory deficits are prominent features of mild cognitive impairment (MCI) and Alzheimer’s disease (AD). The genetic architecture underlying these memory deficits likely involves the combined effects of multiple genetic variants operative within numerous biological pathways. In order to identify functional pathways associated with memory impairment, we performed a pathway enrichment analysis on genome-wide association data from 742 Alzheimer’s Disease Neuroimaging Initiative (ADNI) participants. A composite measure of memory was generated as the phenotype for this analysis by applying modern psychometric theory to item-level data from the ADNI neuropsychological test battery. Using the GSA-SNP software tool, we identified 27 canonical, expertly-curated pathways with enrichment (FDR-corrected p-value < 0.05) against this composite memory score. Processes classically understood to be involved in memory consolidation, such as neurotransmitter receptor-mediated calcium signaling and long-term potentiation, were highly represented among the enriched pathways. In addition, pathways related to cell adhesion, neuronal differentiation and guided outgrowth, and glucose- and inflammation-related signaling were also enriched. Among genes that were highly-represented in these enriched pathways, we found indications of coordinated relationships, including one large gene set that is subject to regulation by the SP1 transcription factor, and another set that displays co-localized expression in normal brain tissue along with known AD risk genes. These results 1) demonstrate that psychometrically-derived composite memory scores are an effective phenotype for genetic investigations of memory impairment and 2) highlight the promise of pathway analysis in elucidating key mechanistic targets for future studies and for therapeutic interventions.
Keywords: memory, psychometrics, Alzheimer’s disease, mild cognitive impairment, pathway analysis, genome-wide association study
INTRODUCTION
Memory deficits are prominent features of Alzheimer’s disease (AD) and of mild cognitive impairment (MCI), a transitional state with significant risk of progression to clinically-diagnostic AD (Aisen et al. 2010). While genomic studies of MCI and AD susceptibility are frequent and ongoing, surprisingly, there has been less large-scale research on the genetic variants specifically related to memory impairment. Genetic studies, especially of quantitative traits (QT) such as memory, are highly dependent on the quality of the phenotypic data. There are numerous extant metrics for assessing memory in amnestic populations, and these metrics can have differential sensitivities to various deficits (Lezak 2004). As a result, studies often attempt to leverage the relative strengths and weaknesses of these metrics by creating composite memory scores from multiple assessments given to study participants (Barbeau et al. 2011; Sloan et al. 2010). In addition, emerging evidence suggests that applying modern psychometric theory to the creation of these composite scores may yield an optimized measure of memory functioning that serves as a more powerful phenotype for genetic studies (see (Crane et al. 2011) and the other companion papers in this issue for more detail).
In addition, it is clear from human and animal studies that the complex processes of memory consolidation and recall involve numerous and diverse cellular and molecular pathways (Sweatt 2009). While genome-wide association (GWA) studies of complex phenotypes have historically focused on identifying individual susceptibility loci, their efficacy has been confounded by three factors: 1) most common alleles implicated by GWA studies have exhibited modest effect sizes (Chee Seng et al. 2010); 2) gene and gene variant associations do not always indicate therapeutic targets (Penrod et al. 2011); and 3) it is well-understood that genes do not exist in isolation, but instead function as sets within biological pathways and networks (Schadt 2009). As a result, GWA studies of complex phenotypes are increasingly being analyzed through statistical methods designed to identify gene sets with significant relationships to those phenotypes (Hirschhorn 2009; Ramanan et al. 2012).
As a particularly prominent approach, pathway enrichment methods analyze genomic data through gene sets representing biological pathways. Pathway enrichment analysis uses the association signals of genomic markers within a pathway to determine whether that pathway is enriched (i.e., has a statistically significant association) against a phenotype. While many tools and strategies for pathway enrichment analysis have been developed in recent years, to date there is no gold standard approach (Cantor et al. 2010; Wang et al. 2010). Nevertheless, despite using diverse strategies, high-profile investigations of numerous complex disorders, including breast cancer (Li et al. 2011), type 2 diabetes (Zhong et al. 2010), and Multiple Sclerosis (Sawcer et al. 2011), suggest that pathway analysis of GWA study data can reveal larger effects that are otherwise concealed from standard single locus analysis.
While GWA-based pathway analysis has been used against complex neurological phenotypes, including brain glutamate levels (Baranzini et al. 2010), cerebrospinal fluid (CSF) amyloid-β42 peptide levels (Han et al. 2010), and information processing speed (Luciano et al. 2011), to our knowledge this strategy has not been previously applied to a quantitative memory phenotype. In this study, we use enrichment analysis of GWA data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort to identify pathways associated with changes in a psychometrically-derived composite episodic memory score. In addition, we identify genes with high representation in these pathways as key targets for future studies of memory deficits. Finally, using transcription factor network analysis and data from a human brain tissue expression atlas, we isolate sets of these gene targets which are co-regulated and/or co-expressed, suggesting that memory-impaired phenotypes are the result of gene variants exerting their effects through complex pathway and network interactions. The collection of enriched pathways, gene targets, and gene networks identified in this analysis 1) represent prime targets for further studies of memory impairment and normal memory processes and 2) further demonstrate the efficacy of pathway-based approaches to analyzing GWA data on complex phenotypes.
METHODS
Alzheimer’s Disease Neuroimaging Initiative (ADNI)
Data used for this study were obtained through the ADNI database (http://adni.loni.ucla.edu/). ADNI was launched in 2003 (PI: Michael W. Weiner, MD, VA Medical Center and University of California-San Francisco) with the goal of evaluating biomarkers of AD-related neuropathology in patients with MCI and early AD. This multi-site longitudinal study is supported by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations. Participants in ADNI include older individuals, aged 55-90 years, who were recruited from 59 sites across the United States and Canada. These subjects include approximately 200 cognitively normal patients (CN), 400 patients diagnosed with MCI, and 200 patients diagnosed with early probable AD. As described elsewhere (Jack et al. 2009; Weiner et al. 2010; Aisen et al. 2010), diagnoses of participants were made on a clinical basis (via neuropsychological assessment data and patient and informant reports of cognitive performance and functioning in activities of daily living) at consensus conferences involving neurologists, neuropsychologists, and study coordinators. Written informed consent was obtained for all participants and prior Institutional Review Board approval was obtained at each participating institution. All demographic information, neuropsychological and clinical assessment data, and diagnostic information used in this study were downloaded from the ADNI clinical data repository (http://adni.loni.ucla.edu/). Genotype data used in this study include genome-wide single nucleotide polymorphism (SNP) data obtained from a GWAS on the full ADNI sample (Saykin et al. 2010) and APOE ε4 allele status. Further information about ADNI can be found in (Weiner et al. 2010) and at http://www.adni-info.org/.
Participants
Seven hundred and forty-two non-Hispanic Caucasian participants (188 AD, 396 MCI, and 226 CN at baseline) from the ADNI cohort with baseline composite episodic memory scores (see later section) and genotype data passing quality control procedures (see later section) were included in the present analyses (Table 1). Sample characteristics were evaluated using the IBM SPSS 19.0 statistical software (SPSS, Inc., Chicago, IL). The sample was analyzed for differences across diagnostic groups using a one-way analysis of variance (ANOVA) for continuous variables and a Pearson chi-square test for categorical variables.
Table 1.
Characteristics for participants included in the analyses
| CN (N = 207) |
MCI (N = 362) |
AD (N = 173) |
p-value* | |
|---|---|---|---|---|
| Age at baseline | 76.1 ± 5.0 | 74.9 ± 7.4 | 75.6 ± 7.5 | 0.139 |
| Gender (male/female) | 113/94 | 234/128 | 92/81 | 0.012 |
| Years of education | 16.2 ± 2.7 | 15.7 ± 3.0 | 14.9 ± 3.0 | < 0.001 |
| Handedness (right/left) | 191/16 | 328/34 | 161/12 | 0.586 |
| APOE ε4 allele (absent/present) | 152/55 | 165/197 | 61/112 | < 0.001 |
| Composite memory scoreϕ | 1.05 ± 0.59 | −0.14 ± 0.64 | −1.01 ± 0.63 | < 0.001 |
Values represented are mean ± SD
For categorical variables, p-values were computed using the Pearson chi-square tests; for continuous variables, p-values were computed using a one-way analysis of variance.
Scores for individual participants were represented as z-scores with a defined mean of 0, and standard deviation of 1, based on the 810 participants with complete item-level memory data at baseline.
Composite Episodic Memory Scores
All ADNI participants (original N = 818) were administered an extensive neuropsychological assessment, including several measures of memory, at each study visit. For each subject, a composite score for episodic memory at the baseline visit was calculated by applying modern psychometric theory to item-level data from the ADNI neuropsychological battery. This psychometrically-derived episodic memory score was developed for the ADNI cohort as part of the 2011 Friday Harbor Workshop on Advanced Psychometrics (see (Crane et al. 2011) and the other companion papers in this special issue for details). Briefly, the authors used psychometric theory to select test battery items which could be considered as indicators of episodic memory functioning. An iterative process of confirmatory factor analysis was used to construct the final, optimized model for describing episodic memory performance at baseline. In particular, the following item-level tests were applied to the final model: the memory sub-scores from the Mini-Mental Status Examination (Folstein et al. 1975; Cockrell and Folstein 1988); the immediate and delayed recall and recognition scores on a word list learning task from the Alzheimer’s Disease Assessment Scale-cognitive subscale (Mohs et al. 1997); all immediate and delayed recall and recognition scores from the Rey Auditory Verbal Learning Test (Rey 1964); and all immediate and delayed recall scores on Logical Memory prose passages from the Wechsler Memory Scale-Revised (Wechsler 1987). The final model exhibited excellent fit based on standard criteria (Confirmatory Fit Index > 0.95, Tucker Lewis Index > 0.95, and Root Mean Squared Error of Approximation < 0.05) (Reeve et al. 2007). Notably, a composite episodic memory score could not be calculated for eight participants due to incomplete item-level data at baseline.
Genotyping and Quality Control
The acquisition and processing of genotype data for the ADNI sample have been previously described in detail (Saykin et al. 2010). All participants analyzed in this study were genotyped according to the manufacturer’s protocol using the Human610-Quad BeadChip (Illumina, Inc., San Diego, CA), which included 620,901 SNPs and copy number variant markers. In addition, given the strong association of its genetic variants with MCI and AD susceptibility (Bertram et al. 2010; Corder et al. 1993; Strittmatter et al. 1993), APOE genotype status was also of interest. Since the two APOE SNPs (rs429358, rs7412) that characterize the ε2, ε and ε4 risk loci alleles were not directly available on the Human 610-Quad BeadChip, these SNPs were genotyped separately and their data were made available in the ADNI database (Potkin et al. 2009; Saykin et al. 2010).
The ADNI genotype data (original N = 818 participants) was subjected to standard quality control procedures as previously described (Shen et al. 2010) using Plink, version 1.07 (Purcell et al. 2007). Genotype markers were excluded if they had a call rate < 90%, Hardy-Weinberg equilibrium test < 10−6, or minor allele frequency < 5%. Samples were excluded if they had a call rate < 90% (1 participant), ambiguous gender identification (2 participants), or failed an identity check (3 participants). In order to select non-Hispanic Caucasian participants, multidimensional scaling analysis was performed on the combined genotype data from ADNI and from reference populations in HapMap phase 3, release 2 (International HapMap 2003); this resulted in the exclusion of 62 participants. Following all quality control procedures, 750 participants and 531,096 SNPs were designated for subsequent analyses and the genotyping rate was > 0.995 among the remaining samples.
Genome-wide Association (GWA) Analysis
Of the 750 subjects who passed sample quality control procedures, 742 subjects were identified as having a composite, psychometrically-defined episodic memory score at baseline. To generate input data for pathway analysis, a GWA analysis against this composite memory score was performed using an additive genetic model with the linear association analysis for quantitative traits in Plink. Demographic factors with known influences on memory or cognition were included as covariates in this analysis: these factors included age at the baseline visit, education, gender, and handedness. The direct and inverse relationships, respectively, of age and education level on memory decline have been well-established (Negash et al. 2011), while putative effects of gender and handedness on cognition are subjects of active exploration (Andreano and Cahill 2009; Gunstad et al. 2007). In addition, APOE ε4 allele status (presence vs. absence) was also used as a covariate in the GWA analyses to account for the largest known genetic influence on memory performance in an MCI- and AD-based clinical population (Bookheimer and Burggren 2009). For all SNPs included in the analysis, a p-value was generated by the model, representing the nominal association of that genotype marker to the composite memory score. Manhattan and Quantile-Quantile (Q-Q) plots for the GWA analysis were generated using Plink, Haploview (Barrett et al. 2005), and the R statistical package, version 2.15.
Genome-wide Pathway Analysis
Pathway analysis of the GWA results was performed to identify functional gene sets with significant association to the composite memory phenotype. All SNPs included in the GWA analysis were mapped to genes using the NCBI Build 36.1 reference sequence (International Human Genome Sequencing Consortium 2004). A gene mapping window of 20kb was used to account for SNPs belonging to putative regulatory regions; notably, this resulted in some SNPs being mapped to more than one gene. In total, 277,615 SNPs were assigned to 17,456 genes. Pathway annotations, representing gene sets defined by membership in biological pathways, were downloaded from the Molecular Signatures Database (Subramanian et al. 2005), version 3.0 (http://www.broadinstitute.org/gsea/msigdb/index.jsp/). This annotation data comprised a collection of canonical, expertly-curated gene sets from three publically-available pathway databases, BioCarta (http://www.biocarta.com/), the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/), and Reactome (http://www.reactome.org/). In total, 818 gene sets were downloaded, included 217 sets from BioCarta, 186 sets from KEGG, and 430 sets from Reactome. Analysis of these gene sets was restricted to those containing between 10 and 200 genes; as a result, 280 total gene sets were analyzed for enrichment.
The GSA-SNP software (Nam et al. 2010) was used to assess for pathways enriched against the composite memory score. This software uses a competitive enrichment algorithm (Goeman and Bühlmann 2007), where the null hypothesis holds that a pathway-phenotype association is not significantly different from all other pathway-phenotype associations under analysis. Competitive enrichment strategies are robust to the effects of genomic inflation due to population stratification or other confounding factors (Holmans 2010; Fridley and Biernacka 2011). In GSA-SNP, the significance score for each gene under analysis was calculated as the -log of the k-th best SNP-level p-value in the gene. Corresponding with the authors’ recommendation (Nam et al. 2010), we selected k = 2 to limit the effects of both single, highly-significant loci and of spurious SNP-level associations on driving pathway enrichment. Each gene set (representing a pathway) was then assessed for phenotype enrichment by the Z-statistic method (Kim and Volsky 2005), which incorporates the gene-wide significance scores and the number of genes within each set. In addition, since small pathways can exhibit spurious phenotype associations due to large single locus effects (Holmans 2010), and since large pathways are more likely to exhibit association by chance alone (Elbers et al. 2009), analysis was restricted to gene sets containing 10-200 genes. To correct for multiple hypothesis testing, the False Discovery Rate (FDR) (Benjamini and Hochberg 1995) was applied to the p-values generated by the enrichment algorithm. For pathways enriched at an FDR-corrected p-value < 0.05, we analyzed their constituent genes to obtain a count of each gene’s occurrences in those enriched pathways. Genes that were highly-represented among the enriched pathways (defined as being constituents of > 15%, i.e., 5, of the enriched pathways) were isolated for two follow-up analyses.
Transcription Factor Network Analysis
We further investigated the list of highly-represented genes from our enriched pathways through network analysis using MetaCore (GeneGo, Inc.). In particular, we applied the transcription factor network analysis algorithm to identify subsets of those genes with coordinate regulation by known transcription factors. In addition, APOE was also included in these network analyses, given its well-characterized association with MCI and AD and their related memory deficits.
Gene Expression Analysis Using the Allen Human Brain Atlas
We also interrogated the list of highly-represented genes from our enriched pathways for their expression profiles in normal brain tissue using the Allen Human Brain Atlas (Allen Institute for Brain Science, Seattle, WA; available from http://www.brain-map.org/). The Allen Human Brain Atlas (AHBA) includes genome-wide microarray-based expression profiles in postmortem brain tissue from subjects with no known neuropsychiatric or neuropathological history. These expression profiles cover the entire brain through systematic sampling of regional tissue, and are integrated with multi-modal brain imaging and other data for visualization and analysis. Detailed information on the AHBA is available on-line (http://human.brain-map.org/docs.html/). We employed the AHBA to examine genes of interest (including highly-represented genes from our enriched pathways) for patterns in their expression profiles. In particular, we used the heat map tool (which visually displays normalized expression for a gene probe across 25 large neuroanatomic regions) and correlational analysis (which calculates a Pearson correlation coefficient between the expression profiles of two gene probes) to identify a set of key genes with high (r > 0.7) co-localization and co-expression.
RESULTS
Demographic characteristics and mean composite memory scores for all diagnostic groups (CN, MCI, and AD) are presented in Table 1. While baseline age and handedness were not significantly different across diagnostic groups, gender exhibited a significant difference (p < 0.05), with males relatively overrepresented among MCI subjects. In addition, as expected, education level and APOE ε4 allele status exhibited significant differences across groups (p < 0.001). Also as expected, composite memory scores differed across all diagnostic groups, including all pairwise group comparisons (p < 0.001).
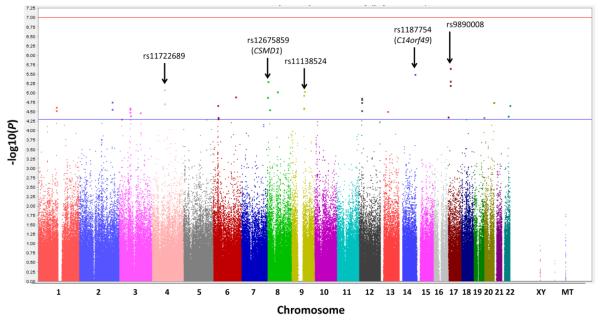
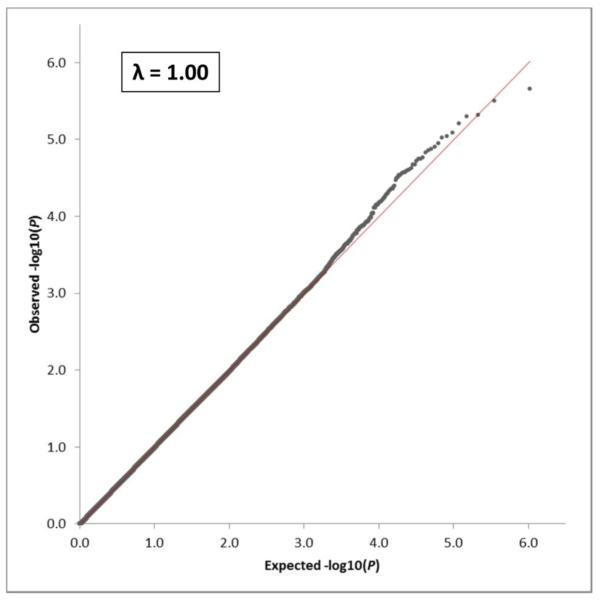
In order to assess SNP associations to the composite memory scores in this sample, we performed a GWA analysis with the addition of five covariates. The GWA analysis failed to identify any SNPs with significant association to the composite memory score at a Bonferroni-determined threshold p-value of 9.42 × 10−8 (i.e., 0.05/531,096). The peak SNP in the association analysis was rs9890008 (Chr 17), which has not been mapped to a known gene and which exhibited an unadjusted p-value of 2.21 × 10−6. Overall, 25,960 SNPs showed nominal p-values < 0.05 (unadjusted). Manhattan (Figure 1) and Q-Q (Figure 2) plots are displayed for the GWA analysis. Notably, when APOE ε4 allele status was removed as a covariate, one SNP (rs2075650) residing in the TOMM40 gene did exhibit significant association (p = 2.19 × 10−9) to the composite memory score. This result was probable, given that the TOMM40 gene is adjacent to, and often considered as one locus with, APOE on chromosome 19. In addition, TOMM40 variants have been associated with late-onset AD (Roses et al. 2010; Wijsman et al. 2011; Potkin et al. 2009) as well as with structural brain and cognitive function changes suggestive of presymptomatic late-onset AD (Johnson et al. 2011).
Fig. 1.
Manhattan plot for the GWA analysis of episodic memory. The x-axis refers to positions along the genome (separated by chromosome) for each SNP (represented by a dot) included in the analysis. The y-axis refers to the negative logarithm of the p-value for the test of association between each SNP and the quantitative memory phenotype. No SNPs exhibited genome-wide significant association (red line) to the composite episodic memory score, while 40 SNPs exhibited suggestive association (p < 5 × 10−5, blue line). The 5 most significant, independent (r2 < 0.2) SNPs are labeled along with their corresponding genes (if known).
Fig. 2.
Quantile-Quantile (Q-Q) plot for the GWA analysis of episodic memory. The genomic inflation factor (λ) for the analysis was 1.
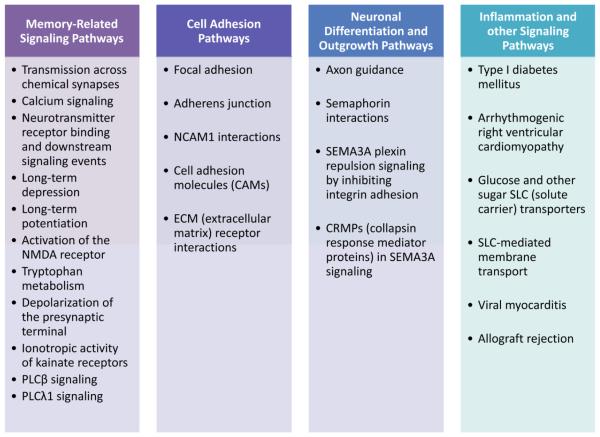
Next, the p-value output from the GWA analysis was used as input for pathway enrichment analysis. Using the GSA-SNP software tool, we identified 27 canonical pathways with enrichment (FDR-corrected p-value < 0.05) against the composite memory score (Table 2). Following these analyses, we examined the enriched pathways in detail in order to better characterize their biological import in memory deficits. First, we used existing knowledge and insight to conceptually categorize the 27 enriched pathways into 4 broader realms of biology (Figure 3). In particular, 11 enriched pathways represented classical cellular and molecular processes essential in normal memory consolidation signaling (Sweatt 2009). These pathways included functions of neurotransmitter receptor activation, downstream calcium-mediated signaling, and long-lasting potentiation of synaptic strength, among other processes. In addition, six pathways related to cell adhesion were enriched, including focal adhesion pathways from both the Reactome and KEGG databases, and interactions involving neuronal cell adhesion molecule 1 (NCAM1). Finally, four enriched pathways were related to neuronal differentiation and guided axonal growth, while a further six enriched pathways were involved in inflammation or other complex signaling processes. Notably, while we restricted analysis to pathways containing 10-200 genes, the enrichment results were nearly identical when upper limits of 300 or 400 genes were used: in those cases, two additional Reactome pathways exhibited enrichment (transmembrane transport of small molecules, FDR-corrected p-value = 0.029; adherens junctions interactions, FDR-corrected p-value = 0.049).
Table 2.
Canonical pathways enriched against the composite memory score
| Pathway (Gene Set) Name | Set Size1 | p-value | FDR p-value |
|---|---|---|---|
| rTransmission across chemical synapses | 136 (122) | 2.14 × 10−7 | 1.77 × 10−4 |
| kCalcium signaling pathway | 184 (165) | 2.82 × 10−7 | 1.17 × 10−4 |
| kType I diabetes mellitus | 50 (41) | 7.90 × 10−6 | 0.002 |
|
rNeurotransmitter receptor binding and downstream transmission |
90 (78) | 2.84 × 10−5 | 0.006 |
| kArrhythmogenic right ventricular cardiomyopathy | 82 (72) | 3.11 × 10−5 | 0.005 |
| rSLC-mediated membrane transport | 175 (162) | 3.87 × 10−5 | 0.005 |
| kFocal adhesion | 207 (187) | 4.68 × 10−5 | 0.006 |
| kAxon guidance | 135 (123) | 4.68 × 10−5 | 0.005 |
| kLong-term depression | 76 (65) | 9.02 × 10−5 | 0.008 |
| rAxon guidance | 167 (154) | 9.58 × 10−5 | 0.008 |
| kAdherens junction | 81 (71) | 1.04 × 10−4 | 0.008 |
| rOther semaphorin interactions | 22 (15) | 1.20 × 10−4 | 0.008 |
| rNCAM1 interactions | 50 (42) | 1.37 × 10−4 | 0.009 |
| kLong-term potentiation | 76 (65) | 3.14 × 10−4 | 0.019 |
|
rActivation of glutamate NMDA receptor and post-synaptic events |
42 (32) | 5.02 × 10−4 | 0.028 |
| kCell adhesion molecules (CAMs) | 140 (123) | 5.60 × 10−4 | 0.029 |
|
rSEMA3A plexin repulsion signaling by inhibiting integrin adhesion |
20 (13) | 6.06 × 10−4 | 0.030 |
| kTryptophan metabolism | 46 (35) | 6.53 × 10−4 | 0.030 |
|
rDepolarization of the presynaptic terminal triggers the opening of calcium channels |
18 (12) | 6.71 × 10−4 | 0.029 |
| rPLCβ-mediated events | 44 (35) | 6.91 × 10−4 | 0.029 |
| kViral myocarditis | 79 (67) | 9.98 × 10−4 | 0.039 |
| kAllograft rejection | 44 (34) | 0.001 | 0.039 |
| rGlucose and other sugar SLC transporters | 88 (80) | 0.001 | 0.039 |
| rIonotropic activity of kainate receptors | 18 (11) | 0.001 | 0.042 |
| kECM receptor interaction | 90 (83) | 0.001 | 0.041 |
| rPLCγ1 signaling | 41 (32) | 0.001 | 0.044 |
| rCRMPs in SEMA3A signaling | 22 (15) | 0.001 | 0.045 |
Entries are displayed as: number of genes in the set (number of genes from the GWA data)
Reactome pathway
KEGG pathway
Fig. 3.
Conceptual classification of 27 pathways enriched in the setting of memory impairment
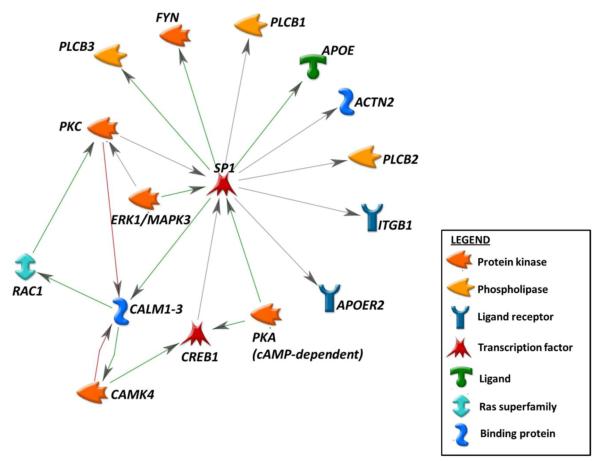
As further follow-up, we identified 44 genes that were highly-represented across the 27 enriched pathways (Table 3). Half (22) of these 44 genes were constituents of 6 or more enriched pathways, suggesting that variants in those genes can have wide-ranging roles in mediating memory impairment due to their diverse functions. We also assessed for underlying transcriptional relationships among these highly-represented genes and APOE, given the latter’s singular association with MCI and AD and their related memory deficits. Using network analysis in MetaCore, we discovered that 14 of the 22 most-represented genes from our analyses were part of a transcriptional regulation network driven by the specificity protein 1 (SP1) transcription factor and involving APOE and the APOE receptor-2 (Figure 4).
Table 3.
Highly-represented genes among the 27 canonical pathways enriched against the composite memory score
| Occurrences in Enriched Pathways |
Gene ID | Gene Name |
|---|---|---|
| 9 | MAPK1 | mitogen-activated protein kinase 1 |
| 8 | CALM1 | calmodulin 1 (phosphorylase kinase, delta) |
| 8 | CALM2 | calmodulin 2 (phosphorylase kinase, delta) |
| 8 | CALM3 | calmodulin 3 (phosphorylase kinase, delta) |
| 8 | HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| 7 | ADCY1 | adenylate cyclase 1 (brain) |
| 7 | ADCY8 | adenylate cyclase 8 (brain) |
| 7 | CAMK4 | calcium/calmodulin-dependent protein kinase IV |
| 7 | FYN | FYN oncogene related to SRC, FGR, YES |
| 7 | ITGB1 | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29) |
| 7 | PRKACB | protein kinase A, cAMP-dependent, catalytic, beta |
| 7 | RAF1 | v-raf-1 murine leukemia viral oncogene homolog 1 |
| 6 | ACTN2 | actinin, alpha 2 |
| 6 | ADCY3 | adenylate cyclase 3 |
| 6 | CREB1 | cAMP responsive element binding protein 1 |
| 6 | MAPK3 | mitogen-activated protein kinase 3 |
| 6 | PLCB1 | phospholipase C, beta 1 (phosphoinositide-specific) |
| 6 | PLCB2 | phospholipase C, beta 2 |
| 6 | PLCB3 | phospholipase C, beta 3 (phosphatidylinositol-specific) |
| 6 | PRKCA | protein kinase C, alpha |
| 6 | PRKCG | protein kinase C, gamma |
| 6 | RAC1 | ras-related C3 botulinum toxin substrate 1 (rho family, GTP binding protein) |
| 5 | CACNA1C | calcium channel, voltage-dependent, L type, alpha 1C subunit |
| 5 | CACNB1 | calcium channel, voltage-dependent, beta 1 subunit |
| 5 | CACNB2 | calcium channel, voltage-dependent, beta 2 subunit |
| 5 | CACNB3 | calcium channel, voltage-dependent, beta 3 subunit |
| 5 | CACNB4 | calcium channel, voltage-dependent, beta 4 subunit |
| 5 | CAMK2A | calcium/calmodulin-dependent protein kinase II alpha |
| 5 | CAMK2B | calcium/calmodulin-dependent protein kinase II beta |
| 5 | CAMK2D | calcium/calmodulin-dependent protein kinase II delta |
| 5 | CAMK2G | calcium/calmodulin-dependent protein kinase II gamma |
| 5 | GRIA1 | glutamate receptor, ionotropic, AMPA 1 |
| 5 | GRIA2 | glutamate receptor, ionotropic, AMPA 2 |
| 5 | GRIN1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| 5 | GRIN2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A |
| 5 | GRIN2C | glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
| 5 | GRIN2D | glutamate receptor, ionotropic, N-methyl D-aspartate 2D |
| 5 | ITGA1 | integrin, alpha 1 |
| 5 | ITPR1 | inositol 1,4,5-trisphosphate receptor, type 1 |
| 5 | ITPR2 | inositol 1,4,5-trisphosphate receptor, type 2 |
| 5 | ITPR3 | inositol 1,4,5-trisphosphate receptor, type 3 |
| 5 | PLXNA1 | plexin A1 |
| 5 | PLXNA2 | plexin A2 |
| 5 | RAC2 | ras-related C3 botulinum toxin substrate 2 (rho family, GTP binding protein) |
Fig. 4.
Transcriptional regulation network centered on SP1 involves many genes of interest from pathways enriched in memory impairment. Green arrows indicate positive regulatory effects, red arrows indicate negative regulatory effects, and gray arrows indicate unspecified regulatory effects.
*Primary image generated with the MetaCore software (GeneGo, Inc.)
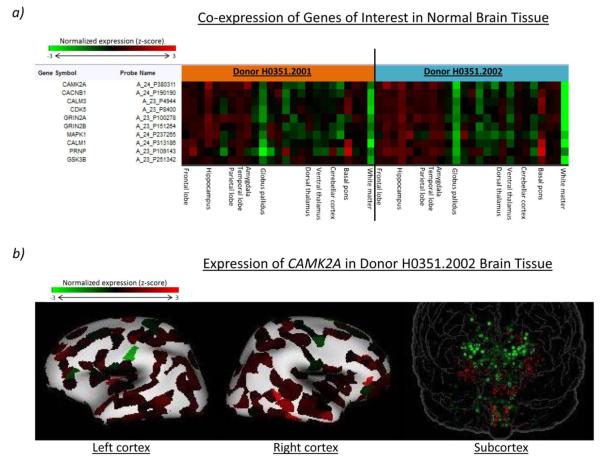
Finally, we used data from the Allen Human Brain Atlas to evaluate if the identified genes of interest exhibited co-expression in normal brain tissue. Through heat map visualization and correlational analysis, we identified a set of 10 key genes with strong co-expression (Pearson r > 0.7) across the major neuroanatomic regions of the brain (Figure 5a). In particular, 6 of these genes (CAMK2A, CACNB1, CALM1, CALM3, GRIN2A, and MAPK1) were highly-represented among our enriched pathways, while the other 4 genes (CDK5, GSK3B, GRIN2B, and PRNP) were constituents of our enriched pathways that were also known AD susceptibility genes found in the AlzGene database (http://www.alzgene.org/) (Bertram et al. 2007). An example of the cortical and subcortical expression patterns common to this gene set was also generated for one of these genes, CAMK2A (Figure 5b). These findings suggest further underlying functional relationships between MCI and AD disease risk and the pathogenesis of memory impairment.
Fig. 5.
Genes of interest in memory impairment exhibit similar expression profiles in postmortem normal brain tissue. a) Normalized, microarray-based expression profiles across 25 major neuroanatomic regions of the brain are provided for 10 key genes of interest. Moving from left to right on the heat map is analogous to moving from anterior to posterior regions first in the cortex, followed by subcortical areas and then the cerebellum and brainstem. The genes represented exhibit strong co-expression (Pearson r > 0.7) across the brain in data from two subjects. b) For the co-expressed gene set, a representative cortical and subcortical expression profile is shown for the CAMK2A gene.
*Images derived from the Allen Human Brain Atlas.
DISCUSSION
In this study, we used a psychometrically-optimized composite measure of overall episodic memory performance as a phenotype for GWAS in a sample of MCI and AD patients and controls. Then, through a genome-wide pathway analysis, we identified 27 canonical pathways with enrichment against this composite memory phenotype. These enriched pathways suggest that the genetic architecture of memory impairment in this sample spans both processes classically understood to be involved in normal memory consolidation as well as processes with broader roles in cognition and aging, such as those involving neuronal cell adhesion and inflammation.
Moreover, the results of this pathway enrichment study have valuable implications for the future. First, these results illuminate prime cellular, molecular, and genetic targets for future studies of normal and impaired memory states. Indeed, it should be emphasized that pathway-based approaches analyze genetic data in the context of its operative functional groups; as a result, pathway analysis findings are uniquely and naturally connected to the functional biology underlying complex phenotypes. This insight is vital for future investigations, given that pathway mechanisms are principal sources for developing strategies to diagnose, treat, and prevent complex disorders. It is also important to note that our analysis elucidated pathways with robust enrichment despite using GWA input data that included a relatively modest distribution of SNP-level phenotype associations. This insight affirms the potential of pathway-based analytical approaches to detect significant relationships that are otherwise concealed within single-SNP or single-gene analysis. Finally, the use of genome-wide pathway analysis in this study facilitated the detection of unexpected relationships with memory performance, including pathways not classically related to memory signaling, and subsequently, interesting transcriptional and expression networks. While targeted “candidate pathway” approaches have advantages, these unexpected relationships would not have been easily predicted as candidates for analysis based on prior knowledge.
At a functional level, the enriched pathways identified in this study present interesting biological implications in relation to memory impairment. In a sense, it might be expected that cellular and molecular processes classically understood as mediating memory consolidation would constitute a major part of the genetic architecture of memory impairment. However, the processes underlying memory consolidation are numerous and diverse, and to date it has not been clear which specific pathways are essential objects of the impact of genetic variants. The results of our enrichment analysis suggest prime potential components of this genetic architecture. In particular, we observed significant enrichment of pathways related to neurotransmitter receptor activation and downstream signaling. These pathways and their resultant calcium-mediated signaling are vital in converting short-term memories, which exist as axonal firing patterns, into long-lasting changes in synaptic strength (Sweatt 2011). As a parallel, it makes sense that a composite long-term potentiation pathway (comprising multiple processes leading to long-lasting increases in synaptic strength) would include genetic determinants related to memory performance. Finally, our results indicate the need for further exploration of long-term depression as a substrate for memory impairment, particularly given its proposed roles in mediating the cognitive effects of acute stress and the elimination of synapses in neurodegenerative diseases (Collingridge et al. 2010).
Meanwhile, other pathways enriched with regard to memory impairment in this study have previously been implicated in other roles involving neuronal development and cognition. There is an extensive literature on the roles of neuronal cell adhesion molecules (NCAMs) in susceptibility for schizophrenia, bipolar disorder, and autism-spectrum disorders (Corvin 2010). Genetic variants of NCAMs have also been associated with CSF biomarkers for AD (Han et al. 2010), and cell adhesion molecule pathways have exhibited enrichment in a GWA-based pathway analysis of AD susceptibility (Liu et al. 2012). In addition, expression of NCAMs in cholinergic neurons appears to increase susceptibility to AD-related neurodegeneration (Aisa et al. 2010), and there is emerging evidence of interactions among NCAMs, the MAPK pathway, and amyloid precursor protein (Chen and Dou 2012). More broadly, these findings suggest a prominent role for cell adhesion pathways in maintaining the processes of synaptic plasticity that are believed to underlie learning and memory (Ho et al. 2011).
Finally, it is interesting that pathways on axon guidance, including those involving functions of ephrins, semaphorins, and rho GTPases, were enriched in this study. Axon guidance pathways are key in forming guided neuronal network connections, and have been previously implicated in early neuronal development and associated genetic conditions (Izzi and Charron 2011). Together with these enrichment results, the proposed interaction between vascular and neuronal factors related to axon guidance (Arese et al. 2011) in relation to memory may be an important direction for further studies. In addition, given the complex interactions among brain cells and immune system functions, the immune-related pathways enriched in this study suggest additional candidates for modulation of memory and synaptic plasticity (Yirmiya and Goshen 2011). It may be particularly fruitful to examine immune mediators of memory dysfunction that exert influences independent from amyloid-related activation of microglia in AD (Emilio 2010).
A related perspective – and additional interesting targets for future investigations – can be achieved by examining the set of genes that were highly-represented across the enriched pathways in this study. Prominent groups of gene products represented in this set are particularly important in memory consolidation. For example, integrins, cadherins, and alpha-actinin are known to regulate neuronal cytoskeletal structure to mediate synaptic plasticity; further, these molecules are proposed to signal through MAPK cascades for localized protein synthesis at the specific dendrites being activated to precisely potentiate their synaptic connections (Sweatt 2009). Another important group of gene products is related to the calcium influx that follows neurotransmitter receptor activation at synapses: this calcium influx leads to activation of a signaling axis involving calmodulin, protein kinases (PKA, PKC-α, CAMKII subtypes, and CAMKIV), and transcription factors (CREB subtypes), among other molecules (Sweatt 2009). Overall, since the highly-represented genes from our data act in numerous pathways, our results demonstrate the importance of studying their gene variants within a pathway-based framework: in this context, variants of moderate individual effect sizes can nevertheless be identified as exerting strong and wide-ranging effects when juxtaposed with other meaningful variants in the same functional unit (Ramanan et al. 2012). Extensions of this pathway-based analytic framework will be extremely valuable in identifying localized effects of specific pathways on particular brain regions, particularly given that imaging correlates have been identified for loci with known effects on memory, such as the impact of KIBRA gene variants on hippocampal activation (Pawlowski and Huentelman 2011). Notably, innovative voxelwise SNP- (Stein et al. 2010a; Stein et al. 2010b) and gene-based (Hibar et al. 2011) imaging genetics approaches have been successfully employed in studies of AD, as has a novel method for generating multivariate “genetic components” for imaging analysis (Meda et al. 2012). These strategies will serve as rich foundations for future pathway-based imaging genetics analyses.
In addition, the network analyses in this study reinforce the notion that key genes related to memory impairment function coordinately. We found that a preponderance of the most highly-represented genes in our enriched pathways were constituents of a transcriptional regulation network driven by the SP1 transcription factor (Figure 4). The SP1 transcription factor has known binding regions in the promoters of genes related to beta-amyloid precursor protein (Yu et al. 2010; Rossner et al. 2006), tau protein (Santpere et al. 2006), and APOE. In particular, SP1 has been proposed as a regulator of APOE promoter activity in relation to two promoter polymorphisms with significant association to AD (Maloney et al. 2010). Given that networks of common regulation represent prime targets for identifying common functions, further investigation of the transcriptional network that we have identified may elucidate the as-yet-unknown mechanistic connections among APOE and other susceptibility loci, AD pathogenesis, and MCI- and AD-related memory impairment.
Finally, expression analysis using the Allen Human Brain Atlas revealed additional functional relationships among key genes. Since strong co-expression of a set of 10 key genes in the brain may indicate common modes of function, further study of this and other similar sets may be of great value. In addition, the co-expression of highly-represented genes from the enriched pathways in this study with known AD susceptibility genes suggests the possibility of significant crosstalk between AD pathogenesis and basic memory processes. While the data in the Allen Human Brain Atlas has several limitations, including a small number of subjects and the inclusion of only postmortem brain tissue from neuropsychologically- and neuropathologically-normal subjects, at present it is the only available resource which integrates multi-modal brain imaging data with whole-brain genome-wide expression data. As such, this and other functional annotation resources will be vital for identifying mechanistic connections between AD pathogenesis and memory impairment, including future efforts to quantitatively assess the significance of overlap between memory pathways and AD pathways. There are some notable limitations to the current study. First, a pathway analysis is only as good as the functional information underlying its pathway definitions. Importantly, some intragenic SNPs may not affect the function or expression of their assigned gene, while other SNPs may functionally impact distant genes or even multiple genes (Kapranov et al. 2007; Portin 2009). As functional annotation of the genome becomes more extensive, the power of pathway analyses will heighten. For this study, we used a collection of canonical pathways curated through expert review. While these pathway annotations are expected to have high accuracy, differences across pathway databases can lead to divergent enrichment analysis results (Elbers et al. 2009). For example, similarly-named pathways can have vastly different gene constituents, while distinctly-named pathways can nevertheless include significant gene overlap. As a result, an early discussion of pathway analysis methods recommended the use of multiple databases for each analysis (Cantor et al. 2010). While we have followed this recommendation for this analysis, future studies may benefit from formally assessing the relationships in biological coverage among the diverse pathways tested. In addition, at this time there is no gold standard for pathway-based study design. Indeed, different enrichment algorithms and different parameters, such as those guiding SNP-to-gene mapping, can impact analytical results (Gui et al. 2011). As such, pathway enrichment results benefit from further study using independent replication data sets and using alternative enrichment strategies. While differences across annotation resources, data sets, and analytical strategies may impact SNP- or gene-level statistics (Luo et al. 2010), legitimately-associated pathways will likely exhibit significant enrichment or strongly-trending signal across a healthy percentage of studies. Finally, while it is beyond the scope of this study, future efforts will benefit from examining key memory-implicated genes and gene sets for epistatic (gene-gene) interactions with each other and with APOE.
In addition, there are several caveats about the clinical setting for this study. First, the ADNI cohort represents a clinical trial population and is not a sample of the general population. As a result, the extent to which the present findings can be extended to account for episodic memory impairment in the general population remains to be determined. In addition, it is probable that the memory deficits in this study’s MCI and AD participants are at least partially driven by AD-related pathology. While using APOE ε4 allele status as a covariate in these analyses likely attenuated this effect, a better understanding of the pathways underlying normal memory and other pathologies than AD may be achieved through studies of normal cohorts and other memory-impaired populations without AD-related pathology. In particular, further exploration of the relationships among APOE genotype status, amyloid-β load and pathology, and cognition in normal adults (Kantarci et al. 2012; Buchman and Bennett 2012) may be especially fruitful. Additionally, meta-analytic approaches to achieve larger study sample sizes may reveal greater SNP-level phenotype associations which could impact the pathway enrichment results. Finally, while this study used a composite episodic memory score optimized on the basis of modern psychometric theory, similar pathway-based studies using other quantitative memory phenotypes may provide different sensitivity and specificity to fine-grained memory deficits and would potentially serve as a validation for the discoveries of pathways enriched against the phenotype used in this study.
Nevertheless, the present results provide several new insights into key functional pathways associated with memory deficits in older adults with MCI or AD and controls. Importantly, these results highlight numerous candidates for further explorations of the SNPs, genes, and gene sets underlying normal memory processes and memory impairment. Overall, these findings encourage further use of pathway-based genetic analyses of quantitative memory phenotypes as statistically-powerful vehicles for discovery and as bridges to underlying biological mechanisms.
ACKNOWLEDGEMENTS
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd.; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research provides funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation. Data management and the specific analyses reported here were supported by NSF IIS-1117335 (Shen), NIA R13 AG030995 (Mungas), NIA R01 AG19771 (Saykin), P30 AG10133 (Saykin/Ghetti), and R01 AG029672 (Crane).
Footnotes
The authors declare that they have no conflict of interest.
REFERENCES
- Aisa B, Gil-Bea FJ, Solas M, García-Alloza M, Chen CP, Lai MK, et al. Altered NCAM Expression Associated with the Cholinergic System in Alzheimer’s Disease. [10.3233/JAD-2010-1398] Journal of Alzheimer’s Disease. 2010;20(2):659–668. doi: 10.3233/JAD-2010-1398. [DOI] [PubMed] [Google Scholar]
- Aisen PS, Petersen RC, Donohue MC, Gamst A, Raman R, Thomas RG, et al. Clinical core of the Alzheimer’s disease neuroimaging initiative: Progress and plans. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2010;6(3):239–246. doi: 10.1016/j.jalz.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano JM, Cahill L. Sex influences on the neurobiology of learning and memory. Learning & Memory. 2009;16(4):248–266. doi: 10.1101/lm.918309. doi:10.1101/lm.918309. [DOI] [PubMed] [Google Scholar]
- Arese M, Serini G, Bussolino F. Nervous vascular parallels: axon guidance and beyond. [Research Support, Non-U.S. Gov’t, Review] Int J Dev Biol. 2011;55(4-5):439–445. doi: 10.1387/ijdb.103242ma. doi:10.1387/ijdb.103242ma. [DOI] [PubMed] [Google Scholar]
- Baranzini SE, Srinivasan R, Khankhanian P, Okuda DT, Nelson SJ, Matthews PM, et al. Genetic variation influences glutamate concentrations in brains of patients with multiple sclerosis. [Research Support, N.I.H., Extramural, Research Support, Non-U.S. Gov’t] Brain. 2010;133(9):2603–2611. doi: 10.1093/brain/awq192. doi:10.1093/brain/awq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeau EJ, Didic M, Joubert S, Guedj E, Koric L, Felician O, et al. Extent and Neural Basis of Semantic Memory Impairment in Mild Cognitive Impairment. [10.3233/JAD-2011-110989] Journal of Alzheimer’s Disease. 2011 doi: 10.3233/JAD-2011-110989. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. doi:10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57(1):289–300. [Google Scholar]
- Bertram L, Lill CM, Tanzi RE. The Genetics of Alzheimer Disease: Back to the Future. Neuron. 2010;68(2):270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. [10.1038/ng1934] Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. doi: http://www.nature.com/ng/journal/v39/n1/suppinfo/ng1934_S1.html. [DOI] [PubMed] [Google Scholar]
- Bookheimer S, Burggren A. APOE-4 Genotype and Neurophysiological Vulnerability to Alzheimer’s and Cognitive Aging. Annual Review of Clinical Psychology. 2009;5(1):343–362. doi: 10.1146/annurev.clinpsy.032408.153625. doi:doi:10.1146/annurev.clinpsy.032408.153625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman AS, Bennett DA. Amyloid pathology in persons with “normal” cognition. Neurology. 2012;78(4):228–229. doi: 10.1212/WNL.0b013e31824367c2. doi:10.1212/WNL.0b013e31824367c2. [DOI] [PubMed] [Google Scholar]
- Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86(1):6–22. doi: 10.1016/j.ajhg.2009.11.017. doi:S0002-9297(09)00532-1 [pii], 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee Seng K, En Yun L, Yudi P, Kee Seng C. The pursuit of genome-wide association studies: where are we now? [Article] Journal of Human Genetics. 2010;55(4):195–206. doi: 10.1038/jhg.2010.19. doi:10.1038/jhg.2010.19. [DOI] [PubMed] [Google Scholar]
- Chen K.-p., Dou F. Selective Interaction of Amyloid Precursor Protein with Different Isoforms of Neural Cell Adhesion Molecule. Journal of Molecular Neuroscience. 2012;46(1):203–209. doi: 10.1007/s12031-011-9578-3. doi:10.1007/s12031-011-9578-3. [DOI] [PubMed] [Google Scholar]
- Cockrell JR, Folstein MF. Mini-Mental State Examination (MMSE) Psychopharmacol Bull. 1988;24(4):689–692. [PubMed] [Google Scholar]
- Collingridge GL, Peineau S, Howland JG, Wang YT. Long-term depression in the CNS. [10.1038/nrn2867] Nat Rev Neurosci. 2010;11(7):459–473. doi: 10.1038/nrn2867. doi: http://www.nature.com/nrn/journal/v11/n7/suppinfo/nrn2867_S1.html. [DOI] [PubMed] [Google Scholar]
- Corder E, Saunders A, Strittmatter W, Schmechel D, Gaskell P, Small G, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. doi:10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Corvin AP. Neuronal cell adhesion genes: Key players in risk for schizophrenia, bipolar disorder and other neurodevelopmental brain disorders? Cell Adh Migr. 2010;4(4):511–514. doi: 10.4161/cam.4.4.12460. doi:12460 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane PK, Carle A, Gibbons LE, Insel P, Mackin RS, Gross A, et al. Development and assessment of a composite score for memory in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Brain Imaging Behav. 2011 doi: 10.1007/s11682-012-9186-z. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbers CC, van Eijk KR, Franke L, Mulder F, van der Schouw YT, Wijmenga C, et al. Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genetic Epidemiology. 2009;33(5):419–431. doi: 10.1002/gepi.20395. doi:10.1002/gepi.20395. [DOI] [PubMed] [Google Scholar]
- Emilio B. Microglia: Activation in acute and chronic inflammatory states and in response to cardiovascular dysfunction. The International Journal of Biochemistry & Cell Biology. 2010;42(10):1580–1585. doi: 10.1016/j.biocel.2010.07.005. doi:10.1016/j.biocel.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Fridley BL, Biernacka JM. Gene set analysis of SNP data: benefits, challenges, and future directions. [Research Support, N.I.H., Extramural, Review] Eur J Hum Genet. 2011;19(8):837–843. doi: 10.1038/ejhg.2011.57. doi:10.1038/ejhg.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman JJ, Bühlmann P. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics. 2007;23(8):980–987. doi: 10.1093/bioinformatics/btm051. doi:10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
- Gui H, Li M, Sham P, Cherny S. Comparisons of seven algorithms for pathway analysis using the WTCCC Crohn’s Disease dataset. BMC Research Notes. 2011;4(1):386. doi: 10.1186/1756-0500-4-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunstad J, Spitznagel MB, Luyster F, Cohen RA, Paul RH. Handedness and cognition across the healthy lifespan. Int J Neurosci. 2007;117(4):477–485. doi: 10.1080/00207450600773483. doi:10.1080/00207450600773483. [DOI] [PubMed] [Google Scholar]
- Han M-R, Schellenberg G, Wang L-S, Initiative, t. A. s. D. N. Genome-wide association reveals genetic effects on human Abeta42 and tau protein levels in cerebrospinal fluids: a case control study. BMC Neurology. 2010;10(1):90. doi: 10.1186/1471-2377-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Kohannim O, Jahanshad N, Saykin AJ, Shen L, et al. Voxelwise gene-wide association study (vGeneWAS): Multivariate gene-based association testing in 731 elderly subjects. Neuroimage. 2011;56(4):1875–1891. doi: 10.1016/j.neuroimage.2011.03.077. doi:10.1016/j.neuroimage.2011.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN. Genomewide Association Studies — Illuminating Biologic Pathways. New England Journal of Medicine. 2009;360(17):1699–1701. doi: 10.1056/NEJMp0808934. doi:doi:10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- Ho VM, Lee J-A, Martin KC. The Cell Biology of Synaptic Plasticity. Science. 2011;334(6056):623–628. doi: 10.1126/science.1209236. doi:10.1126/science.1209236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P. Statistical methods for pathway analysis of genome-wide data for association with complex genetic traits. Advances in genetics. 2010;72:141–179. doi: 10.1016/B978-0-12-380862-2.00007-2. doi:citeulike-article-id:8423729. [DOI] [PubMed] [Google Scholar]
- International HapMap, C. The International HapMap Project. [Multicenter Study, Research Support, Non-U.S. Gov’t, Research Support, U.S. Gov’t, P.H.S.] Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. doi:10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. [10.1038/nature03001] Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. doi: http://www.nature.com/nature/journal/v431/n7011/suppinfo/nature03001_S1.html. [DOI] [PubMed] [Google Scholar]
- Izzi L, Charron F. Midline axon guidance and human genetic disorders. Clinical Genetics. 2011;80(3):226–234. doi: 10.1111/j.1399-0004.2011.01735.x. doi:10.1111/j.1399-0004.2011.01735.x. [DOI] [PubMed] [Google Scholar]
- Jack CR, Lowe VJ, Weigand SD, Wiste HJ, Senjem ML, Knopman DS, et al. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer’s disease: implications for sequence of pathological events in Alzheimer’s disease. Brain. 2009 doi: 10.1093/brain/awp062. doi:10.1093/brain/awp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, La Rue A, Hermann BP, Xu G, Koscik RL, Jonaitis EM, et al. The effect of TOMM40 poly-T length on gray matter volume and cognition in middle-aged persons with APOE ε3/ε3 genotype. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2011;7(4):456–465. doi: 10.1016/j.jalz.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarci K, Lowe V, Przybelski SA, Weigand SD, Senjem ML, Ivnik RJ, et al. APOE modifies the association between Aβ load and cognition in cognitively normal older adults. Neurology. 2012;78(4):232–240. doi: 10.1212/WNL.0b013e31824365ab. doi:10.1212/WNL.0b013e31824365ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. [Research Support, N.I.H., Extramural, Research Support, Non-U.S. Gov’t, Review] Nat Rev Genet. 2007;8(6):413–423. doi: 10.1038/nrg2083. doi:10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kim SY, Volsky DJ. PAGE: parametric analysis of gene set enrichment. [Comparative Study, Research Support, N.I.H., Extramural, Research Support, U.S. Gov’t, P.H.S.] BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. doi:10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezak MD. Neuropsychological assessment. Oxford University Press; 2004. [Google Scholar]
- Li J, Humphreys K, Heikkinen T, Aittomäki K, Blomqvist C, Pharoah P, et al. A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Research and Treatment. 2011;126(3):717–727. doi: 10.1007/s10549-010-1172-9. doi:10.1007/s10549-010-1172-9. [DOI] [PubMed] [Google Scholar]
- Liu G, Jiang Y, Wang P, Feng R, Jiang N, Chen X, et al. Cell adhesion molecules contribute to Alzheimer’s disease: multiple pathway analyses of two genome-wide association studies. Journal of Neurochemistry. 2012;120(1):190–198. doi: 10.1111/j.1471-4159.2011.07547.x. doi:10.1111/j.1471-4159.2011.07547.x. [DOI] [PubMed] [Google Scholar]
- Luciano M, Hansell NK, Lahti J, Davies G, Medland SE, Räikkönen K, et al. Whole genome association scan for genetic polymorphisms influencing information processing speed. Biological Psychology. 2011;86(3):193–202. doi: 10.1016/j.biopsycho.2010.11.008. doi:10.1016/j.biopsycho.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Peng G, Zhu Y, Dong H, Amos CI, Xiong M. Genome-wide gene and pathway analysis. Eur J Hum Genet. 2010;18(9):1045–1053. doi: 10.1038/ejhg.2010.62. doi:ejhg201062 [pii], 10.1038/ejhg.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney B, Ge YW, Petersen RC, Hardy J, Rogers JT, Perez-Tur J, et al. Functional characterization of three single-nucleotide polymorphisms present in the human APOE promoter sequence: Differential effects in neuronal cells and on DNA-protein interactions. [Research Support, N.I.H., Extramural, Research Support, Non-U.S. Gov’t] Am J Med Genet B Neuropsychiatr Genet. 2010;153B(1):185–201. doi: 10.1002/ajmg.b.30973. doi:10.1002/ajmg.b.30973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda SA, Narayanan B, Liu J, Perrone-Bizzozero NI, Stevens MC, Calhoun VD, et al. A large scale multivariate parallel ICA method reveals novel imaging-genetic relationships for Alzheimer’s disease in the ADNI cohort. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2011.12.076. (0), doi:10.1016/j.neuroimage.2011.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohs RC, Knopman D, Petersen RC, Ferris SH, Ernesto C, Grundman M, et al. Development of cognitive instruments for use in clinical trials of antidementia drugs: additions to the Alzheimer’s Disease Assessment Scale that broaden its scope. The Alzheimer’s Disease Cooperative Study. [Research Support, U.S. Gov’t, P.H.S.] Alzheimer Dis Assoc Disord. 1997;11(Suppl 2):S13–21. [PubMed] [Google Scholar]
- Nam D, Kim J, Kim S-Y, Kim S. GSA-SNP: a general approach for gene set analysis of polymorphisms. Nucleic Acids Research. 2010;38(suppl 2):W749–W754. doi: 10.1093/nar/gkq428. doi:10.1093/nar/gkq428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negash S, Bennett DA, Wilson RS, Schneider JA, Arnold SE. Cognition and neuropathology in aging: multidimensional perspectives from the Rush Religious Orders Study and Rush Memory And Aging Project. [Review] Curr Alzheimer Res. 2011;8(4):336–340. doi: 10.2174/156720511795745302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski TL, Huentelman MJ. Identification of a Common Variant Affecting Human Episodic Memory Performance Using a Pooled Genome-Wide Association Approach: A Case Study of Disease Gene Identification. In: DiStefano JK, editor. Methods in Molecular Biology. Vol. 700. Humana Press; 2011. pp. 261–269. [DOI] [PubMed] [Google Scholar]
- Penrod NM, Cowper-Sal-lari R, Moore JH. Systems genetics for drug target discovery. [Research Support, N.I.H., Extramural] Trends Pharmacol Sci. 2011;32(10):623–630. doi: 10.1016/j.tips.2011.07.002. doi:10.1016/j.tips.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portin P. The elusive concept of the gene. [Review] Hereditas. 2009;146(3):112–117. doi: 10.1111/j.1601-5223.2009.02128.x. doi:10.1111/j.1601-5223.2009.02128.x. [DOI] [PubMed] [Google Scholar]
- Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. [Research Support, N.I.H., Extramural, Research Support, Non-U.S. Gov’t, Research Support, U.S. Gov’t, P.H.S.] PLoS One. 2009;4(8):e6501. doi: 10.1371/journal.pone.0006501. doi:10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. [Research Support, N.I.H., Extramural, Research Support, Non-U.S. Gov’t] Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet. 2012;28(7):323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve BB, Hays RD, Bjorner JB, Cook KF, Crane PK, Teresi JA, et al. Psychometric evaluation and calibration of health-related quality of life item banks: plans for the Patient-Reported Outcomes Measurement Information System (PROMIS). [Research Support, N.I.H., Extramural] Med Care. 2007;45(5 Suppl 1):S22–31. doi: 10.1097/01.mlr.0000250483.85507.04. doi:10.1097/01.mlr.0000250483.85507.04. [DOI] [PubMed] [Google Scholar]
- Rey A. L’examen clinique en psychologie. Presses Universitaires de France; Paris: 1964. [Google Scholar]
- Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS, et al. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease. Pharmacogenomics J. 2010;10(5):375–384. doi: 10.1038/tpj.2009.69. doi: http://www.nature.com/tpj/journal/v10/n5/suppinfo/tpj200969s1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossner S, Sastre M, Bourne K, Lichtenthaler SF. Transcriptional and translational regulation of BACE1 expression--implications for Alzheimer’s disease. [Research Support, Non-U.S. Gov’t, Review] Prog Neurobiol. 2006;79(2):95–111. doi: 10.1016/j.pneurobio.2006.06.001. doi:10.1016/j.pneurobio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Santpere G, Nieto M, Puig B, Ferrer I. Abnormal Sp1 transcription factor expression in Alzheimer disease and tauopathies. [Comparative Study, Research Support, Non-U.S. Gov’t] Neurosci Lett. 2006;397(1-2):30–34. doi: 10.1016/j.neulet.2005.11.062. doi:10.1016/j.neulet.2005.11.062. [DOI] [PubMed] [Google Scholar]
- Sawcer S, Hellenthal G, Pirinen M, Spencer C, Patsopoulos N, Moutsianas L, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. [10.1038/nature10251] Nature. 2011;476(7359):214–219. doi: 10.1038/nature10251. doi:10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 2010;6(3):265–273. doi: 10.1016/j.jalz.2010.03.013. doi:S1552-5260(10)00082-8 [pii], 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE. Molecular networks as sensors and drivers of common human diseases. [Review] Nature. 2009;461(7261):218–223. doi: 10.1038/nature08454. doi:10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. [Research Support, N.I.H., Extramural, Research Support, Non-U.S. Gov’t, Research Support, U.S. Gov’t, P.H.S.] Neuroimage. 2010;53(3):1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. doi:10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan CD, Shen L, West JD, Wishart HA, Flashman LA, Rabin LA, et al. Genetic pathway-based hierarchical clustering analysis of older adults with cognitive complaints and amnestic mild cognitive impairment using clinical and neuroimaging phenotypes. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2010;153B(5):1060–1069. doi: 10.1002/ajmg.b.31078. doi:10.1002/ajmg.b.31078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Hua X, Lee S, Ho AJ, Leow AD, Toga AW, et al. Voxelwise genome-wide association study (vGWAS). [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] Neuroimage. 2010a;53(3):1160–1174. doi: 10.1016/j.neuroimage.2010.02.032. doi:10.1016/j.neuroimage.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Hua X, Morra JH, Lee S, Hibar DP, Ho AJ, et al. Genome-wide analysis reveals novel genes influencing temporal lobe structure with relevance to neurodegeneration in Alzheimer’s disease. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] Neuroimage. 2010b;51(2):542–554. doi: 10.1016/j.neuroimage.2010.02.068. doi:10.1016/j.neuroimage.2010.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proceedings of the National Academy of Sciences. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. doi:10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. Mechanisms of Memory. 2nd Edition Academic Press; 2009. [Google Scholar]
- Sweatt JD. Neuroscience. Creating stable memories. Science. 2011;331(6019):869–870. doi: 10.1126/science.1202283. doi:331/6019/869 [pii], 10.1126/science.1202283. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. [10.1038/nrg2884] Nat Rev Genet. 2010;11(12):843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- Wechsler D. Wechsler Memory Scale -- Revised. Psychological Association; New York: 1987. [Google Scholar]
- Weiner MW, Aisen PS, Jack CR, Jr., Jagust WJ, Trojanowski JQ, Shaw L, et al. The Alzheimer’s disease neuroimaging initiative: progress report and future plans. Alzheimers Dement. 2010;6(3):202–211. e207. doi: 10.1016/j.jalz.2010.03.007. doi:S1552-5260(10)00067-1 [pii], 10.1016/j.jalz.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman EM, Pankratz ND, Choi Y, Rothstein JH, Faber KM, Cheng R, et al. Genome-Wide Association of Familial Late-Onset Alzheimer’s Disease Replicates BIN1 and CLU and Nominates CUGBP2 in Interaction with APOE. PLoS Genet. 2011;7(2):e1001308. doi: 10.1371/journal.pgen.1001308. doi:10.1371/journal.pgen.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yirmiya R, Goshen I. Immune modulation of learning, memory, neural plasticity and neurogenesis. Brain, Behavior, and Immunity. 2011;25(2):181–213. doi: 10.1016/j.bbi.2010.10.015. doi:10.1016/j.bbi.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Yu HT, Chan WW, Chai KH, Lee CW, Chang RC, Yu MS, et al. Transcriptional regulation of human FE65, a ligand of Alzheimer’s disease amyloid precursor protein, by Sp1. [Research Support, Non-U.S. Gov’t] J Cell Biochem. 2010;109(4):782–793. doi: 10.1002/jcb.22457. doi:10.1002/jcb.22457. [DOI] [PubMed] [Google Scholar]
- Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet. 2010;86(4):581–591. doi: 10.1016/j.ajhg.2010.02.020. doi:S0002-9297(10)00102-3 [pii], 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]