Abstract
The Rat Genome Database (RGD) was started >10 years ago to provide a core genomic resource for rat researchers. Currently, RGD combines genetic, genomic, pathway, phenotype and strain information with a focus on disease. RGD users are provided with access to structured and curated data from the molecular level through the organismal level. Those users access RGD from all over the world. End users are not only rat researchers but also researchers working with mouse and human data. Translational research is supported by RGD’s comparative genetics/genomics data in disease portals, in GBrowse, in VCMap and on gene report pages. The impact of RGD also goes beyond the traditional biomedical researcher, as the influence of RGD reaches bioinformaticians, tool developers and curators. Import of RGD data into other publicly available databases expands the influence of RGD to a larger set of end users than those who avail themselves of the RGD website. The value of RGD continues to grow as more types of data and more tools are added, while reaching more types of end users.
Keywords: database, genome, rat, disease, human
INTRODUCTION
The Rat Genome Database (RGD, http://rgd.mcw.edu) is the model organism database for the laboratory rat, Rattus norvegicus. The rat is a widely studied animal model for physiology, pharmacology, toxicology, nutrition, behavior, immunology and disease [1, 2]. The primary goal of RGD is to provide support for researchers using the rat as a model organism to understand human physiology and disease. To provide data for translational research, RGD manually curates disease and molecular pathway literature for rat, mouse and human and mammalian phenotype literature for rat and human. Imported data for gene ontology, drug–gene interactions, pathways and mammalian phenotype fill out the database to give broad coverage of relevant information across all three species. Behind the data, RGD represents an infrastructure of software tools and ontology development that supports both the display and analysis of the data.
WHAT IS THE VALUE OF HAVING DATA AVAILABLE IN RGD?
The inherent value of information in a database is the convenience of having accumulated data in an easy-to-access location and format. The realized value to the database user is the time savings because the data is collected and displayed by a trained dedicated team of curators and developers. Information in the database is presented in an organized manner with controlled vocabularies and ontologies. The result is the ability of the database user to access condensed, filtered data sets that would take far more time to accumulate and assimilate if curators and automated pipelines had not collected them into the database [3]. Having rat, mouse and human data in the same place makes it easy to compare disease phenotypes, genes and functional annotations across the three species.
A secondary, but important, access to RGD data is the display of RGD-generated annotations on other publicly available websites. Major databases that store and display RGD annotations are the Gene Ontology Consortium [4], NCBI Gene [5], Ensembl [6], UniProt-GOA [7] and UCSC Genome Bioinformatics [8]. Sharing data with other databases exposes more researchers to RGD annotations and offers alternate routes to the RGD website as noted earlier in the text. Conversely, RGD imports data from and displays links to an expanding array of other databases, such as the Comparative Toxicogenomics Database (CTD) [9], the Kyoto Encyclopedia of Genes and Genomes (KEGG) [9] and the Pathway Interaction Database (PID) [10].
Beyond the convenience of data presentation and access, RGD provides software tools for data analysis. Efficient data search, ontology browser and genome browser tools [11, 12] are provided to find the information that the database user seeks. Analysis tools (Gene Annotator, RatMine, SNPlotyper and VCMap) [13 14] allow manipulation of data sets gathered at RGD to make qualitative and quantitative assessments.
WHO USES THE RAT GENOME DATABASE AND FOR WHAT PURPOSES?
For many years, RGD has been and continues to be the key repository for rat gene, quantitative trait locus (QTL) and strain data. As a measure of what type of researchers use these data, where the researchers are located and how they use the database, we have used a Google Analytics analysis of the RGD website and a manual analysis of publications in PubMed Central that cite RGD. Demographics, website use behavior, website traffic sources and context of citations have all been used to evaluate the use of RGD.
RGD users reside all over the globe, with the largest number being in the United States (∼45%), followed by Europe (∼25%) and Asia (∼25%). Use increases at an annual rate of ∼7%. Half of all visits to RGD originate in a general internet search engine like Google. In all, 15–20% of visits consist of users who come directly to the RGD website by way of bookmarks or manual URL entry. The majority of the remaining visits to RGD come via referring links at the NCBI (National Center for Biotechnology Information) [15], HGNC (HUGO Gene Nomenclature Committee) [16] and Ensembl [6] websites.
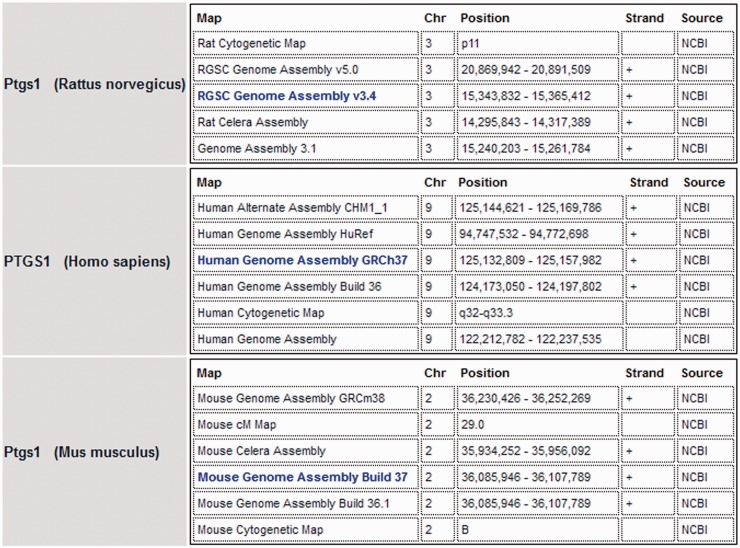
One might assume that researchers using rat as an experimental model would be the primary users of RGD. Based on articles in PubMed Central that cite ‘Rat Genome Database’, this is true for ∼40% of the returned titles. It is also true that 40% of the genetics articles involve studies with some combination of rat, mouse and human genes [17–19]. Those articles with human and rat and/or mouse data are examples of basic translational research. The authors reporting rat and mouse data provide insights and discoveries that may be applied to human research or clinical practice. Likewise, articles with human data may provide information that provokes new ideas or approaches in the study of disease or physiology in animal models. The genetic rat data at RGD allows comparison with both mouse and human without leaving the database. Every rat gene report page at RGD has comparative genomics data for the corresponding mouse and human orthologs (Figure 1). At a glance the database user can see the base pair coordinates or cytogenetic positions on the specific chromosomes that contain the gene and orthologs in question. Similarly, the mouse and human gene report pages at RGD have comparative genomics data for the corresponding two orthologs.
Figure 1:
Comparative map data. Genomic map data for prostaglandin synthase 1 in rat, human and mouse. The assemblies shown in bold font are the current reference assemblies. Alternate assemblies (e.g. previous assemblies and Celera assemblies) and alternate map data (cytogenetic and genetic maps) are also shown for each species.
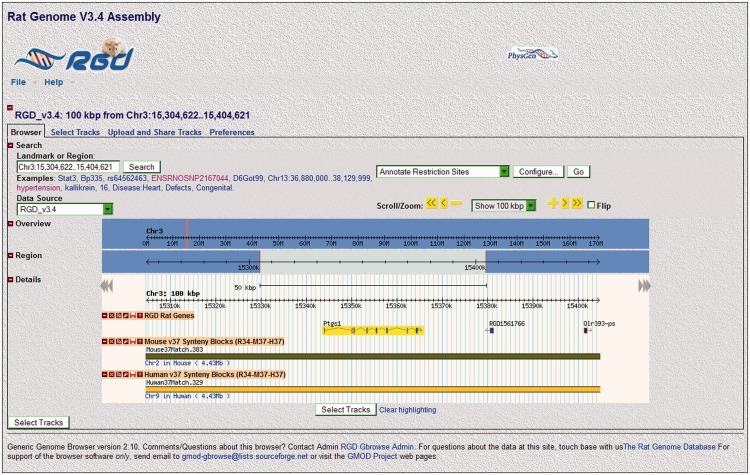
The genome tool GBrowse [12, 20–22] affords another opportunity to analyze and compare the genetics/genomics of rat, mouse and human orthologs. RGD has three interconnected GBrowse tools specifically for rat, mouse and human genome assemblies. The rat version of GBrowse can show subsets of rat genes, QTLs and single-nucleotide polymorphisms (SNPs). Rat GBrowse also has human and mouse synteny blocks to allow quick comparison of homologous segments between rat, mouse and human chromosomes (Figure 2). The user can select one of the synteny tracks and switch the viewer to mouse or human GBrowse via a pop-up window and then view the other two species as synteny blocks.
Figure 2:
Rat GBrowse. The rat Ptgs1 gene and two other genes are shown on chromosome 3 with the mouse and human synteny blocks from chromosomes 2 and 9, respectively, which indicates where the human and mouse orthologs of these particular rat genes are located.
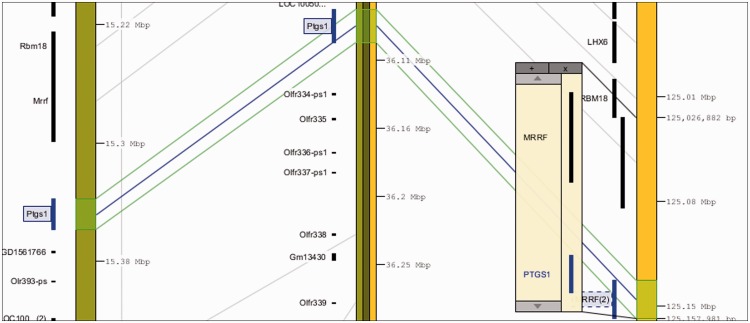
A third comparative genomics feature at RGD is VCMap (Figure 3) [14], which is a tool that compares genomic positions of genes simultaneously between different species, including rat, mouse and human. It gives a finer view than GBrowse of syntenic regions between species. The relative locations of numerous genes can be viewed in one window and compared across species.
Figure 3:
VCMap tool. A view of homologous regions of rat, mouse and human chromosomes containing the Ptgs1/PTGS1 gene and its syntenic neighbors. VCMap also contains genetic/genomic maps of other vertebrates (cow, chicken, horse and pig).
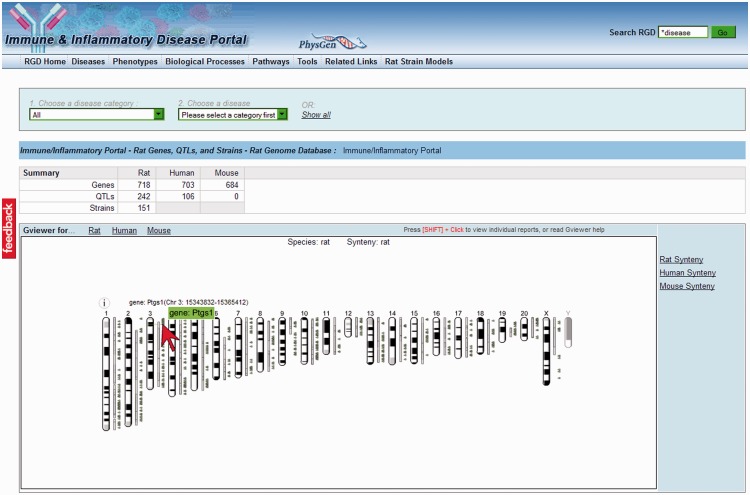
Traditionally, the laboratory rat has been used as a model system for human disease. That continues to be the case and is the driving force behind the creation of RGD disease portals. Currently, RGD has seven disease portals: Cancer, Cardiovascular Disease, Diabetes, Immune and Inflammatory Disease, Neurological Disease, Obesity/Metabolic Syndrome and Respiratory Disease Portals. The disease curation done at RGD covers disease models in rat and mouse, as well as disease data generated by research with human subjects. Disease–gene annotations are not only made to the species from which the data originated but also to the orthologous genes in the other two species. That allows users to find significant relevant data from the other two species while looking specifically at data in rat, mouse or human. Disease data for rat QTLs, human QTLs and rat strains are also found in the disease portals. The GViewer within the portal home page shows the genomic locations of all genes and QTLs associated with the chosen disease (Figure 4) [23]. The view toggles between rat, mouse and human genomes, with an option to see homologous syntenic views in the other two species.
Figure 4:
The Immune & Inflammatory Disease Portal is the latest of the RGD disease portals. Rat, human and mouse genes, rat and human QTLs and rat strains associated with immune and inflammation-related disease can be accessed through this portal home page. The GViewer shows chromosomal locations of all genes (gray triangles) and QTLs (light gray bars) in the portal. The location of an individual gene (Ptgs1) is shown here by mousing over (arrowhead) the appropriate triangle adjacent to chromosome 3.
Appropriate to RGD’s focus on disease data for genes, QTLs and strains, the articles that cite RGD most often are those concerned with disease research (33% of articles in PubMed Central from January 2010 to December 2012, examples: [24–26]). Those users cover a range of disease research from QTLs to strain models to gene targets. They use RGD as a source of SNP, simple sequence length polymorphisms (SSLP), QTL and gene data.
An additional important use of RGD data occurs via the FTP site [27]. All of the data found on the RGD website can be downloaded in bulk from the FTP site. The files available for download include descriptive information for genes, QTLs and strains, functional gene annotations, SSLPs, SNPs, ontology term files and more. Bulk downloads of data allow bioinformaticians to make analyses with their own software tools or other tools that are not available at RGD. Similar to widespread use of the rest of the RGD website, users from all over the world download files from the FTP site. Some of the downloaded data are used for public display of RGD data at sites such as NCBI, Ensembl and RIKEN [28]. Other users of the FTP site include many universities, research institutes and pharmaceutical companies in the United States, Europe and Asia.
One of the tasks of RGD, in addition to data collection and presentation, is the responsibility for naming rat genes, QTLs and strains in a systematic manner. The fact that, during the last year and a half, references to nomenclature of rat genes, QTLs and strains [29–32] almost equal references to RGD QTL or gene data emphasizes the importance of official nomenclature. The importance of consistency in the naming of genetic elements and other data objects requires a stable source of that nomenclature, and RGD provides that service. The nomenclature of genes is particularly important in translational research because it helps keep track of orthologs between species. Gene nomenclature of rat, mouse and human genes are kept aligned by the coordinated work of the nomenclature committees of rat (RGD), mouse (MGD [33]) and human (HGNC [16]).
SUMMARY
For >10 years, RGD has been, and continues to be, a core genomic resource for rat researchers. During that time, RGD has expanded several elements. It has increased its data coverage [13], developed ontologies [34, 35] and has developed increasingly sophisticated software tools for both end users and curators [11, 36]. RGD provides numerous tools and data for rat, mouse and human to provide support for translational research. FTP files provide access to bulk RGD data for personalized analysis by users. The number of users of the database increases annually, accompanied by a broadening of user identity. The growing value of RGD comes not only from its data but also from its involvement in biological nomenclature, ontology development and software tool development.
Key points.
RGD is a global resource of biomedical data and analysis tools.
RGD serves a diverse user base.
RGD data support translational research.
Funding
The National Heart, Lung, and Blood Institute on behalf of the National Institutes of Health (HL64541). Funding for open access charge: The National Heart, Lung and Blood Institute on behalf of the National Institutes of Health (HL64541).
Biographies
Stanley J. F. Laulederkind is a research scientist in the Human & Molecular Genetics Center at the Medical College of Wisconsin. He is currently involved in disease-gene literature curation and ontology development for RGD.
G. Thomas Hayman is a research scientist in the Human & Molecular Genetics Center at the Medical College of Wisconsin. He is presently engaged in disease-gene and physiological literature curation for RGD.
Shur-Jen Wang is a research scientist in the Human & Molecular Genetics Center at the Medical College of Wisconsin. She is currently involved in disease-gene and phenotype-QTL-strain literature curation.
Jennifer R. Smith is a curator in the Human & Molecular Genetics Center at the Medical College of Wisconsin. She currently has primary responsibility for ontology development and education/outreach at RGD.
Timothy F. Lowry is a research scientist in the Human & Molecular Genetics Center at the Medical College of Wisconsin. He is presently engaged in disease-gene curation and gene nomenclature for RGD.
Rajni Nigam is a curator in the Human & Molecular Genetics Center at the Medical College of Wisconsin. She is responsible for the incorporation and nomenclature of rat strains and QTLs at RGD.
Victoria Petri is a research scientist in the Human & Molecular Genetics Center at the Medical College of Wisconsin. She is currently involved in Pathway Portal development for RGD.
Jeff de Pons is a bioinformatics manager in the Human & Molecular Genetics Center at the Medical College of Wisconsin. He currently leads the software development team for RGD.
Melinda R. Dwinell is an Assistant Professor of Physiology in the Human & Molecular Genetics Center at the Medical College of Wisconsin. Her current focus is on integration of physiological data using multiple ontologies.
Mary Shimoyama is an Assistant Professor of Surgery-Cardiothoracic in the Human & Molecular Genetics Center at the Medical College of Wisconsin. She is currently co-investigator of the Rat Genome Database.
Diane H. Munzenmaier is an Assistant Professor of Physiology in the Human & Molecular Genetics Center at the Medical College of Wisconsin. Her current focus is on integration of physiological pathways with genomics.
Elizabeth A. Worthey is an Assistant Professor of Pediatrics in the Human & Molecular Genetics Center at the Medical College of Wisconsin. Her current focus is the clinical and translational use of genomic data.
Howard J. Jacob is a Professor of Physiology and Director of the Human & Molecular Genetics Center at the Medical College of Wisconsin. He is principal investigator of the Rat Genome Database.
References
- 1.Jacob HJ. Functional genomics and rat models. Genome Res. 1999;9:1013–16. doi: 10.1101/gr.9.11.1013. [DOI] [PubMed] [Google Scholar]
- 2.Aitman TJ, Critser JK, Cuppen E, et al. Progress and prospects in rat genetics: a community view. Nat Genet. 2008;40:516–22. doi: 10.1038/ng.147. [DOI] [PubMed] [Google Scholar]
- 3.Shimoyama M, Hayman GT, Laulederkind SJ, et al. The rat genome database curators: who, what, where, why. PLoS Comput Biol. 2009;5:e1000582. doi: 10.1371/journal.pcbi.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. GO Consortium. http://www.geneontology.org/ (15 February 2013, date last accessed)
- 5. NCBI Gene. http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene (15 February 2013, date last accessed)
- 6. Ensembl. http://www.ensembl.org/index.html (15 February 2013, date last accessed)
- 7. UniProt-GOA. http://www.ebi.ac.uk/GOA/rat_release.html (15 February 2013, date last accessed)
- 8. UCSC Genome Browser. http://genome.ucsc.edu/cgi-bin/hgGateway?hgsid=286942711&clade=mammal&org=Rat&db=0 (15 February 2013, date last accessed)
- 9. CTD. http://ctdbase.org/ (15 February 2013, date last accessed)
- 10. PID. http://pid.nci.nih.gov/index.shtml (15 February 2013, date last accessed)
- 11.Laulederkind SJ, Tutaj M, Shimoyama M, et al. Ontology searching and browsing at the Rat Genome Database. Database. 2012;2012:bas016. doi: 10.1093/database/bas016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laulederkind SJ, Hayman GT, Wang SJ, et al. Exploring genetic, genomic, and phenotypic data at the rat genome database. Curr Protoc Bioinformatics. 2012 doi: 10.1002/0471250953.bi0114s40. Chapter 1: Unit 1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimoyama M, Smith JR, Hayman T, et al. RGD: a comparative genomics platform. Hum Genomics. 2011;5:124–9. doi: 10.1186/1479-7364-5-2-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. RGD Genome Tools. http://rgd.mcw.edu/wg/tool-menu?100 (15 February 2013, date last accessed)
- 15. NCBI. http://www.ncbi.nlm.nih.gov/guide/ (15 February 2013, date last accessed)
- 16. HGNC. http://www.genenames.org/ (15 February 2013, date last accessed)
- 17.Martin-Rufian M, Tosina M, Campos-Sandoval JA, et al. Mammalian glutaminase gls2 gene encodes two functional alternative transcripts by a surrogate promoter usage mechanism. PLoS One. 2012;7:e38380. doi: 10.1371/journal.pone.0038380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDermott-Roe C, Ye J, Ahmed R, et al. Endonuclease G is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature. 2011;478:114–18. doi: 10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fulton DL, Denarier E, Friedman HC, et al. Towards resolving the transcription factor network controlling myelin gene expression. Nucleic Acids Res. 2011;39:7974–91. doi: 10.1093/nar/gkr326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Twigger SN, S Smith J, Zuniga-Meyer A, et al. Exploring phenotypic data at the rat genome database. Curr Protoc Bioinformatics. 2006 doi: 10.1002/0471250953.bi0114s14. Chapter 1:Unit 1.14. [DOI] [PubMed] [Google Scholar]
- 21.Donlin MJ. Using the generic genome browser (GBrowse) Curr Protoc Bioinformatics. 2009 doi: 10.1002/0471250953.bi0909s28. Chapter 9:Unit 9.9. [DOI] [PubMed] [Google Scholar]
- 22. Rat GBrowse. http://rgd.mcw.edu/fgb2/gbrowse/rgd_904/ (15 February 2013, date last accessed)
- 23. Immune & Inflammatory Disease Portal. http://rgd.mcw.edu/rgdCuration/?module=portal&func=show&name=immune (15 February 2013, date last accessed)
- 24.Northrup E, Zschemisch NH, Eisenblatter R, et al. The ter mutation in the rat Dnd1 gene initiates gonadal teratomas and infertility in both genders. PLoS One. 2012;7:e38001. doi: 10.1371/journal.pone.0038001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chabod M, Pedros C, Lamouroux L, et al. A spontaneous mutation of the rat Themis gene leads to impaired function of regulatory T cells linked to inflammatory bowel disease. PLoS Genet. 2012;8:e1002461. doi: 10.1371/journal.pgen.1002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders J, Haag JD, Samuelson DJ. Physical confirmation and mapping of overlapping rat mammary carcinoma susceptibility QTLs, Mcs2 and Mcs6. PLoS One. 2011;6:e19891. doi: 10.1371/journal.pone.0019891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. RGD FTP site. ftp://rgd.mcw.edu/pub/ (15 February 2013, date last accessed)
- 28. RIKEN. https://database.riken.jp/sw/en/Home/cria254s94i/ (15 February 2013, date last accessed)
- 29.Fundel K, Zimmer R. Gene and protein nomenclature in public databases. BMC Bioinformatics. 2006;7:372. doi: 10.1186/1471-2105-7-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonner MK, Poole DS, Xu T, et al. Mitotic spindle proteomics in Chinese hamster ovary cells. PLoS One. 2011;6:e20489. doi: 10.1371/journal.pone.0020489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmes RS, Cox LA, VandeBerg JL. Comparative studies of mammalian acid lipases: evidence for a new gene family in mouse and rat (Lipo) Comp Biochem Physiol Part D Genomics Proteomics. 2010;5:217–26. doi: 10.1016/j.cbd.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blankenhorn EP, Cort L, Greiner DL, et al. Virus-induced autoimmune diabetes in the LEW.1WR1 rat requires Iddm14 and a genetic locus proximal to the major histocompatibility complex. Diabetes. 2009;58:2930–8. doi: 10.2337/db09-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouse Genome Informatics. http://www.informatics.jax.org/ (15 February 2013, date last accessed)
- 34.Shimoyama M, Nigam R, McIntosh LS, et al. Three ontologies to define phenotype measurement data. Front Genet. 2012;3:87. doi: 10.3389/fgene.2012.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petri V, Shimoyama M, Hayman GT, et al. The rat genome database pathway portal. Database. 2011;2011:bar010. doi: 10.1093/database/bar010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laulederkind SJ, Shimoyama M, Hayman GT, et al. The rat genome database curation tool suite: a set of optimized software tools enabling efficient acquisition, organization, and presentation of biological data. Database. 2011;2011:bar002. doi: 10.1093/database/bar002. [DOI] [PMC free article] [PubMed] [Google Scholar]