Abstract
Memory deficits in Drosophila nalyot mutants suggest that the Myb family transcription factor Adf-1 is an important regulator of developmental plasticity in the brain. However, the cellular functions for this transcription factor in neurons or molecular mechanisms by which it regulates plasticity remain unknown. Here, we use in vivo 3D reconstruction of identifiable larval motor neuron dendrites to show that Adf-1 is required cell autonomously for dendritic development and activity-dependent plasticity of motor neurons downstream of CaMKII. Adf-1 inhibition reduces dendrite growth and neuronal excitability, and results in motor deficits and altered transcriptional profiles. Surprisingly, analysis by comparative chromatin immunoprecipitation followed by sequencing (ChIP-Seq) of Adf-1, RNA Polymerase II (Pol II), and histone modifications in Kc cells shows that Adf-1 binding correlates positively with high Pol II-pausing indices and negatively with active chromatin marks such as H3K4me3 and H3K27ac. Consistently, the expression of Adf-1 targets Staufen and Fasciclin II (FasII), identified through larval brain ChIP-Seq for Adf-1, is negatively regulated by Adf-1, and manipulations of these genes predictably modify dendrite growth. Our results imply mechanistic interactions between transcriptional and local translational machinery in neurons as well as conserved neuronal growth mechanisms mediated by cell adhesion molecules, and suggest that CaMKII, Adf-1, FasII, and Staufen influence crucial aspects of dendrite development and plasticity with potential implications for memory formation. Further, our experiments reveal molecular details underlying transcriptional regulation by Adf-1, and indicate active interaction between Adf-1 and epigenetic regulators of gene expression during activity-dependent neuronal plasticity.
Introduction
Normal dendrite development and plasticity are indispensible for basal and adaptive functions of the nervous system (Cline, 2001; Tavosanis, 2012). Additionally, pathophysiological changes in dendrites might underlie neurological disorders such as autism spectrum disorders, Alzheimer's disease, and schizophrenia (van Spronsen and Hoogenraad, 2010; Penzes et al., 2011). As sites for signal integration, changes in dendritic architecture have profound effects on information processing in neuronal circuits (Yuste, 2011). As a result, molecular mechanisms that modify dendrite architecture are likely to fundamentally regulate activity-dependent plasticity and behavioral adaptation (Steward and Worley, 2002; Sutton and Schuman, 2006; Bramham and Wells, 2007).
Transcription is an integral part of protein synthesis-dependent neuronal plasticity (West and Greenberg, 2011). Several transcription factors, including CREB, CREST, AP-1 (activator protein-1), MEF-2 (myocyte enhancer factor-2), and NFAT (nuclear transcription factor of activated T cells), respond to changes in neural activity and calcium influx to activate plasticity-related transcriptional programs (Greenberg et al., 1985; Morgan and Curran, 1986; Chrivia et al., 1993; Davis et al., 1996; Sanyal et al., 2002; Graef et al., 2003; Aizawa et al., 2004; Redmond and Ghosh, 2005; Flavell et al., 2006, 2008; Shalizi et al., 2006; Wu et al., 2007; Schwartz et al., 2009; Freeman et al., 2011). Such programs, often transduced by signaling proteins such as MAP kinases and calcium/calmodulin-dependent protein kinases (CaMKs), include post-translational modifications, transcriptional activation, nuclear translocation and recruitment of chromatin modifying factors (West et al., 2002; Burgoyne, 2007; West and Greenberg, 2011). Several studies have also demonstrated the importance of rapid, synapse-specific translational activation in plasticity (Steward, 1987; Kang and Schuman, 1996; Martin et al., 1997; Aakalu et al., 2001; Miller et al., 2002; Ju et al., 2004; Jakawich et al., 2010).
Molecular regulation of dendritic plasticity can be studied in Drosophila motor neurons that are glutamatergic, display activity-dependent plasticity and possess stereotypic dendritic arbors (Consoulas et al., 2002; Landgraf et al., 2003; Sánchez-Soriano et al., 2005; Hartwig et al., 2008; Kim et al., 2009; Mauss et al., 2009; Vonhoff and Duch, 2010; Vonhoff et al., 2013). 3D reconstruction methods, assessment of motor output and whole-cell recordings provide additional reasons for using motor neurons to study dendritic plasticity (Choi et al., 2004; Schmitt et al., 2004; Hartwig et al., 2008). Here, we examine the role of the Myb family transcription factor Adf-1, mutations in which lead to long-term memory deficits in Drosophila, in dendrite growth regulation (Heberlein et al., 1985; DeZazzo et al., 2000; Kacsoh et al., 2013). We find that Adf-1 controls the growth of larval RP2 motor neuron dendrites downstream of neural activity and CaMKII signaling, and regulates both membrane properties and motor behavior. chromatin immunoprecipitation followed by sequencing (ChIP-Seq) from larval brains identifies gene promoters that bind Adf-1 in vivo including two that we assess mechanistically [Fasciclin II (FasII) and Staufen]. Additionally, we discover strong correlations between Adf-1 binding at promoters and RNA Polymerase II (Pol II) pausing, and the relative absence of marks of active chromatin (H3K4me3 and H3K27ac). Our results suggest that Adf-1 interacts with epigenetic machinery to control dendritic plasticity by modulating two key processes, cell adhesion and mRNA transport, downstream of neural activity and CaMKII signaling.
Materials and Methods
Drosophila stocks, rearing, and genetics.
Drosophila strains were raised on standard media in a 25°C incubator throughout life (unless otherwise noted). nalyot mutants (DeZazzo et al., 2000); UAS-FasIIA-RNAi and UAS-FasII A PEST+ (Beck et al., 2012); UAS-CaMKII [T287D] and UAS-CaMKII ala peptide inhibitor (Jin et al., 1998); futschC380-GAL4 (Budnik et al., 1996; Sanyal, 2009); Cha-GAL4 and Cha-GAL80 (Kitamoto, 2002); UAS-DenMark (Nicolaï et al., 2010); UAS-Kir2.1 (Baines et al., 2001); UAS-Sh[DN] and UAS-eag[DN], which together make up EKI (Broughton et al., 2004; Mosca et al., 2005; Hartwig et al., 2008); Tub-GAL80ts flies that are part of the TARGET system (McGuire et al., 2003); and the RU486-sensitive elav-GeneSwitch system (Osterwalder et al., 2001) have all been described previously. To visualize dendritic structure in individual RP2 motor neurons, we made use of the RN2 flipout line, previously characterized by our laboratory (Hartwig et al., 2008). The Adf-1 [DN] line was generated by replacing 18 aa in the transactivation domain (amino acids 131–148: D I F A Q P F N G S A T T S A Q A L) with 5 alanine residues using standard cloning procedures and then inserting this deletion into the pUASt vector. The Adf-1 Gal4 line was generated by transposase-mediated P-element replacement of the lacZ enhancer trap P{PZ}Adf101349 (Sepp and Auld, 1999). The Adf-1 [S64/184] and Adf-1 [S64/184D] substituted lines were generated by site-directed mutagenesis of the wild-type Adf-1 cDNA (obtained from the Drosophila Genomics Resource Center) and cloning into the Gateway-compatible pTWF vector using p-ENTR/D as the entry vector (Invitrogen). Staufen EP and Staufen RNAi (from the TRiP collection, Harvard University, Cambridge, MA) were obtained from the Bloomington Stock Center. The other fly stocks used are part of the Sanyal laboratory stock collection. Only female flies were used for all experiments.
Antibody generation, immunohistochemistry, and Western blotting.
Rabbit anti-Adf-1 polyclonal antibodies were generated by Alpha diagnostics against two KLH (keyhole limpet hemocyanin)-tagged antigenic peptides, GSQSANQVADPSQQ and GKDQKPYFYEPPLK, and affinity purified against these immobilized peptides. Larval immunohistochemistry was performed following established protocols for confocal microscopy (Sanyal, 2009). Animals were fixed in 4% paraformaldehyde followed by washing in PBS. Preparations were incubated in primary antibody in PBS with 0.1% Triton X-100 (PBT) overnight at 4°C, then washed in PBT followed by incubation in specific Alexa fluorophore-conjugated secondary antibody (Invitrogen). After final washes in PBT, brains were dissected out and mounted on poly-lysine-coated slides in Vectashield (Vector Laboratories). Confocal microscopy was performed on a Zeiss 510 confocal microscope. Antibody dilutions are as follows: rabbit anti-Adf-1 (1:100); rabbit anti-lacZ (1:1000; Abcam); rabbit/mouse anti-GFP (1:1000; Invitrogen); mouse anti-Elav (1:100; Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City, IA); rabbit anti-Staufen (1:500, St Johnston et al., 1991); and guinea pig anti-Zfh-1 (1:100, Postigo et al., 1999). Western blots were performed as previously described (Vrailas-Mortimer et al., 2011). Antibody dilutions for Western blot are as follows: rabbit anti-Adf1 antibody (1:5000); rabbit anti-H3 antibody (1:10,000; Abcam); mouse anti-actin (1:500,000; Millipore); and mouse anti-FLAG (1:1000; Sigma).
Microscopy and 3D reconstructions.
Dissected and fixed larval nerve chords were imaged using a Zeiss 510 confocal microscope with a 40× oil-immersion lens as previously described (Hartwig et al., 2008). Dendrites were reconstructed using a custom semiautomated AMIRA (4.1.1) plug-in, as previously described (Evers et al., 2005; Vonhoff et al., 2013). Quantitative measures of dendrite growth were analyzed and plotted using Microsoft Excel.
Electrophysiology.
Whole-cell tight seal recordings were performed on motoneuron RP2 (MNISN1s) using external salines (in mm: 118 NaCl, 2 NaOH, 2 KCl, 4 MgCl2, 4 CaCl2, 40 sucrose, 5 trehalose, 5 HEPES, pH 7.1; osmolality, 305 mmol/kg) and internal salines (in mm[scap]: 130 K-gluconate, 2 NaCl, 10 HEPES, 1 EGTA, 2 MgCl2, 0.1 CaCl2, 10 KOH, pH 7.2; osmolality, 285 mmol/kg), as described previously (Choi et al., 2004; Sandstrom, 2008; Freeman et al., 2011). RP2 somata were visualized with the synthetic GAL4 line (Hartwig et al., 2008), driving membrane-bound GFP. For recording, the CNS was removed from the larva, immobilized on a small chip of coverslip coated with poly-dl-ornithine (Sigma), and placed in a recording chamber under continuous superfusion. The sheath was softened and removed using a small-bore pipette filled with 0.1% collagenase type XIV (Sigma). Neurons were visualized with a 63× water-immersion objective, using Nomarski optics and GFP fluorescence (BX51WI; Olympus America). Whole-cell recordings were performed with an AxoPatch 200B controlled by pClamp 8.2. Electrodes were fabricated to 5–10 MΩ from thick-walled capillary glass (World Precision Instruments) on a vertical puller (PP-830, Narashige International USA). F–I curves were measured in current-clamp at −60 mV, while input resistance was measured from the slope of the I–V relation in voltage-clamp at a holding potential of −70 mV.
Larval neuromuscular recordings were carried using HL 3 external saline (in mm: NaCl 70, KCl 5, MgCl2 20, CaCl2 1, NaHCO3 10, sucrose 115, trehalose 5, BES [2 2-[bis(2-hydroxyethyl)amino]ethanesulfonic acid 5], pH 7.2), as described previously (Stewart et al., 1994; Timmerman and Sanyal, 2012). Intracellular recording electrodes with tip resistances between 25 and 50 MΩ were filled with 3 m KCl. Only those recordings were used where the resting membrane potential was more polarized than −60 mV and the muscle input resistance was >10 MΩ. Muscle 6 (VL3) in abdominal segment A2 was used for all recordings. For sensori-motor transmission, the axon bundle contralateral to the recording segment was cut close to the muscle entry point and introduced into a stimulating electrode and stimulated at 10 Hz for 1.5 s (Dasari and Cooper, 2004).
Behavioral assays.
Larval locomotion was measured by videotaping individual larvae for a period of 5 min and tracking their motion using a Spot Tracker plugin in ImageJ as described previously (Vrailas-Mortimer et al., 2011). The Buridan assay was performed as described previously (Freeman et al., 2012), and at least 10 animals were tested for each genotype. Briefly, flies aged 3–5 d had their wings clipped off close to their thorax and were allowed to recover for 3 d before behavioral analysis. Single flies were placed onto an arena (10 cm in diameter) with the only visual cues being two black bars on opposite ends of the arena. At the edge of the arena is a moat (2 cm in width of water surrounding the platform). Wild-type flies will normally respond by repetitively walking between the two visual cues. This motor behavior was videotaped for 5 min. Locomotor tracks were analyzed using custom-designed software in the statistical package R (Colomb et al., 2012).
ChIP-Seq analysis and determination of molecular networks.
ChIP was performed with 500 hand-dissected larval brains or ∼4 × 107 Kc167 cells. Cells were cross-linked with 1% formaldehyde for 10 min at room temperature. Nuclear lysates were sonicated to generate 200–1000 bp DNA fragments. ChIP was then performed with 6 μl of Drosophila α-Adf-1 antibody. Libraries were prepared using the Illumina TruSeq DNA Sample Preparation Kit. Fragments in the 200–300 bp range were selected and sequenced in an Illumina HiSeq Sequencer at the HudsonAlpha Institute for Biotechnology (Huntsville, AL). Sequences were aligned to Drosophila dm3 using Bowtie, and binding intensity files (.wig) were generated using MACS. In addition to the Drosophila Adf-1 data obtained in this study, we used several datasets obtained from public sources including Pol II ChIP-chip (modENCODE_328) and ChIP-Seq datasets for H3K4me3 and H3K27ac (GSE36374). To build heatmaps, values for each ChIP-Seq dataset were extracted for the 2000 bp region around transcription start sites (TSSs) of genes in dm3 annotation using custom R scripts (available upon request), and heatmap graphs were created using TreeView. In Figure 6E, regions were sorted according to the intensity of Adf-1 signal at TSSs. Group1 TSSs were selected to represent genes associated with Adf-1. Group 2 TSSs were selected to represent genes with low intensity of Adf-1. Group 1 and Group 2 TSSs have approximately the same level of RNA polymerase to eliminate the effect of Pol II intensity when calculating pausing indices. The pausing index of genes was calculated using ChIP-chip datasets of Pol II in Kc cells obtained from modENCODE. Pol II levels at TSSs (PTSS) were calculated as the mean enrichment of Pol II in the 200 bp region around each TSS. Pol II in the gene body (Pbody) was calculated as the mean enrichment of Pol II from +200 bp to the end of the gene. The pausing index is defined as the difference between PTSS and Pbody.
Figure 6.
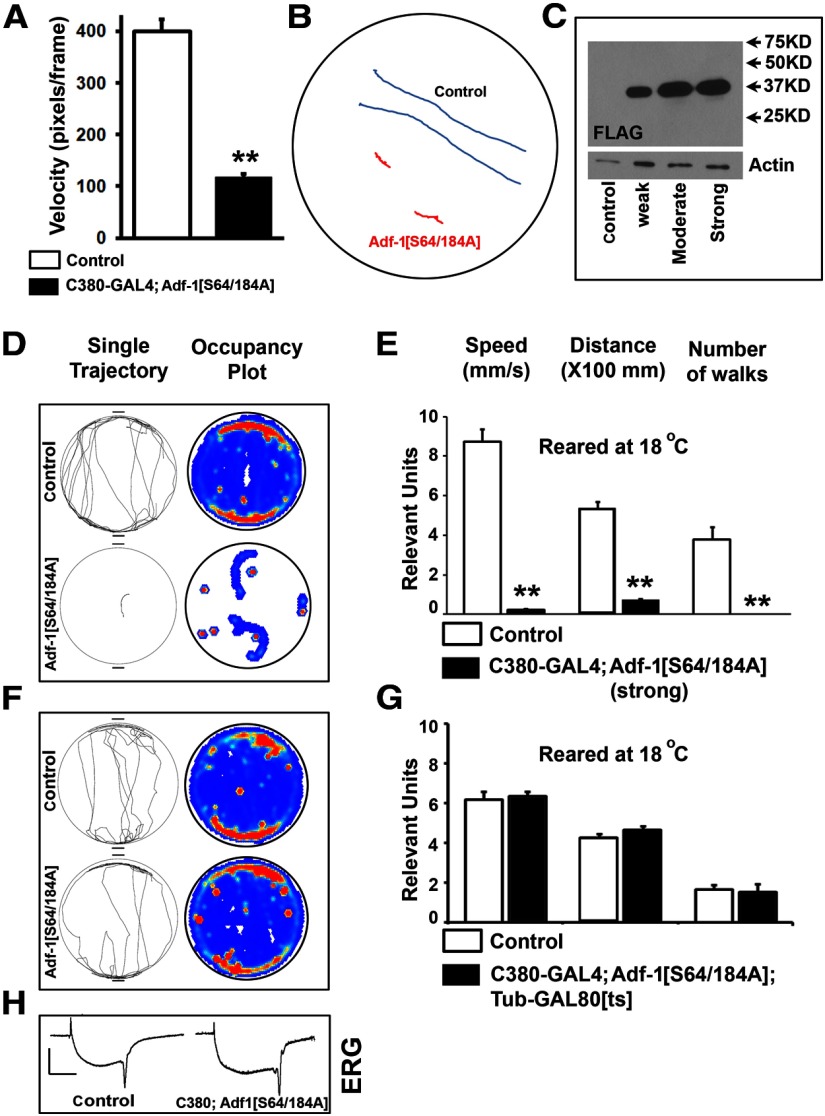
Behavioral consequences of Adf-1 inhibition in motor neurons. A, Expression of Adf-1[S64/184A] in larval motor neurons using the futschC380-GAL4 driver leads to a dramatic reduction in larval locomotion. Average velocities are reduced by almost fourfold. B, Two representative traces of crawling larvae from control (blue) and Adf-1[S64/184A] animals (red) over a 3 min recording period. C, Western blot of protein from larval brains in which different Adf-1[S64/184A] transgenic lines are used to express FLAG-tagged Adf-1[S64/184A] protein. D, E, Representative individual fly tracks (top) and cumulative occupancy plots of controls compared with animals in which strong expression of Adf-1[S64/184A] is driven from the futschC380-GAL4 driver at 18°C. F, G, The strongly impaired locomotor behavior shown above is dependent on the expression of the transgene and is not observed when a temperature-sensitive GAL80 is used to represses GAL4 activity at 18°C. F, G, Representative individual tracks, cumulative occupancy plots (F), and quantification of locomotor parameters (G). H, Representative electroretinogram traces from control and animals strongly expressing Adf-1[S64/184A] in all neurons. **p < 0.01.
Network analysis of genes that bind Adf-1 close to their promoters in the larval brain was performed using GeneMANIA as an executable plugin within Cytoscape (Mostafavi et al., 2008; Montojo et al., 2010). Cluster analysis of gene ontology terms was performed using BiNGO (Maere et al., 2005) and the enrichment map plugin for Cytoscape (Merico et al., 2010).
RU486 feeding, RNA extraction, and qRT-PCR.
RU486 (M8046; Sigma) was reconstituted in EtOH. Animals were starved overnight at 25°C. After starvation, animals were transferred to vials containing a Whatman filter disc saturated with a 2% sucrose solution with either RU486 to final concentration of 1 mm or a 2% sucrose solution with EtOH (unfed controls). Flies were left on this food for 24 h and then transferred to standard corn meal fly food for 6 h to recover. RNA was extracted from adult heads using the standard TRIzol protocol. Following isopropanol precipitation and quantification, RNA was reverse transcribed with the Applied Biosystems High Capacity cDNA Reverse Transcription Kit. Quantitative PCR was performed using the Thermo Scientific Maxima Sybr Green kit and reactions were run on the Roche Lightcycler 480. Each reaction was run in triplicate, and each genotype was tested a minimum of three times. Samples were normalized to the housekeeping gene rp49. Primer sequences obtained from published data: fasIIA (Beck et al., 2012); and rp49 (LaLonde et al., 2006). Other primer sequences were designed using the Roche Universal Probe Library assay design center and are as follows: adh (forward, 5′ attttcgttgccggtctg; reverse, 5′cgaggatcaccaggttcttc); and staufen (forward, 5′agtcttcagcagcaacagca; reverse, 5′gattcagacgtgcgtggag).
Statistical methods.
Student's t test or Mann–Whitney U test was used for pairwise comparisons. One-way ANOVA was used for multiple comparisons, followed by Bonferroni's post hoc testing.
Results
Adf-1 is expressed in motor neurons in the Drosophila larval nerve cord
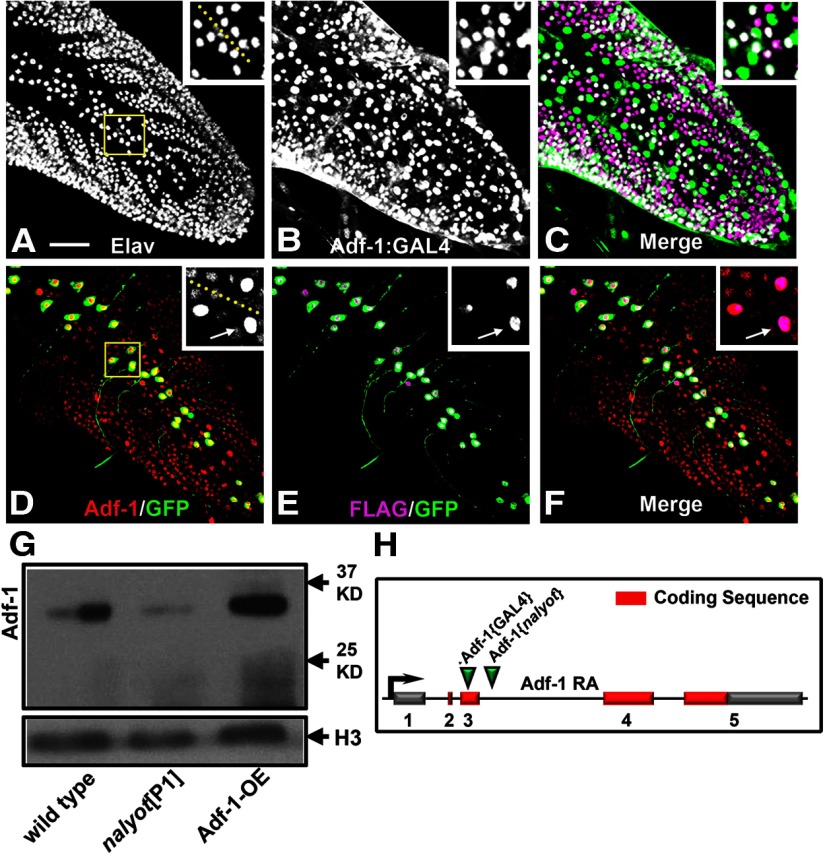
We used the Drosophila larval motor neuron RP2 as a model to examine Adf-1-dependent regulation of dendrite development and plasticity since this neuron displays activity-dependent changes in growth (Hartwig et al., 2008). To test whether Adf-1 is expressed endogenously in these neurons, we generated affinity-purified polyclonal antibodies against two Adf-1-derived peptides. These antibodies detected both basal and transgenically overexpressed Adf-1 in the nuclei of larval motor neurons (Fig. 1D–F; see insets for dorsal midline motor neurons RP2 and aCC). Wild-type FLAG-tagged Adf-1, when expressed transgenically using a synthetic GAL4 that is only active in RP2 and aCC neurons in a mosaic fashion (a system that we have described previously; Hartwig et al., 2008), is elevated selectively in the nuclei of motor neurons where GAL4 activity is present (Fig. 1D–F, compare GAL4-driven GFP expression with Adf-1 and transgenic FLAG expression in). Anti-Adf-1 antibodies also detected a band of the predicted size (∼30 kDa) on Western blots of proteins extracted from larval and adult brains (Fig. 1G) that was increased in intensity when wild-type Adf-1 is overexpressed pan-neuronally and reduced in Adf-1 hypomorphic nalyot mutants. Nervous system expression of Adf-1 was also evident from the expression pattern of a new Adf-1-GAL4 enhancer trap line, which revealed extensive neuronal staining in addition to some non-neuronal cells in the ventral nerve cord (Fig. 1A–C, compare Elav staining of postmitotic neuronal nuclei with GFP expression driven by the Adf-1-GAL4 line). We generated this line by replacing a lacZ enhancer trap P-element (P{PZ}Adf-101349) inserted in an the first exon 408 bp downstream of the transcription start site with a P{GawB} element inserted close to the futsch gene (Sepp and Auld, 1999; Sanyal, 2009; C380-GAL4; Fig. 1H). Together, these data clearly demonstrate that Adf-1 is an essential gene that is expressed in the Drosophila nervous system.
Figure 1.
Adf-1 is expressed in larval motor neurons. A–C, An Adf-1 enhancer trap line (Adf-1-GAL4) expresses β-galactosidase from a UAS-nls-lacZ reporter in larval neurons in the larval nerve cord (B), double-labeled with anti-Elav to mark all postmitotic neuronal nuclei (A). Dorsomedial motor neurons, including RP2 motor neurons, also express Adf-1 (shown in inset and merged images in C). D–F, Overexpression of a FLAG-tagged wild-type Adf-1 using our synthetic GAL4 system is also detectable using staining for the FLAG epitope mCD8::GFP or increased Adf-1 staining. Inset shows a closeup of neuronal nuclei that shows expression of FLAG, GFP, and elevated levels of Adf-1 driven by GAL4. The larval nerve cord is stained for Adf-1 as a reference. Scale bar, 50 μm. G, Western blot of protein extracted from adult brain tissue probed using our peptide antibodies to Adf-1. The previously described nalP1 allele shows reduced expression as expected, while transgenic overexpression of Adf-1 increases Adf-1 protein levels. Anti-Histone H3 is used as a loading control. H–J, Polytene chromosome immunofluorescence shows Adf-1 staining at discrete regions (I). Sytox 24 is used as a general DNA stain to reveal the banding pattern in a standard polytene chromosome preparation (H). K, Schematic of the Adf-1 gene region showing sites of insertion for the pGawB enhancer trap and the P[ry+] nalyot P-element transposons.
Inhibition of Adf-1 in the RP2 motor neuron leads to severely reduced dendrite growth and excitability
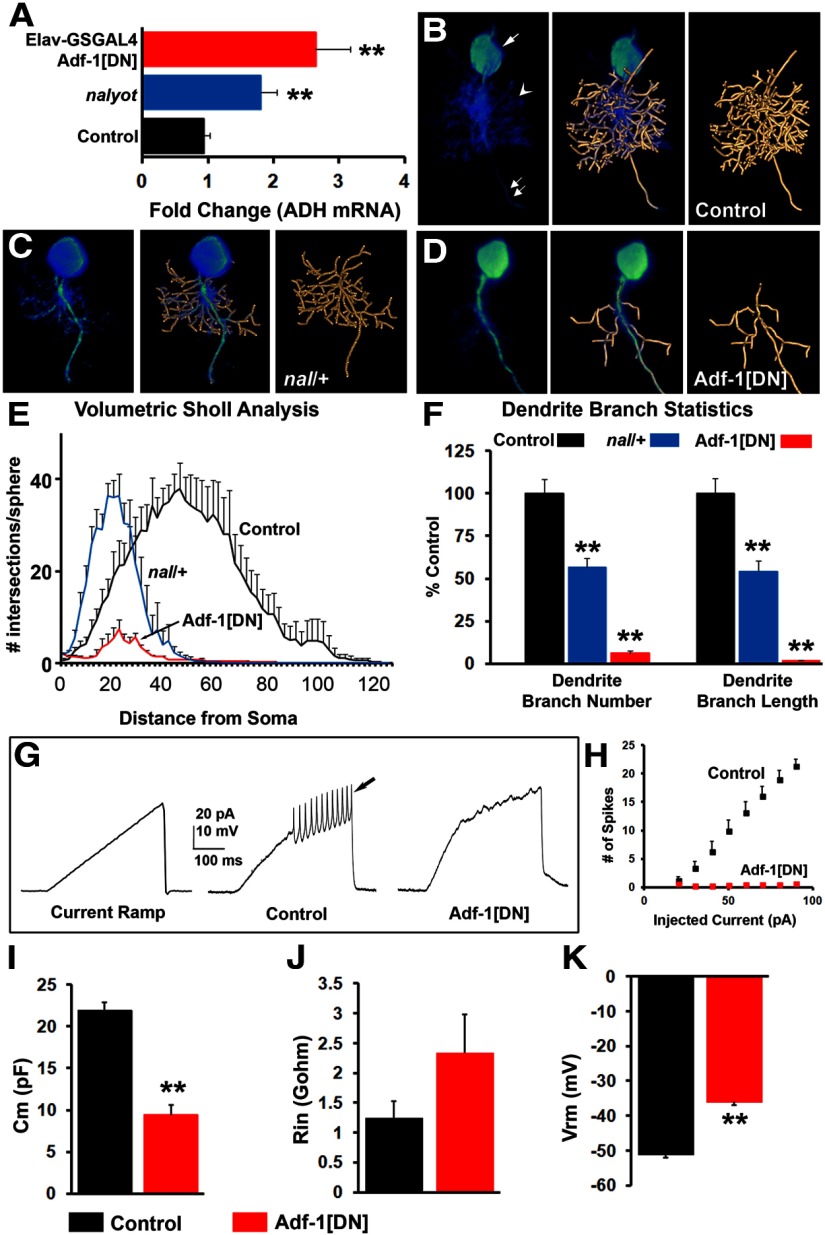
nalyot mutants have been shown to result in marginally smaller presynaptic termini at larval synapses with fewer boutons (DeZazzo et al., 2000). Here, we asked whether the loss of Adf-1 affects dendrite growth in larval motor neurons. We first examined the effect of a mild reduction in Adf-1 levels in a nalyot heterozygous animal. RP2 neuron dendrites were reconstructed using the mcd8::GFP label (mcd8::GFP colocalizes with dendritic markers such as DenMark, data not shown; Sánchez-Soriano et al., 2005; Evers et al., 2005; Schmitt et al., 2004; Vonhoff et al., 2013). These reconstructions revealed a 50% reduction in both the number of dendritic branches and the total dendrite length in nalyot heterozygotes compared with controls (Fig. 2B,C; E,F, present volumetric Sholl analysis and quantification of dendrite branches and length). Multiple attempts to excise this P-element or the Gal4 enhancer trap did not result in any verifiable null allele for Adf-1, owing perhaps to the close proximity of this gene to the second chromosome centromere (Adf-1 maps to 42C3). For this reason, we were also unable to generate a recombinant of the nalyot allele with an FRT-containing insertion on 2R and carry out MARCM-based mosaic analysis of Adf-1. Despite these limitations, we attempted to determine the cell-autonomous origin of this phenotype by targeting expression of a modified Adf-1 protein (in which 18 aa in the putative trans-activating domain have been replaced with 5 alanine residues) to RP2 neurons using our synthetic GAL4 driver line (this GAL4 line crossed to w1118 was used as the control strain for this and all subsequent experiments). Expression of this Adf-1-dominant negative transgene (Adf-1[DN]) resulted in a more severe loss of dendrites (Fig. 2, compare B, D) and a dramatic reduction in dendritic complexity (Fig. 2E), branch number, and length (Fig. 2F). Whole-cell patch-clamp measurements from RP2 motor neurons expressing Adf-1[DN] revealed healthy neurons with robust input resistance and resting membrane potentials, albeit with lower cell capacitance (likely due to reduced dendritic membrane; Fig. 2I–K; Cm (membrane capacitance), p < 0.001, Student's t test; Vm (resting membrane potential), p < 0.01, Mann–Whitney U test; Rin (input resistance) p > 0.05, Mann–Whitney U test). However, these cells were largely refractory to the injection of depolarizing current, which typically elicited robust action potentials from control RP2 neurons (Fig. 2G,H). Together, these results suggest that Adf-1 activity is required in motor neurons for normal dendrite growth and complexity.
Figure 2.
Normal dendrite growth in larval RP2 motor neurons requires Adf-1. A, qRT-PCR from adult brains showing the effect of Adf-1 inhibition on the expression of the target gene ADH. B–D, 3D reconstruction of RP2 neuron dendrite branches in vivo using a semiautomated AMIRA-based method in control (n = 9; B), nalyot heterozygotes (n = 5; C), and animals expressing a dominant-negative Adf-1 transgene selectively in RP2 neurons (n = 5; D). E, Volumetric Sholl analysis shows a strong reduction in dendrite complexity that results from Adf-1 inhibition. F, Both the number of dendrite branches and the total dendrite length are reduced in nalyot heterozygotes and animals expressing an Adf-1 dominant-negative transgene in RP2 neurons (**p < 0.01). G, Whole-cell patch-clamp recordings from control RP2 neurons and those expressing the Adf-1[DN] transgene. Current ramps elicit action potentials in control neurons (arrow), whereas Adf-1-inhibited neurons remain silent. H, Quantification of spike frequency with increasing current injection. I–K, Average capacitance, input resistance, and resting membrane potential from control RP2 neurons and those expressing Adf-1[DN].
Adf-1 controls dendrite growth downstream of CaMKII signaling
While Adf-1 is known to regulate alcohol dehydrogenase (ADH) transcription (Heberlein et al., 1985) and is involved in memory formation (DeZazzo et al., 2000; Kacsoh et al., 2013), virtually nothing is known about upstream regulatory elements that control Adf-1 activity. An in silico scan of Adf-1 with NetPhosK (Blom et al., 2004) highlighted two serine residues, Ser-64 and Ser-184, which are putative CaMKII targets. Indeed, a previous study had shown that Drosophila CaMKII could phosphorylate purified recombinant Adf-1 with a preference for Ser-64 (GuptaRoy et al., 2000). Based on these findings and the widely appreciated role for CaMKII in dendritic plasticity across species (Wu and Cline, 1998; Rongo and Kaplan, 1999; Zou and Cline, 1999; Miller et al., 2002; Gaudillière et al., 2004; Lee et al., 2009; Vonhoff et al., 2013), we tested the idea that CaMKII-dependent phosphorylation of Ser-64/184 regulates Adf-1 activity, and thereby dendrite growth, in RP2 motor neurons.
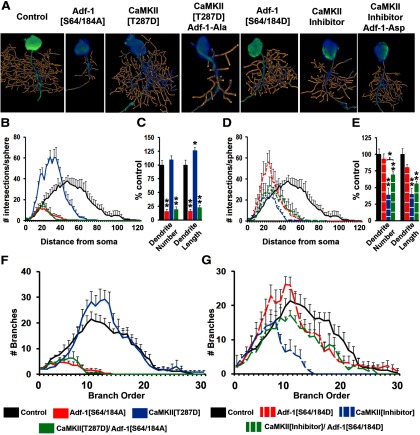
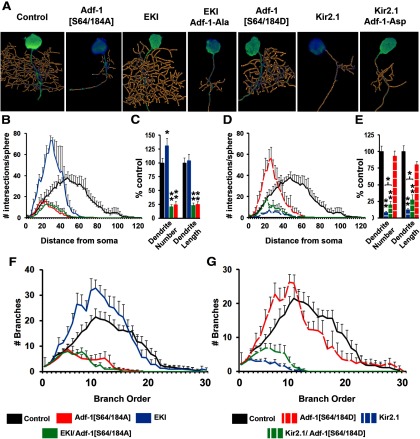
To test the importance of this CaMKII phosphorylation site on Adf-1 function, we constructed two UAS-driven, Adf-1 transgenes, one in which Ser-64/184 were mutated to alanine (Adf-1-[S64/184A], a phospho-null substitution) and another in which the residues were changed to aspartate (Adf-1-[S64/184D], a phospho-mimic substitution). We expressed these transgenes in RP2 neurons either singly or in combination with reagents that increase or inhibit CaMKII (Jin et al., 1998). Our results show that mutating Ser-64/184 to Ala leads to a dramatic reduction in dendritic complexity, number of dendritic branches and overall dendrite length (Fig. 3A–C). Since this phenotype resembles both nalyot mutants and Adf-1[DN], it suggests a crucial role for Ser-64/184 phosphorylation in normal Adf-1 function. Consistent with the idea that CaMKII is the kinase responsible for this phosphorylation (GuptaRoy et al., 2000), expression of the Adf-1 [S64/184A] transgene completely abrogates the stimulatory effect of a constitutively activate calcium-independent CaMKII transgene (CaMKII-T287D) on dendrite growth (Fig. 3A–C; Jin et al., 1998; Vonhoff et al., 2013). Note that CaMKII[T287D] results in a significant increase in overall dendrite length, but not the number of branches (Fig. 3C). Volumetric Sholl analysis indicates, however, that the overall pattern of branching changed such that there were a greater number of branches closer to the neuronal soma (Fig. 3B). A similar shift occurred when we expressed the Asp-substituted Adf-1 (Adf-1[S64/a84D]), although both overall dendrite branch number and length remained unchanged compared with controls (Fig. 3D,E). Significantly, the expression of Adf-1[S64/184D] increased both branch number and total dendrite length in a CaMKII-inhibited background (a CaMKII peptide inhibitor, ala; Jin et al., 1998), which results in a strong decrease in dendrite growth (Fig. 3D,E; Fig. 3F,G, branch-order analysis). In sum, these results support the importance of the two putative CaMKII target sites in Adf-1 in the regulation of Adf-1 activity and dendrite growth.
Figure 3.
Adf-1 functions downstream to CaMKII signaling to regulate dendrite growth in RP2 motor neurons. A, Representative RP2 dendrite reconstructions for Adf-1 and CaMKII genetic interactions. B, C, Volumetric Sholl analysis and the measurement of dendrite branch number and total dendrite length for CaMKII[T287D], Adf-1[S64/184A], and their combination, compared with control neurons. D, E, Volumetric Sholl analysis and measurements of dendrite branch number and length for the CaMKII peptide inhibitor Adf-1[S64/184D] and their combination compared with control neurons. The expression of Adf-1[S64/184D] significantly increases dendrite branch number in a CaMKII-inhibited background (*p < 0.05 for pairwise comparison after Bonferroni's correction). F, G, Branch-order analysis in experiments to test genetic interaction between CaMKII and Adf-1.
Adf-1 regulates activity-dependent developmental plasticity of dendrites in RP2 motor neurons
Drosophila motor neurons display activity-dependent changes in dendrite growth (Hartwig et al., 2008; Vonhoff et al., 2013), and we next tested whether Adf-1 is required in this process. Elevating neuronal firing in RP2 motor neurons through the coexpression of two dominant-negative voltage-gated potassium channels Eag and Shaker (EKI) resulted in increased dendritic complexity near the soma (Fig. 4A,B) and increased dendrite branch number (Fig. 4C). In this background, the expression of Adf-1[S64/184A] completely inhibited dendrite growth (Fig. 4A–C), suggesting that normal Adf-1 activity is necessary for activity-dependent growth of RP2 dendrites. Conversely, silencing RP2 neurons by expressing the inward rectifying Kir2.1 channel strongly reduced dendrite growth as expected (Fig. 4A,D,E). Consistent with a role downstream of neural activity, Adf-1[S64/184D] expression in an activity-inhibited background was able to modestly rescue dendrite growth such that both dendrite branch number and total length were significantly greater than in Kir2.1. Thus, Adf-1[S64/184D] was only able to increase dendrite growth in a background where neural activity, and by extension CaMKII and Adf-1 activity, were severely limiting (Fig. 4F,G, branch-order analysis for these genotypes). Overall, these results suggest that normal Adf-1 phosphorylation downstream of CaMKII is required for activity-dependent changes in dendrite growth.
Figure 4.
Adf-1 is required for activity-dependent change in dendrite growth. A, Representative RP2 dendrite reconstructions to test the interaction between Adf-1 and changes in neuronal activity. B, C, Volumetric Sholl analysis and the measurement of dendrite branch number and length following an increase in neural activity and Adf-1 inhibition. D, E, Volumetric Sholl analysis and measurement of dendrite branch number and length following neuronal silencing and Adf-1 activation. Adf-1[S64/184D] increases both dendrite number and length in an activity silenced background (*p < 0.05 for pairwise comparison after Bonferroni's correction). **p < 0.01. F, G, Branch-order analysis in experiments that test Adf-1 function downstream of activity manipulations in RP2 motor neurons.
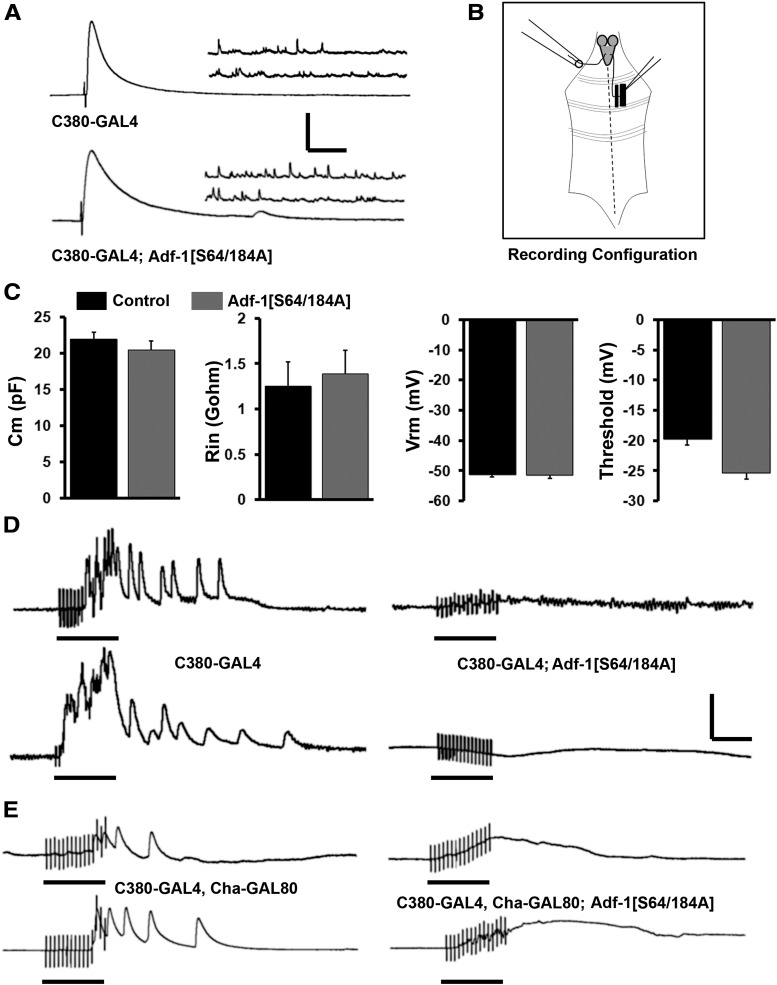
Adf-1 inhibition in motor neurons impairs sensori-motor transmission
Does Adf-1-dependent alteration in dendrite architecture affect synaptic excitation of motor neurons and their output at the neuro-muscular junction? To test this, we used the Adf-1[S64/184A] transgene, as pan-neuronal expression of Adf-1[DN] is lethal. The results shown in Figure 5 suggest that synaptic transmission from these motor neurons at the neuro-muscular junction is normal, consistent with a previous report (DeZazzo et al., 2000; Fig. 5A). The amplitudes of both the excitatory junction potential (EJP) and miniature EJP (mEJP) are unchanged, as are mEJP frequency and the quantal content of transmitter release (data not shown). Basic membrane properties of these neurons were also not affected by the expression of Adf-1[S64/184A], since cell capacitance, input resistance, resting membrane potential, and the threshold for action potential firing were all comparable with control neurons (Fig. 5C). Next, we tested whether synaptic stimulation of motor neurons is altered following Adf-1 inhibition. Since precise presynaptic partners for larval motor neurons are currently unknown, we made use of an assay where high-frequency (10 Hz) stimulation of the sensory afferent results in recruitment of motor units in the contralateral segment (Dasari and Cooper, 2004; Fig. 5B). When a sensory afferent was stimulated, we invariably observed motor neuron firing in the contralateral segment that continued beyond the stimulus duration (motor neuron firing was monitored indirectly by recording muscle EPSPs from muscle 6; 10 independent animals were tested; Fig. 5D). By contrast, 50% of animals in which Adf-1[S64/184A] was expressed in a motor neuron-enriched manner using the futschC380-GAL4 driver failed to display motor neuron firing under these conditions (Fig. 5D). Since futschC380-GAL4 also expresses in sensory neurons (Sanyal, 2009), we limited expression to motor neurons by suppressing GAL4 expression in cholinergic neurons with the Cha-GAL80 transgene (Salvaterra and Kitamoto, 2001; Hartwig et al., 2008). Even under these conditions, we observe the same deficiency in sensori-motor transmission, suggesting that the inhibition of Adf-1 in motor neurons is sufficient to cause this transmission defect (Fig. 5E). These results are consistent with phenotypic consequences that are expected from a reduction in dendrite branching (reduced presynaptic input), and alteration in excitability and membrane properties at the soma.
Figure 5.
Adf-1-mediated dendrite phenotypes affect sensori-motor transmission in larvae. A, Neuro-muscular synaptic transmission (both evoked and spontaneous release) is unaffected following Adf-1 inhibition in motor neurons compared with control neuromuscular junctions. B, Schematic of recording configuration for testing sensori-motor transmission in larvae. C, Membrane capacitance, input resistance, resting membrane potential, and threshold of firing are comparable in control neurons and those expressing Adf-1[S64/184A], as measured using whole-cell patch-clamp. D, Representative muscle recordings reflecting motor neuron firing following stimulation of contralateral sensory afferents at 10 Hz for 1.5 s. Traces are shown from control animals (futschC380-GAL4/w1118) and animals expressing Adf-1[S64/184A] from the futschC380-GAL4 driver. E, Representative traces of muscle recordings, as in D, from animals that selectively express Adf-1[S64/184A] in glutamatergic motor neurons (futschC380-GAL4; Cha-GAL80; UAS-Adf-1[S64/184A] compared with appropriate genetic controls. Calibration: vertical = 10 mV; horizontal, (in A) EJP = 50 ms; mEJP = 500 ms; (in D and E) EJP and mEJP = 1 s. Black bars under traces denote the duration of sensory stimulation.
Developmental perturbation of Adf-1 in motor neurons alters locomotor behavior
To test whether dendrite growth phenotypes in larval motor neurons cause aberrant motor behavior, we measured larval crawling in control and Adf-1-inhibited genotypes. As shown in Figure 6, A and B, the expression of the Adf-1[S64/184A] mutant transgene in motor neurons through the futschC380-GAL4 driver (Sanyal, 2009) results in reduced larval locomotion (Fig. 6B). Since we observe aberrant development of motor neuron dendrites upon Adf-1 inhibition, we next asked whether locomotor phenotypes in these animals derive from developmental abnormalities. To this end, we turned to adult flies, which allow us to inhibit Adf-1 after developmental changes in the nervous system are largely completed. We first confirmed that Adf-1 is expressed in adult motor neurons in the thoracic–abdominal ganglion using a marker for motor neurons, the transcription factor Zfh1 (Layden et al., 2006; data not shown). The expression of the Adf-1 [S64/184A] transgene in motor neurons throughout development at 25°C resulted in widespread adult lethality with few escaper animals that display striking defects in locomotion (data not shown). Interestingly, nalyot mutants also displayed sluggish locomotion compared with age-matched controls, though with less severity, an observation that is consistent with a less severe loss of Adf-1 in these mutants. To circumvent lethality, and to focus on the motor neuron-derived phenotypes of Adf-1, we identified two other transgenic lines that had weaker expression (Fig. 6C). The weakest line, when crossed to futschC380-GAL4 and reared at 25°C, resulted in adults that displayed prominent locomotor phenotypes in the Buridan assay (Götz, 1980; Strauss and Heisenberg, 1993; Freeman et al., 2012; data not shown). Similarly, the strong line, when crossed to C380-GAL4 and grown at 18°C, also resulted in similar locomotor defects (Fig. 6D,E). Interestingly, these phenotypes could be completely rescued by preventing GAL4 expression during development using the TARGET system (McGuire et al., 2004; Fig. 6F,G). Together, these results demonstrate that Adf-1 is critically required in motor neurons during development for normal functional output from motor neurons.
Neuronal targets of Adf-1 identified through genome-wide ChIP-Seq analysis
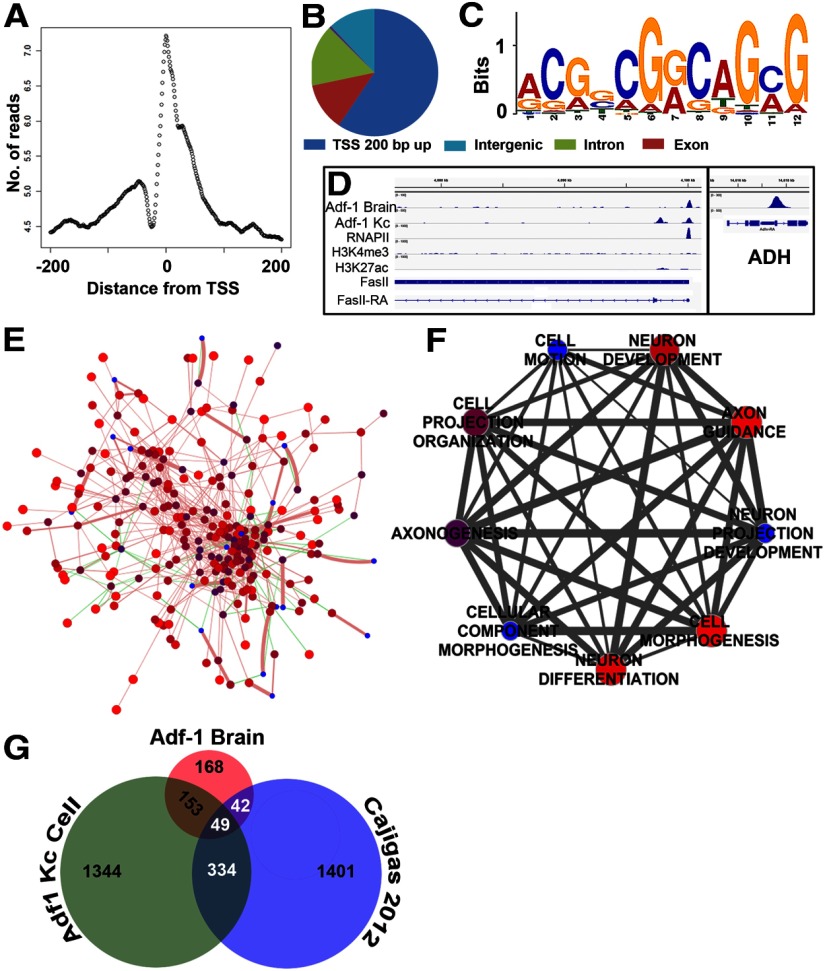
To identify genes whose expression is regulated by Adf-1 in the brain, we performed ChIP from dissected larval brains using anti-Adf-1 antibodies followed by sequencing (ChIP-Seq). We identified robust binding of Adf-1 to the ADH enhancer, as described previously, and to the promoters of several other genes such as FasII (Fig. 7D). Further analysis showed that, as expected for a transcription factor, >50% of Adf-1 binding sites occur within 200 bp of TSSs (Fig. 7A,B). These binding sites were used to calculate a primary Adf-1 binding consensus sequence (Fig. 7C; Machanick and Bailey, 2011). Interestingly, this primary consensus sequence (ACGG/CCGG/ACA/TGC/AG) contains the core (in italics) of a recently reported, computationally derived, high-affinity Adf-1 binding consensus (GT/CGG/AC; Lang and Juan, 2010) and is dissimilar to the binding sequence reported previously [GC/TC/T]4–5 (Heberlein et al., 1985). More than 80% of the genes that bound Adf-1 close to the TSS in the brain also fell into a network of proteins (Fig. 7E), created using GeneMania on the basis of published reports of genetic and physical interactions among Drosophila proteins (Mostafavi et al., 2008; Guruharsha et al., 2011). Moreover, gene ontology analysis revealed a high degree of enrichment for genes with neuronal functions (Fig. 7F), as can be expected for a transcription factor with a prominent neural function.
Figure 7.
Adf-1 transcriptional targets in the brain identified by ChIP-Seq analysis. A, Adf-1 DNA binding peaks in the brain are enriched close to the TSS. B, Distribution of Adf-1 binding peaks within the genome from brain ChIP-Seq. C, Primary consensus Adf-1 binding sequence derived from brain and Kc cell ChIP-Seq experiments using MEME-ChIP. D, Gene track of FasII showing Adf-1 occupancy in the brain and Kc cells, compared with RNA Pol II, H3K4me3, and H3K27ac chromatin marks in Kc cells. Box shows Adf-1 binding at the ADH gene locus in brains. E, Protein interaction network of genes that contain Adf-1 within 200 bp of their TSS in larval brains created using GeneMania, and published genetic and physical interaction studies in Drosophila. Sphere color and size reflect GeneMania scores (red and larger are higher scores), green lines denote genetic interactions, and pink lines denote physical interactions. Line thickness denotes interaction strength. A total of 82% of input genes (i.e., those that bind Adf-1 in their promoters in the brain) show physical interaction, while 18% have known genetic interactions. F, Clustering of highly significant neuron-related GO terms represented by genes that bind Adf-1 close to the TSS. The size and color of spheres represent p values, and line thickness denotes the number of shared genes between two GO terms. G, Venn diagram representing overlap between genes that bind Adf-1 in brains and Kc cells compared with dendrite-enriched mRNAs in the rodent brain.
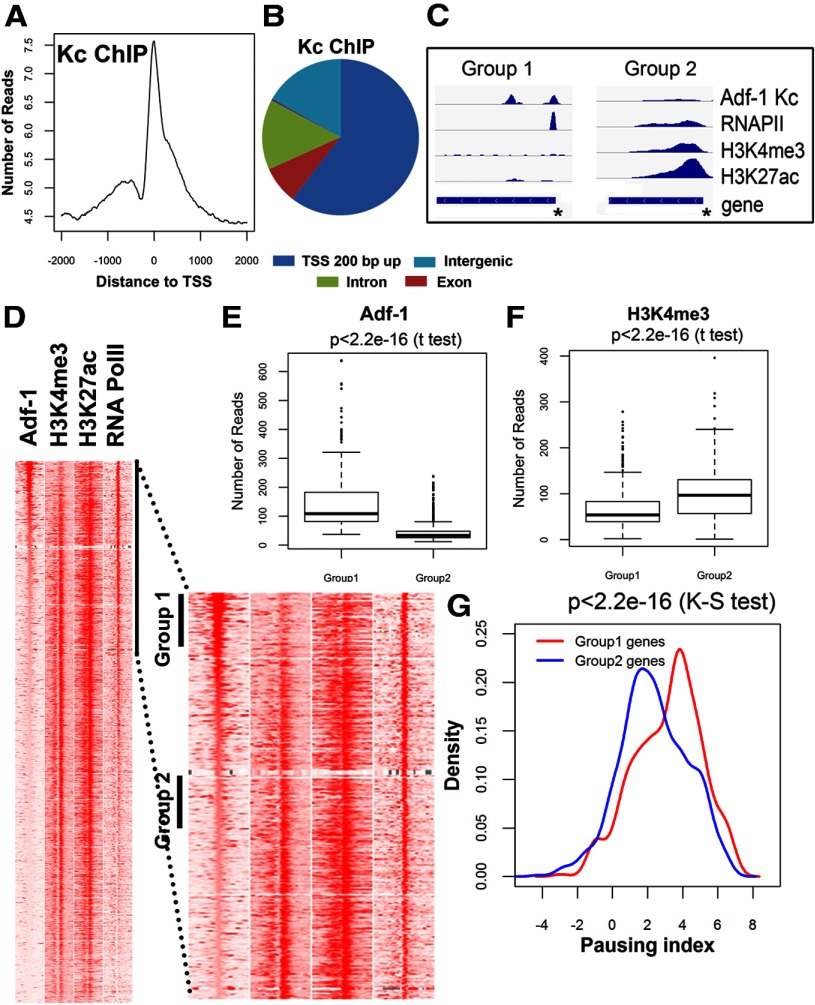
We had noticed previously that Adf-1 inhibition in the brain resulted in increased transcription of ADH (Fig. 2A). To derive a mechanistic understanding of Adf-1-dependent gene expression, we performed an additional Adf-1 ChIP-Seq experiment in Kc cells. This allowed us to generate a dataset that could be compared with a library of ChIP-Seq data for RNA Pol II binding and the distribution of covalent histone modifications. Kc cell ChIP-Seq with anti-Adf-1 antibodies resulted in a larger number of binding peaks that, however, showed a similar profile of binding compared with the brain ChIP-Seq experiment (Fig. 8A–C). Comparison to Pol II binding sites and active marks of chromatin also showed that Adf-1 is enriched at promoters that show a higher Pol II pausing index (Fig. 8C). The pausing index is a measure of how long Pol II pauses after initiation and before elongation. If the pausing index is low, Pol II continues elongating after initiation, so genes with a low pausing index are likely to be regulated at the initiation step. If the pausing index is high, then Pol II pauses for a long time after initiation, and it needs to be released to proceed into elongation. Visual inspection of specific genomic sites suggests a negative correlation between Adf-1 binding and H3K4me3 and H3K27ac (for representative gene tracks, see Fig. 8C). To determine the general significance of this observation, we arranged all TSSs in the genome in descending order based on Adf-1 levels, and we examined the distributions of Pol II, H3K4me3, and H3K27ac (Fig. 8D). Genes with high levels of Adf-1 (Group 1 genes) contain Pol II at their TSS, but they have low levels of H3K4me3 and H3K27ac, suggesting low transcriptional activity. On the other hand, genes with low Adf-1 have similar amounts of Pol II but higher levels of these two histone modifications (Fig. 8E,F). The presence of Pol II at the TSSs of both groups of genes with different transcriptional activity suggest a possible role for Adf-1 in transcription pausing. To further dissect the role of Adf-1 in gene expression, we measured the pausing index of Group 1 and Group 2 genes. Figure 8G shows that Adf-1-bound genes have a high pausing index, supporting a role for this protein in promoter-proximal pausing.
Figure 8.
Adf-1 binding to gene promoters correlates with increased RNA Pol II pausing. A, Adf-1 DNA binding peaks in Kc cells are enriched close to the TSS. B, Distribution of genome-wide Adf-1 binding in Kc cells. C, Two independent genes showing high (left) and low (right) Adf-1 occupancy at the gene promoter (asterisks). RNA Pol II occupancy and the presence of two chromatic marks, H3K4me3 and H3K27ac, are also shown for these two gene regions. D, Heatmaps showing chromatin features around TSSs with different Adf-1 levels. Each panel represents 2 kb upstream and downstream of the TSSs. The sites are ordered by the intensity of Adf-1 at TSSs. Part of the left panel is enlarged to allow detailed visualization. Group 1 and Group 2 TSSs were selected for the examination of pausing index in G. E, F, Box plot of Adf-1 (E) or H3K4me3 (F) levels at TSSs of genes in Group 1 or Group 2 genes from D. G, Distribution of genes in Groups 1 and 2 with respect to pausing index. Group 1 genes (that have a higher enrichment of Adf-1) have a higher pausing index compared with Group 2 genes (that are less enriched for Adf-1 binding).
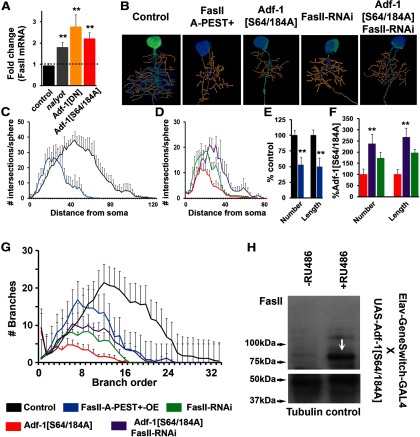
Adf-1 controls FasII and Staufen expression to regulate dendrite growth
We tested a number of genes that were identified as binding Adf-1 close to the TSS as potential Adf-1 targets in the brain, and confirmed Adf-1-dependent regulation of FasII, a Drosophila homolog of NCAM (neural cell adhesion molecule; Lin et al., 1994). Adf-1 inhibition resulted in increased FasII mRNA and protein in adult heads (Fig. 9A,H), and Adf-1 binding to the FasII enhancer region in Kc cells also correlated with paused Pol II and minimal occupancy for H3K4me3 and H3K27ac (Fig. 7D). We next asked whether FasII functions downstream of Adf-1 to regulate dendrite growth in RP2 motor neurons in a predictable manner. If Adf-1 inhibits FasII expression, then we would predict the following: (1) reduced dendrite growth following overexpression of FasII in RP2 neurons; and (2) suppression of Adf-1 inhibition-dependent dendrite growth phenotypes by FasII knockdown. The results shown in Figure 9A–F support these predictions. Thus, overexpression of full-length FasII (using a transgene that expresses the FasIIA-PEST+ sequence; Beck et al., 2012) in RP2 neurons results in reduced dendritic complexity (Fig. 9B,C) and reduced dendritic branching and total length (Fig. 9E,G). Conversely, RNAi-mediated FasII knockdown in an Adf-1-inhibited background (expression of Adf-1[S64/184A]) partially rescued dendritic complexity, branch number, and total length compared with Adf-1 inhibition alone (Fig. 9B,D,F,G). Note that, compared with control neurons, both the overexpression and knockdown of FasII resulted in fewer dendrites. This is consistent with prior observations that have suggested an optimum requirement for FasII in neuronal growth (Schuster et al., 1996; Beck et al., 2012).
Figure 9.
Adf-1 regulates RP2 dendrite growth by controlling FasII expression. A, qRT-PCR analysis from adult brains showing Adf-1-dependent regulation of FasII. B, Representative images of 3D reconstructed RP2 neurons from genotypes that modify Adf-1 and FasII. C, Volumetric Sholl analysis of FasII-overexpressing neurons compared with control. D, Volumetric Sholl analysis to test genetic interaction between Adf-1 and FasII. E, Quantification of dendrite branch number and total dendrite length in FasII-overexpressing neurons. F, Quantification of dendrite branch number and length for Adf-1–FasII interaction. G, Branch-order analysis for Adf-1–FasII interaction. H, Western blot from adult brain showing increased expression of FasII (arrow) 18 h after the induction of Adf-1[S64/184A] in neurons with RU486. Tubulin is used as a loading control.
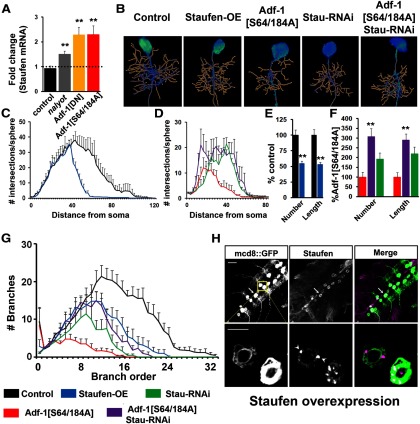
We also noticed in independent experiments that Adf-1 perturbations affected mRNA levels of the RNA-binding and transporting protein Staufen. Although we did not detect Adf-1 binding close to the Staufen gene region, Adf-1 inhibition increased Staufen expression (Fig. 10A), perhaps through an indirect mechanism. Phenotypically, Staufen overexpression using an “EP (Enhancer-Promoter)” insertion (verified in Fig. 10H) resulted in reduced dendritic complexity, branch number, and length (Fig. 10B,C,E,G). Additionally, RNAi-mediated Staufen knockdown partially rescued dendrite growth phenotypes resulting from Adf-1 inhibition in RP2 neurons (Fig. 10B,D,F,G) compared with Adf-1 inhibition alone. Since both FasII and Staufen have been implicated in memory formation and in axonal or dendrite development (Schuster et al., 1996; Kiebler et al., 1999; Cheng et al., 2001; Tang et al., 2001; Dubnau et al., 2003; Kristiansen et al., 2005; Barbee et al., 2006; Vessey et al., 2008; Beck et al., 2012; Miyashita et al., 2012), our results support a model in which FasII and Staufen are two target genes that function downstream of Adf-1 in the regulation of behavioral and cellular forms of activity-dependent plasticity (Fig. 11, models).
Figure 10.
Adf-1 controls dendrite growth by regulating Staufen transcription. A, qRT-PCR analysis of Staufen expression from adult brains following Adf-1 perturbation. B, Representative RP2 neuron dendrite reconstructions from genotypes that modify Adf-1 and Staufen. C, Volumetric Sholl analysis of Staufen-overexpressing neurons compared with control. D, Volumetric Sholl analysis to test the genetic interaction between Adf-1 and Staufen. E, Quantification of dendrite number and length in Staufen-overexpressing neurons. F, Quantification of dendrite branch number and length for Adf-1–Stau interactions. **p < 0.01. G, Branch-order analysis for Adf-1–Staufen interaction. H, Ventral nerve cords of an EP insertion upstream of Staufen crossed to the RN2-flipout tester GAL4 line stained for mcd8::GFP and Staufen. The bottom row shows close up of RP2 neurons. Arrowheads mark Staufen-positive granules. Scale bars: top row, 50 μm; bottom row, 20 μm.
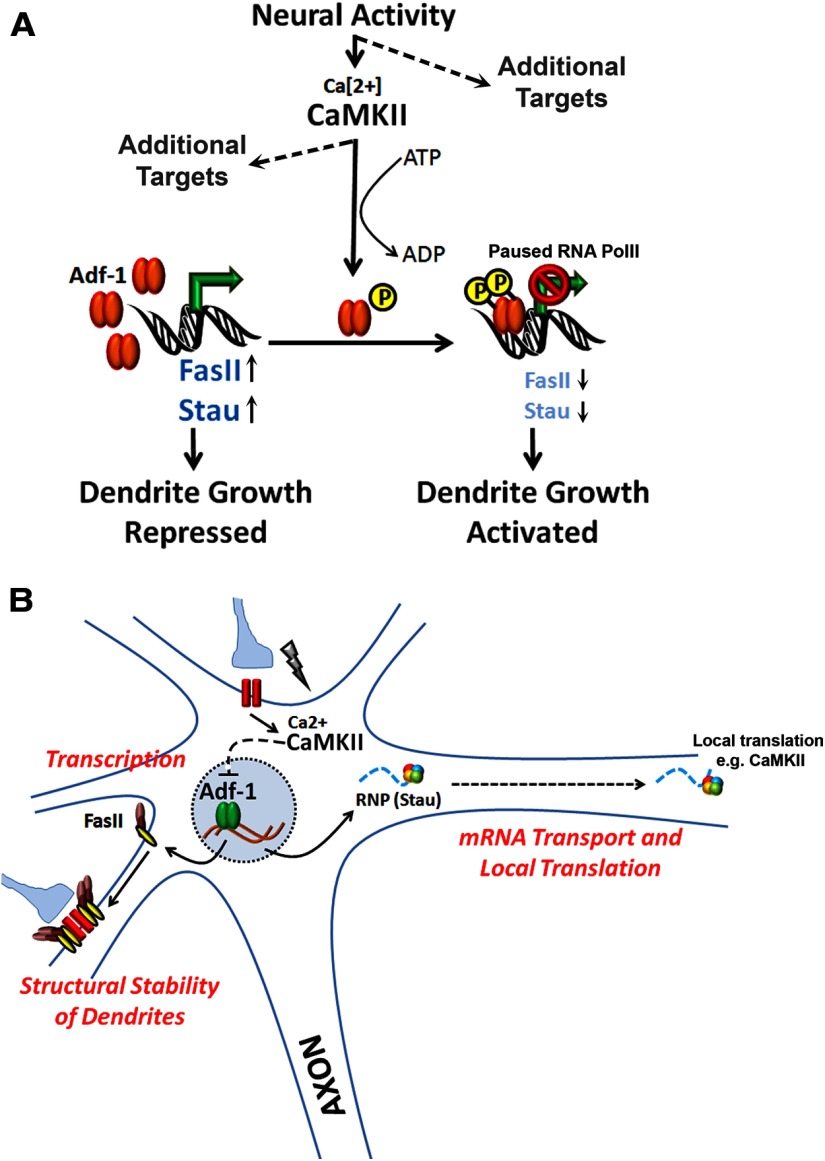
Figure 11.
Regulation of dendrite plasticity in Drosophila by the transcription factorAdf-1. A, Model of Adf-1-dependent regulation of dendrite growth. Adf-1 functions downstream of neuronal activity and CaMKII signaling. Adf-1 binding at promoters of genes such as FasII negatively regulates their transcription, thereby optimizing their expression to suit demands for dendrite growth. B, By regulating genes such as FasII and Staufen, Adf-1might be in a position to couple mechanisms underlying transcription and local translation in neurons during growth and plasticity of dendrites.
Discussion
Our study reveals novel functions for the Drosophila Myb family transcription factor Adf-1 in the regulation of dendrite growth and plasticity, and, by connecting Adf-1 with CaMKII signaling and transcriptional regulation of FasII and Staufen, provides a likely cellular basis for long-term memory defects seen in nalyot mutants (DeZazzo et al., 2000; Kacsoh et al., 2013). Since Adf-1 is not the sole transcription factor in vivo that regulates dendrite growth and plasticity, it is to be expected that Adf-1-dependent rescue of dendrite growth phenotypes resulting from the inhibition of CaMKII or activity will be partial. Thus, Adf-1 is necessary, but not wholly sufficient, for dendrite development and activity- and CaMKII-dependent plasticity. It is known that a number of transcription factors such as Fos, Creb, Mef2, and NFAT (Jessen et al., 2001; Redmond et al., 2002; Aizawa et al., 2004; Flavell et al., 2006; Shalizi et al., 2006; Wayman et al., 2006; Hartwig et al., 2008; Tai et al., 2008; Desfrere et al., 2009; Li et al., 2009; Schwartz et al., 2009; Kwon et al., 2011; Vonhoff et al., 2013) regulate dendrite development, and further studies are needed to determine the relative importance of Adf-1 in relation to these transcription factors. It is also formally possible that Adf-1 functions in a pathway that is parallel to, or distinct from, one that involves CaMKII signaling. In that case, it might be that a kinase other than CaMKII phosphorylates Adf-1 on these serine residues as well as at other amino acids. Nonetheless, the strength of dendrite growth phenotypes following Adf-1 perturbation underscores the salience of this transcription factor as well as these phosphorylation events in dendrite growth regulation.
The Adf-1[DN] transgene likely leads to the inhibition of endogenous Adf-1 (and not increased Adf-1 activity) since: (1) Adf-1[DN] and nalyot have similar phenotypes; (2) the Adf-1[DN] and [S64/184A] versions of Adf-1 have similar phenotypes (Fig. 3A–E); (3) pulsed expression of Adf-1[DN] in the adult brain influences the expression of ADH in a manner similar to that in nalyot mutants (Fig. 2A); (4) Adf-1[DN] and Adf-1[S64/184A] affect the expression of two Adf-1-regulated genes, FasII and Staufen, similarly (Figs. 9, 10); (5) a truncated Adf-1 that retains a dimerization domain is likely to function as a dominant negative; and (6) chronic pan-neuronal expression of Adf-1[DN] results in lethality similar to Adf-1-null homozygotes. Motor neuron inhibition of Adf-1 leads to discernible negative outcomes in the membrane properties of neurons (Fig. 2). We speculate that this is due to a significant reduction in the thickness of the primary neurite, which makes it difficult for the current to spread from the electrode to the spike-initiating zone, or due to disrupted expression of the Para voltage-gated Na channel or a critical subunit, leading to the absence of functional channels on the plasma membrane. Synaptic stimulation of motor neurons is also strongly attenuated when Adf-1 function is selectively impaired in motor neurons (Fig. 5). It is likely that a reduction in the number and branching of dendrites limits the number of presynaptic inputs on to these motor neurons. Alternatively, synaptic stimulation of motor neurons might also be impaired due to the reduced diameter of dendrites that would increase axial resistance, and therefore reduce the spread of synaptic currents to the spike-initiating zone, thereby reducing the transfer function (Gray and Weeks, 2003). Animals with motor neuron inhibition of Adf-1 also show strong deficits in motor behavior (Fig. 6) that are developmental in origin, consistent with a developmental requirement for Adf-1 in long-term memory formation (DeZazzo et al., 2000). We think that a reduction in the excitability of motor neurons and, therefore, a loss in the efficacy of sensori-motor integration, provides the most parsimonious explanation for the behavioral deficits seen in Adf-1 mutants and in animals with a motor neuron-specific knockdown of Adf-1.
Plasticity-related transcription factors might regulate very specific subsets of genes in the brain, few of which have been identified and validated phenotypically (McClung and Nestler, 2003; Etter et al., 2005). Recent work suggests that the transcriptional state of a given gene can only be accurately derived from transcription factor binding correlated with gene-specific chromatin states in a cell-specific manner (Kirilly et al., 2011; Busser et al., 2012; Wilczynski et al., 2012). Based on this observation, our ChIP-Seq analysis confirms that Adf-1 binds close to the transcription start site of many genes in the brain. Clustering of these genes based on gene ontology reveals a tight collection of gene ontology (GO) terms that relate to neuronal function and development (Fig. 7F), consistent with neural functions for Adf-1. Additionally, the primary consensus motif for Adf-1 binding derived from our ChIP-Seq studies matches a more recent analysis of Adf-1 binding sites (Lang and Juan, 2010). The importance of carrying out ChIP-Seq in a tissue-specific manner is also underscored by the observation that an equivalent ChIP-Seq analysis performed in Drosophila Kc cells reveals Adf-1 binding sites that only display a minor overlap with the brain dataset (Fig. 7G), suggesting tissue-specific gene regulation by Adf-1. Does Adf-1 regulate a subset of genes whose mRNA is highly enriched in dendrites? Although a dataset for dendritically enriched mRNAs is not available for Drosophila, a recent study has reported a set of 2550 mRNAs that are enriched in either axons or dendrites in the synaptic neuropil of area CA1 in the adult rat hippocampus (Cajigas et al., 2012). Comparison of the Adf-1 binding genes in the larval brain with this dataset shows that 91 genes (including L1 CAM and Stau2) are shared between them (Fig. 7G)—a group that might represent Adf-1-regulated genes that are especially relevant to dendrite development.
We were surprised to find that Adf-1 inhibition led to increased expression of a bona fide transcriptional target of Adf-1, ADH. Pan-neuronal inhibition of Adf-1 also increased expression of FasII mRNA (a direct Adf-1 target gene) and Staufen mRNA (an indirectly regulated gene; Figs. 9A, 10A). Interestingly, comparison of Adf-1-bound promoters with RNA Pol II distribution in the promoter and gene body, and active chromatin marks such as H3K4me3 and H3K27ac in Kc cells suggested a pattern in which Adf-1-bound genes correlated positively with higher proximal pausing indices for RNA Pol II (Fig. 8D–G), and negatively with H3K4me3 and H3K27ac. In Drosophila, the release of paused Pol II requires phosphorylation of H3S10, which serves as a mark for recruiting 14–3-3, which in turn recruits various histone acetyltransferases to acetylate different residues in histones H3 and H4, including H3K27ac (Kellner et al., 2012). These observations suggest that Adf-1 may act early in the pausing process, since Adf-1-bound genes have low levels of H3K27ac at their promoters.
Finally, we examined two candidate genes whose mRNA levels are regulated by Adf-1 in the brain, FasII and Staufen. Both genes are negatively regulated by Adf-1, and phenotypic analysis positions them downstream to Adf-1 in dendrite growth regulation (Figs. 9, 10). These results suggest that Adf-1 regulates dendrite growth by modulating the expression of FasII. An optimum requirement for FasII is also reminiscent of FasII-dependent presynaptic growth regulation that has been described previously at larval motor synapses (Schuster et al., 1996; Beck et al., 2012). It also suggests that signaling mechanisms that are either proximal or distal to FasII/NCAM might be conserved during postsynaptic growth. Since essential roles for NCAMs in neurite outgrowth have been reported widely (for review, see Corty et al., 2009; Schmid and Maness, 2008), our findings implicate a conserved mechanism downstream of Adf-1 in dendrite growth regulation. We favor the idea that Adf-1 regulates Staufen indirectly since we did not detect Adf-1 binding sites close to the Staufen TSS. Nonetheless, the observation that Adf-1 can modulate expression of a key regulator of mRNA transport in dendrites is intriguing. It might suggest a crucial interplay between nuclear transcription and mechanisms that govern local mRNA transport, sequestration, and translation in dendrites. Staufen-positive ribonucleoprotein (RNP) granules in these larval neurons have been shown to contain regulatory components of yeast P-granules (Barbee et al., 2006), and we speculate that part of the mechanism by which Adf-1 regulates dendrite growth is by controlling Staufen levels in vivo. While other studies have revealed how the local translational machinery can impact transcription (Thompson et al., 2004; Cox et al., 2008; Lai et al., 2008; Ch'ng et al., 2012) our results indicate a complementary pathway by which transcription factors can directly impact components of RNP granules that could affect mRNA transport and translation in dendrites (Fig. 11, models).
In sum, our study provides evidence for a model in which Adf-1 functions downstream of neural activity and CaMKII signaling to control dendrite growth by interacting with the chromatin machinery to regulate transcription of two proteins that are involved in neurite outgrowth (FasII) and mRNA transport (Staufen). Our results suggest that these proteins, all of which have been implicated in plasticity and long-term memory formation, function together to precisely modulate dendrite growth during development and plasticity, potentially through interplay among transcription, cell adhesion, and local translation.
Footnotes
This work was supported by grants from the Brain and Behavior Research Foundation (formerly the National Alliance for Research on Schizophrenia and Depression), and by National Institutes of Health Grants 1R03DA027979 and 5R21MH091520 to S. Sanyal, and 2R01GM035463 to V.G.C. We thank Carsten Duch, Felix Evers, and Fernando Vonhoff for help with AMIRA and 3D dendrite reconstruction; and Pearl Ryder and Victor Faundez for help with Cytoscape. The anti-Elav (Gerald Rubin) antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development at the University of Iowa, Department of Biological Sciences, Iowa City, IA.
The authors declare no competing financial interests.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/S0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- Baines RA, Uhler JP, Thompson A, Sweeney ST, Bate M. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci. 2001;21:1523–1531. doi: 10.1523/JNEUROSCI.21-05-01523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, Brand AH, Newbury SF, Wilhelm JE, Levine RB, Nakamura A, Parker R, Ramaswami M. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck ES, Gasque G, Imlach WL, Jiao W, Jiwon Choi B, Wu PS, Kraushar ML, McCabe BD. Regulation of Fasciclin II and synaptic terminal development by the splicing factor beag. J Neurosci. 2012;32:7058–7073. doi: 10.1523/JNEUROSCI.3717-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Kitamoto T, Greenspan RJ. Excitatory and inhibitory switches for courtship in the brain of Drosophila melanogaster. Curr Biol. 2004;14:538–547. doi: 10.1016/j.cub.2004.03.037. [DOI] [PubMed] [Google Scholar]
- Budnik V, Koh YH, Guan B, Hartmann B, Hough C, Woods D, Gorczyca M. Regulation of synapse structure and function by the Drosophila tumor suppressor gene dlg. Neuron. 1996;17:627–640. doi: 10.1016/S0896-6273(00)80196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne RD. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat Rev Neurosci. 2007;8:182–193. doi: 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busser BW, Huang D, Rogacki KR, Lane EA, Shokri L, Ni T, Gamble CE, Gisselbrecht SS, Zhu J, Bulyk ML, Ovcharenko I, Michelson AM. Integrative analysis of the zinc finger transcription factor lame duck in the Drosophila myogenic gene regulatory network. Proc Natl Acad Sci U S A. 2012;109:20768–20773. doi: 10.1073/pnas.1210415109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Endo K, Wu K, Rodan AR, Heberlein U, Davis RL. Drosophila fasciclinII is required for the formation of odor memories and for normal sensitivity to alcohol. Cell. 2001;105:757–768. doi: 10.1016/S0092-8674(01)00386-5. [DOI] [PubMed] [Google Scholar]
- Ch'ng TH, Uzgil B, Lin P, Avliyakulov NK, O'Dell TJ, Martin KC. Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell. 2012;150:207–221. doi: 10.1016/j.cell.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JC, Park D, Griffith LC. Electrophysiological and morphological characterization of identified motor neurons in the Drosophila third instar larva central nervous system. J Neurophysiol. 2004;91:2353–2365. doi: 10.1152/jn.01115.2003. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Cline HT. Dendritic arbor development and synaptogenesis. Curr Opin Neurobiol. 2001;11:118–126. doi: 10.1016/S0959-4388(00)00182-3. [DOI] [PubMed] [Google Scholar]
- Colomb J, Reiter L, Blaszkiewicz J, Wessnitzer J, Brembs B. Open source tracking and analysis of adult Drosophila locomotion in Buridan's paradigm with and without visual targets. PLoS One. 2012;7:e42247. doi: 10.1371/journal.pone.0042247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consoulas C, Restifo LL, Levine RB. Dendritic remodeling and growth of motoneurons during metamorphosis of Drosophila melanogaster. J Neurosci. 2002;22:4906–4917. doi: 10.1523/JNEUROSCI.22-12-04906.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corty MM, Matthews BJ, Grueber WB. Molecules and mechanisms of dendrite development in Drosophila. Development. 2009;136:1049–1061. doi: 10.1242/dev.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasari S, Cooper RL. Modulation of sensory-CNS-motor circuits by serotonin, octopamine, and dopamine in semi-intact Drosophila larva. Neurosci Res. 2004;48:221–227. doi: 10.1016/j.neures.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Davis GW, Schuster CM, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. III. CREB is necessary for presynaptic functional plasticity. Neuron. 1996;17:669–679. doi: 10.1016/S0896-6273(00)80199-3. [DOI] [PubMed] [Google Scholar]
- Desfrere L, Karlsson M, Hiyoshi H, Malmersjö S, Nanou E, Estrada M, Miyakawa A, Lagercrantz H, El Manira A, Lal M, Uhlén P. Na, K-ATPase signal transduction triggers CREB activation and dendritic growth. Proc Natl Acad Sci U S A. 2009;106:2212–2217. doi: 10.1073/pnas.0809253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZazzo J, Sandstrom D, de Belle S, Velinzon K, Smith P, Grady L, DelVecchio M, Ramaswami M, Tully T. nalyot, a mutation of the Drosophila myb-related Adf1 transcription factor, disrupts synapse formation and olfactory memory. Neuron. 2000;27:145–158. doi: 10.1016/S0896-6273(00)00016-7. [DOI] [PubMed] [Google Scholar]
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, Broger C, Tully T. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/S0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- Etter PD, Narayanan R, Navratilova Z, Patel C, Bohmann D, Jasper H, Ramaswami M. Synaptic and genomic responses to JNK and AP-1 signaling in Drosophila neurons. BMC Neurosci. 2005;6:39. doi: 10.1186/1471-2202-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers JF, Schmitt S, Sibila M, Duch C. Progress in functional neuroanatomy: precise automatic geometric reconstruction of neuronal morphology from confocal image stacks. J Neurophysiol. 2005;93:2331–2342. doi: 10.1152/jn.00761.2004. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A, Franciscovich A, Bowers M, Sandstrom DJ, Sanyal S. NFAT regulates pre-synaptic development and activity-dependent plasticity in Drosophila. Mol Cell Neurosci. 2011;46:535–547. doi: 10.1016/j.mcn.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A, Pranski E, Miller RD, Radmard S, Bernhard D, Jinnah HA, Betarbet R, Rye DB, Sanyal S. Sleep fragmentation and motor restlessness in a Drosophila model of Restless Legs Syndrome. Curr Biol. 2012;22:1142–1148. doi: 10.1016/j.cub.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudillière B, Konishi Y, de la Iglesia N, Yao Gl, Bonni A. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/S0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
- Götz KG. Visual guidance in Drosophila. Basic Life Sci. 1980;16:391–407. doi: 10.1007/978-1-4684-7968-3_28. [DOI] [PubMed] [Google Scholar]
- Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M, Crabtree GR. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell. 2003;113:657–670. doi: 10.1016/S0092-8674(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Gray JR, Weeks JC. Steroid-induced dendritic regression reduces anatomical contacts between neurons during synaptic weakening and the developmental loss of a behavior. J Neurosci. 2003;23:1406–1415. doi: 10.1523/JNEUROSCI.23-04-01406.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Greene LA, Ziff EB. Nerve growth factor and epidermal growth factor induce rapid transient changes in proto-oncogene transcription in PC12 cells. J Biol Chem. 1985;260:14101–14110. [PubMed] [Google Scholar]
- GuptaRoy B, Marwaha N, Pla M, Wang Z, Nelson HB, Beckingham K, Griffith LC. Alternative splicing of Drosophila calcium/calmodulin-dependent protein kinase II regulates substrate specificity and activation. Brain Res Mol Brain Res. 2000;80:26–34. doi: 10.1016/S0169-328X(00)00115-7. [DOI] [PubMed] [Google Scholar]
- Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, McKillip E, Shah S, Stapleton M, Wan KH, Yu C, Parsa B, Carlson JW, Chen X, Kapadia B, VijayRaghavan K, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig CL, Worrell J, Levine RB, Ramaswami M, Sanyal S. Normal dendrite growth in Drosophila motor neurons requires the AP-1 transcription factor. Dev Neurobiol. 2008;68:1225–1242. doi: 10.1002/dneu.20655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein U, England B, Tjian R. Characterization of Drosophila transcription factors that activate the tandem promoters of the alcohol dehydrogenase gene. Cell. 1985;41:965–977. doi: 10.1016/S0092-8674(85)80077-5. [DOI] [PubMed] [Google Scholar]
- Jakawich SK, Nasser HB, Strong MJ, McCartney AJ, Perez AS, Rakesh N, Carruthers CJ, Sutton MA. Local presynaptic activity gates homeostatic changes in presynaptic function driven by dendritic BDNF synthesis. Neuron. 2010;68:1143–1158. doi: 10.1016/j.neuron.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen U, Novitskaya V, Pedersen N, Serup P, Berezin V, Bock E. The transcription factors CREB and c-Fos play key roles in NCAM-mediated neuritogenesis in PC12–E2 cells. J Neurochem. 2001;79:1149–1160. doi: 10.1046/j.1471-4159.2001.00636.x. [DOI] [PubMed] [Google Scholar]
- Jin P, Griffith LC, Murphey RK. Presynaptic calcium/calmodulin-dependent protein kinase II regulates habituation of a simple reflex in adult Drosophila. J Neurosci. 1998;18:8955–8964. doi: 10.1523/JNEUROSCI.18-21-08955.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, Garner CC, Tsien RY, Ellisman MH, Malenka RC. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci. 2004;7:244–253. doi: 10.1038/nn1189. [DOI] [PubMed] [Google Scholar]
- Kacsoh BZ, Lynch ZR, Mortimer NT, Schlenke TA. Fruit flies medicate offspring after seeing parasites. Science. 2013;339:947–950. doi: 10.1126/science.1229625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Kellner WA, Ramos E, Van Bortle K, Takenaka N, Corces VG. Genome-wide phosphoacetylation of histone H3 at Drosophila enhancers and promoters. Genome Res. 2012;22:1081–1088. doi: 10.1101/gr.136929.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Hemraj I, Verkade P, Köhrmann M, Fortes P, Marión RM, Ortín J, Dotti CG. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MD, Wen Y, Jan YN. Patterning and organization of motor neuron dendrites in the Drosophila larva. Dev Biol. 2009;336:213–221. doi: 10.1016/j.ydbio.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilly D, Wong JJ, Lim EK, Wang Y, Zhang H, Wang C, Liao Q, Wang H, Liou YC, Wang H, Yu F. Intrinsic epigenetic factors cooperate with the steroid hormone ecdysone to govern dendrite pruning in Drosophila. Neuron. 2011;72:86–100. doi: 10.1016/j.neuron.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Kitamoto T. Conditional disruption of synaptic transmission induces male-male courtship behavior in Drosophila. Proc Natl Acad Sci U S A. 2002;99:13232–13237. doi: 10.1073/pnas.202489099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen LV, Velasquez E, Romani S, Baars S, Berezin V, Bock E, Hortsch M, Garcia-Alonso L. Genetic analysis of an overlapping functional requirement for L1- and NCAM-type proteins during sensory axon guidance in Drosophila. Mol Cell Neurosci. 2005;28:141–152. doi: 10.1016/j.mcn.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Kwon M, Fernández JR, Zegarek GF, Lo SB, Firestein BL. BDNF-promoted increases in proximal dendrites occur via CREB-dependent transcriptional regulation of cypin. J Neurosci. 2011;31:9735–9745. doi: 10.1523/JNEUROSCI.6785-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KO, Zhao Y, Ch'ng TH, Martin KC. Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc Natl Acad Sci U S A. 2008;105:17175–17180. doi: 10.1073/pnas.0803906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaLonde M, Janssens H, Yun S, Crosby J, Redina O, Olive V, Altshuller YM, Choi SY, Du G, Gergen JP, Frohman MA. A role for phospholipase D in Drosophila embryonic cellularization. BMC Dev Biol. 2006;6:60. doi: 10.1186/1471-213X-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf M, Jeffrey V, Fujioka M, Jaynes JB, Bate M. Embryonic origins of a motor system: motor dendrites form a myotopic map in Drosophila. PLoS Biol. 2003;1:E41. doi: 10.1371/journal.pbio.0000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M, Juan E. Binding site number variation and high-affinity binding consensus of Myb-SANT-like transcription factor Adf-1 in Drosophilidae. Nucleic Acids Res. 2010;38:6404–6417. doi: 10.1093/nar/gkq504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layden MJ, Odden JP, Schmid A, Garces A, Thor S, Doe CQ. Zfh1, a somatic motor neuron transcription factor, regulates axon exit from the CNS. Dev Biol. 2006;291:253–263. doi: 10.1016/j.ydbio.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature. 2009;458:299–304. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang C, Takemori H, Zhou Y, Xiong ZQ. TORC1 regulates activity-dependent CREB-target gene transcription and dendritic growth of developing cortical neurons. J Neurosci. 2009;29:2334–2343. doi: 10.1523/JNEUROSCI.2296-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DM, Fetter RD, Kopczynski C, Grenningloh G, Goodman CS. Genetic analysis of Fasciclin II in Drosophila: defasciculation, refasciculation, and altered fasciculation. Neuron. 1994;13:1055–1069. doi: 10.1016/0896-6273(94)90045-0. [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/S0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- Mauss A, Tripodi M, Evers JF, Landgraf M. Midline signalling systems direct the formation of a neural map by dendritic targeting in the Drosophila motor system. PLoS Biol. 2009;7:e1000200. doi: 10.1371/journal.pbio.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Roman G, Davis RL. Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 2004;20:384–391. doi: 10.1016/j.tig.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/S0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Oda Y, Horiuchi J, Yin JC, Morimoto T, Saitoe M. Mg(2+) block of Drosophila NMDA receptors is required for long-term memory formation and CREB-dependent gene expression. Neuron. 2012;74:887–898. doi: 10.1016/j.neuron.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JI, Curran T. Role of ion flux in the control of c-fos expression. Nature. 1986;322:552–555. doi: 10.1038/322552a0. [DOI] [PubMed] [Google Scholar]
- Mosca TJ, Carrillo RA, White BH, Keshishian H. Dissection of synaptic excitability phenotypes by using a dominant-negative Shaker K+ channel subunit. Proc Natl Acad Sci U S A. 2005;102:3477–3482. doi: 10.1073/pnas.0406164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaï LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, Hassan BA. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc Natl Acad Sci U S A. 2010;107:20553–20558. doi: 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo AA, Ward E, Skeath JB, Dean DC. zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol Cell Biol. 1999;19:7255–7263. doi: 10.1128/mcb.19.10.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond L, Ghosh A. Regulation of dendritic development by calcium signaling. Cell Calcium. 2005;37:411–416. doi: 10.1016/j.ceca.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. doi: 10.1016/S0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Rongo C, Kaplan JM. CaMKII regulates the density of central glutamatergic synapses in vivo. Nature. 1999;402:195–199. doi: 10.1038/46065. [DOI] [PubMed] [Google Scholar]
- Salvaterra PM, Kitamoto T. Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res Gene Expr Patterns. 2001;1:73–82. doi: 10.1016/S1567-133X(01)00011-4. [DOI] [PubMed] [Google Scholar]
- Sánchez-Soriano N, Bottenberg W, Fiala A, Haessler U, Kerassoviti A, Knust E, Löhr R, Prokop A. Are dendrites in Drosophila homologous to vertebrate dendrites? Dev Biol. 2005;288:126–138. doi: 10.1016/j.ydbio.2005.09.026. [DOI] [PubMed] [Google Scholar]
- Sandstrom DJ. Isoflurane reduces excitability of Drosophila larval motoneurons by activating a hyperpolarizing leak conductance. Anesthesiology. 2008;108:434–446. doi: 10.1097/ALN.0b013e318164cfda. [DOI] [PubMed] [Google Scholar]
- Sanyal S. Genomic mapping and expression patterns of C380, OK6 and D42 enhancer trap lines in the larval nervous system of Drosophila. Gene Expr Patterns. 2009;9:371–380. doi: 10.1016/j.gep.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Sandstrom DJ, Hoeffer CA, Ramaswami M. AP-1 functions upstream of CREB to control synaptic plasticity in Drosophila. Nature. 2002;416:870–874. doi: 10.1038/416870a. [DOI] [PubMed] [Google Scholar]
- Schmid RS, Maness PF. L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Curr Opin Neurobiol. 2008;18:245–250. doi: 10.1016/j.conb.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Evers JF, Duch C, Scholz M, Obermayer K. New methods for the computer-assisted 3-D reconstruction of neurons from confocal image stacks. Neuroimage. 2004;23:1283–1298. doi: 10.1016/j.neuroimage.2004.06.047. [DOI] [PubMed] [Google Scholar]
- Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron. 1996;17:641–654. doi: 10.1016/S0896-6273(00)80197-X. [DOI] [PubMed] [Google Scholar]
- Schwartz N, Schohl A, Ruthazer ES. Neural activity regulates synaptic properties and dendritic structure in vivo through calcineurin/NFAT signaling. Neuron. 2009;62:655–669. doi: 10.1016/j.neuron.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Auld VJ. Conversion of lacZ enhancer trap lines to GAL4 lines using targeted transposition in Drosophila melanogaster. Genetics. 1999;151:1093–1101. doi: 10.1093/genetics/151.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A, Gaudillière B, Yuan Z, Stegmüller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Steward O. Regulation of synaptogenesis through the local synthesis of protein at the postsynaptic site. Prog Brain Res. 1987;71:267–279. doi: 10.1016/S0079-6123(08)61830-0. [DOI] [PubMed] [Google Scholar]
- Steward O, Worley P. Local synthesis of proteins at synaptic sites on dendrites: role in synaptic plasticity and memory consolidation? Neurobiol Learn Mem. 2002;78:508–527. doi: 10.1006/nlme.2002.4102. [DOI] [PubMed] [Google Scholar]
- Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1994;175:179–191. doi: 10.1007/BF00215114. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nüsslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-O. [DOI] [PubMed] [Google Scholar]
- Strauss R, Heisenberg M. A higher control center of locomotor behavior in the Drosophila brain. J Neurosci. 1993;13:1852–1861. doi: 10.1523/JNEUROSCI.13-05-01852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Tai Y, Feng S, Ge R, Du W, Zhang X, He Z, Wang Y. TRPC6 channels promote dendritic growth via the CaMKIV-CREB pathway. J Cell Sci. 2008;121:2301–2307. doi: 10.1242/jcs.026906. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32:463–475. doi: 10.1016/S0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Tavosanis G. Dendritic structural plasticity. Dev Neurobiol. 2012;72:73–86. doi: 10.1002/dneu.20951. [DOI] [PubMed] [Google Scholar]
- Thompson KR, Otis KO, Chen DY, Zhao Y, O'Dell TJ, Martin KC. Synapse to nucleus signaling during long-term synaptic plasticity; a role for the classical active nuclear import pathway. Neuron. 2004;44:997–1009. doi: 10.1016/j.neuron.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Timmerman C, Sanyal S. Behavioral and electrophysiological outcomes of tissue-specific Smn knockdown in Drosophila melanogaster. Brain Res. 2012;1489:66–80. doi: 10.1016/j.brainres.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spronsen M, Hoogenraad CC. Synapse pathology in psychiatric and neurologic disease. Curr Neurol Neurosci Rep. 2010;10:207–214. doi: 10.1007/s11910-010-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JP, Macchi P, Stein JM, Mikl M, Hawker KN, Vogelsang P, Wieczorek K, Vendra G, Riefler J, Tübing F, Aparicio SA, Abel T, Kiebler MA. A loss of function allele for murine Staufen1 leads to impairment of dendritic Staufen1-RNP delivery and dendritic spine morphogenesis. Proc Natl Acad Sci U S A. 2008;105:16374–16379. doi: 10.1073/pnas.0804583105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonhoff F, Duch C. Tiling among stereotyped dendritic branches in an identified Drosophila motoneuron. J Comp Neurol. 2010;518:2169–2185. doi: 10.1002/cne.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonhoff F, Kuehn C, Blumenstock S, Sanyal S, Duch C. Temporal coherency between receptor expression, neural activity and AP-1-dependent transcription regulates Drosophila motoneuron dendrite development. Development. 2013;140:606–616. doi: 10.1242/dev.089235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrailas-Mortimer A, del Rivero T, Mukherjee S, Nag S, Gaitanidis A, Kadas D, Consoulas C, Duttaroy A, Sanyal S. A muscle-specific p38 MAPK/Mef2/MnSOD pathway regulates stress, motor function, and life span in Drosophila. Dev Cell. 2011;21:783–795. doi: 10.1016/j.devcel.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Impey S, Marks D, Saneyoshi T, Grant WF, Derkach V, Soderling TR. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron. 2006;50:897–909. doi: 10.1016/j.neuron.2006.05.008. [DOI] [PubMed] [Google Scholar]
- West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci. 2002;3:921–931. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3:a005744. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynski B, Liu YH, Yeo ZX, Furlong EE. Predicting spatial and temporal gene expression using an integrative model of transcription factor occupancy and chromatin state. PLoS Comput Biol. 2012;8:e1002798. doi: 10.1371/journal.pcbi.1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GY, Cline HT. Stabilization of dendritic arbor structure in vivo by CaMKII. Science. 1998;279:222–226. doi: 10.1126/science.279.5348.222. [DOI] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Yuste R. Dendritic spines and distributed circuits. Neuron. 2011;71:772–781. doi: 10.1016/j.neuron.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou DJ, Cline HT. Postsynaptic calcium/calmodulin-dependent protein kinase II is required to limit elaboration of presynaptic and postsynaptic neuronal arbors. J Neurosci. 1999;19:8909–8918. doi: 10.1523/JNEUROSCI.19-20-08909.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]