Abstract
Oxalobacter formigenes is a unique intestinal organism that relies on oxalate degradation to meet most of its energy and carbon needs. A lack of colonization is a risk factor for calcium oxalate stone disease. Protection against calcium oxalate stone disease appears to be due to the oxalate degradation that occurs in the gut on low calcium diets with a possible further contribution from intestinal oxalate secretion. Much remains to be learned about how the organism establishes and maintains gut colonization and the precise mechanisms by which it modifies stone risk. The sequencing and annotation of the genomes of a Group 1 and a Group 2 strain of O. formigenes should provide the informatic tools required for the identification of the genes and pathways associated with colonization and survival. In this review we have identified genes that may be involved and where appropriate suggested how they may be important in calcium oxalate stone disease. Elaborating the functional roles of these genes should accelerate our understanding of the organism and clarify its role in preventing stone formation.
INTRODUCTION
O. formigenes is part of the bacterial flora in the large intestine of many humans and other mammalian species. It is unique in that it requires oxalate both as an energy and carbon source. Its existence was first recognized from its role in acclimating livestock to the ingestion of high-oxalate diets and preventing oxalate toxicity [1, 2]. There is relatively very little known about the biology of the organism, particularly on how it copes with the stresses of the hostile environments it encounters and how it is able to establish a niche in the large intestine. The release of the genome sequence of a Group 1 (OXCC13) and a Group 2 strain (HOxBLS) as part of the Human Microbiome Project has provided a genetic framework for investigating important biological properties of the organism. From the genes identified it is clear that O. formigenes contains a repertoire of genes expected of an intestinal microbe that permit it to survive and effectively compete for its niche in the intestinal environment. General genome statistics are shown in Table 1. The approximate 2100 genes they contain are at the lower end of the range of 1500 – 7500 genes found in free-living bacteria [10].
Table 1.
Genome statistics of O. formigenes strains OXCC13 and HOxBLS.
| HOxBLS | OXCC13 | |
|---|---|---|
| DNA coding number of bases | 2063277 | 2088012 |
| G/C content | 52.68% | 49.59% |
| Protein coding genes | 2125 | 2076 |
| RNA genes | 46 | 48 |
| Proteins coding with a functional prediction | 1403 | 1443 |
| Protein coding as enzymes | 638 | 662 |
Because of the contribution of dietary oxalate to calcium oxalate stone disease [29], the potential relationship of this organism to intestinal oxalate balance and urinary oxalate excretion has attracted considerable attention. Whether high oral doses of this organism can promote sufficient intestinal oxalate secretion to diminish the oxalate burden on the kidney in individuals with Primary Hyperoxaluria is also being tested [30]. What is known about the biology of O. formigenes and its relationship to human health will be examined in this review. Considerations include the identification of the lack of colonization with O. formigenes as a risk factor for recurrent calcium oxalate stone formation; the observation in rodent models that colonization with O. formigenes can modify the oxalate transport properties of intestinal epithelium; the detection of a presumptive secretagogue in lysates of O. formigenes that modifies intestinal oxalate transport in the rat; and some of the most important aspects of the genomic sequence of O. formigenes. As well as covering these recent developments, we also identify the major unknown features of the organism. More information is required, for instance, on how the organism handles periods of oxalate deprivation, how it responds to antibiotics and other antibacterial agents, how they communicate as a group to enhance survival during periods of stress, and what mechanisms are employed to colonize the large intestine. Little is known on the temporal and spatial dynamics of O. formigenes as intestinal oxalate and calcium levels change and how this affects colonic oxalate absorption and secretion. We hypothesize that there is a mutualistic relationship between the host and this organism, with O. formigenes benefiting the host by releasing soluble calcium from ingested calcium oxalate crystals, and the host secreting oxalate into the gut, ensuring an adequate food supply for O. formigenes to sustain its growth during periods of dietary oxalate deprivation. While colonization with O. formigenes may provide some protection against idiopathic calcium oxalate stone disease, a complete characterization of the protective mechanisms is required as well as the exploration of ways to enhance this protective pathway. In addition, methods for establishing or re-establishing gut colonization and maintaining it need to be developed.
DESCRIPTION OF THE ORGANISM
O. formigenes is a Gram-negative, obligately anaerobic, rod or curve shaped, non-motile, non-spore forming bacterium that belongs to the Betaproteobacteria class and Burkholderiales order. Comparisons of 16S rRNA sequences from O. formigenes with sequences from a diversity of other Betaproteobacteria support the concept that O. formigenes is a distinct group of bacteria, and also that it shares a specific relationship with other genera (Telluria, Janthinobacterium and Duganella) that are currently in the Oxalobacteraceae family. Comparisons of the profiles of cellular fatty acids of 17 strains of O. formigenes, including strains isolated from gastrointestinal contents from humans, sheep, cattle, pigs, guinea pigs, rats and from fresh water lake sediments, supports the concept of separating these strains into two main groups (currently designated as Group I and II). In Group 1 strains, a cyclic 17 carbon fatty acid predominates whereas in Group 2 a cyclic 19 carbon acid is dominant [44]. These cyclic fatty acids are believed to help organisms better withstand environmental stress, such as acid tolerance [45]. The O. formigenes genomes contain a key gene essential for their synthesis that codes for cyclopropane-fatty-acyl-phospholipid synthase (Gene Locus tags: OFBG_01193, OFAG_00246). The relationship between fatty acid composition and the different sensitivities of the strains to antibiotics, bile salts, air and pH remains to be determined [4, 20].
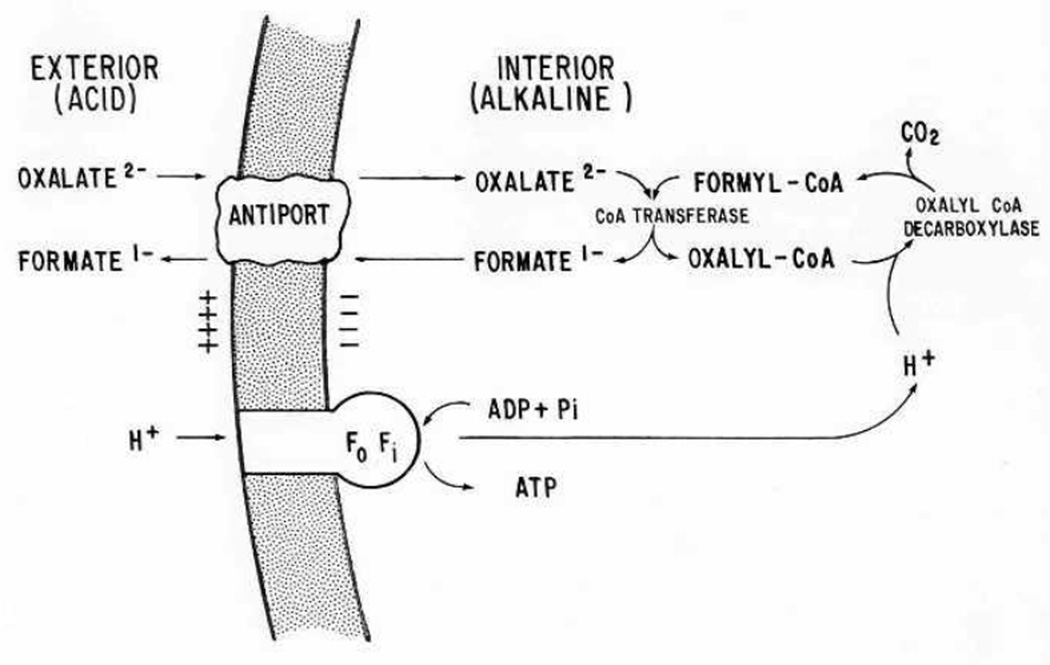
Growth of O. formigenes in culture occurs under anaerobic conditions, with optimal growth at pH between 6 and 7 in a carbonate-bicarbonate buffered medium that contains minerals, oxalate, acetate, and a small amount of yeast extract. Oxalate serves as both the energy yielding substrate and the major source of carbon for growth [16, 17]. The energy yield from this step is low, but sufficient to support growth. The products from oxalate metabolism are carbon-dioxide and formate, with approximately one mole of each produced per mole of oxalate metabolized. Energy generation is centered on the development of a proton motive force through the electrogenic exchange of oxalate (in) and formate (out) across the cell membrane together with the consumption of a proton inside the cell (Fig. 1) when the CoA-ester of oxalate is decarboxylated by oxaly-CoA-decarboxylase [3, 39]. This process can be maintained over a fairly wide external pH range (5.5 – 7.5), enabling it to survive in an environment where pH may fluctuate [39]. Both oxalyl-CoA-decarboxylase and the formyl-CoA transferase have been purified and characterized [6, 7]. An antiporter protein (OxlT) facilitates the oxalate-formate exchange [35, 52].
Figure 1.
Oxalate catabolism and ATP synthesis in O. formigenes. Electrogenic oxalate2-:formate1- exchange forms the basis for sustaining a proton-motive force. Taken from Anantharam et. al., 1989 [3].
GROWTH ON OXALATE
To date most of the information on the growth of the organism has been confined to the use of soluble oxalate in liquid culture. In liquid cultures with a high oxalate concentration, the buildup of formate in the medium can inhibit its growth. This response to formate restricts the growth of O. formigenes in chemostat cultures and poses a problem for the cultivation of large amounts of the bacteria for probiotic use. Different strains appear to have differing capacities to degrade oxalate with the maximal concentration supporting growth reported to be 111 mM [18]. No other substrate has yet been identified that it can grow on. It requires a low concentration of acetate (0.5 mM) to grow, but acetate alone cannot support growth [44]. Genes encoding a malonate decarboxylase (OFBG_00828, OFAG_00704) and a malonate transporter (OFBG_00815) have been identified in the O. formigenes genome, but malonate, a 3-carbon dicarboxylic acid, has not yet been demonstrated to support its growth [18]. The expression of the 3 proteins, oxaly-CoA-decarboxylase (OFAG_01484, OFBG_01523), formyl-Co A transferase (OFAG_02109, OFBG_02073), and OxlT (OFAG_01473, OFBG_01510), is central to oxalate metabolism in this organism (Fig. 1). However, it is not known how the expression of these enzymes is affected by the concentration of oxalate in the environment of the organism or by other potential modifiers. Understanding the regulation of expression is important as it could lead to ways to upregulate the expression of these enzymes and accelerate the breakdown of dietary oxalate in the gut.
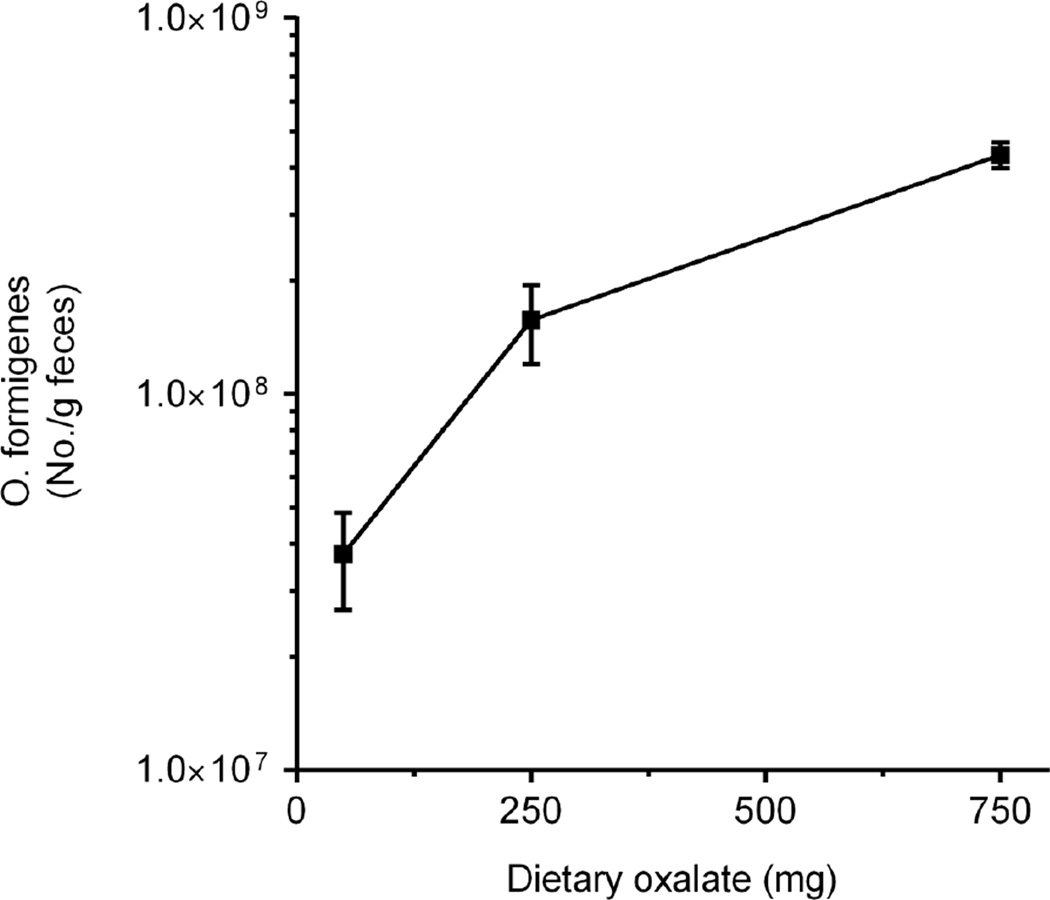
The dependency of O. formigenes on oxalate for growth has the potential to cause dramatic shifts in its population size. This was highlighted in a recent study where O. formigenes numbers were measured in the stool of healthy subjects equilibrated to diets controlled in oxalate, calcium and other nutrients as shown in Fig. 2 [34]. Numbers increased 12-fold on average as dietary oxalate increased 15-fold. The availability of the oxalate also influenced numbers as a 5-fold increase in dietary calcium decreased numbers approximately 5-fold. When the response to the diet was compared in colonized and non-colonized individuals, a significant difference in urinary oxalate excretion was observed only on the low calcium (400 g/day) diet (34.9 ± 2.6 vs. 43.6 ± 2.6 mg/day; P = 0.026). Of note also was the variability in numbers and how it was influenced by the amount of dietary oxalate (Table 2). The greatest variability was observed with the low oxalate diet (50 mg/day). This variability may reflect a greater sensitivity of Oxalobacter to the other gut flora as it struggles to maintain its population size, to some other host factor, or to variability in the availability of dietary oxalate or some other dietary component. The robustness of O. formigenes to survive such dramatic population shifts warrants further examination.
Figure 2.
Number of fecal O. formigenes with changes in dietary oxalate. Daily calcium intake was 1000 mg. Real-time PCR was used to quantitate O. formigenes numbers. Reprinted from Jiang et. al. [34], with permission from Elsevier
Table 2.
Range of O. formigenes fecal number with changes in dietary oxalate. Daily dietary calcium intake was 1000 mg. Real-time PCR was used to quantitate O. formigenes numbers. The limit of detection for this assay is 0.0001 ng of O. formigenes DNA, which equates to approximately 2.41 × 105 O. formigenes per g feces. Data previously published by Jiang et. al. [34].
| Daily Dietary Oxalate Intake (mg) | Range of O. formigenes fecal number | Fold difference |
|---|---|---|
| 50 | 3.9 × 105 − 9.9 × 107 | 2500 |
| 250 | 5 × 106 −4.2 × 108 | 800 |
| 750 | 2.6 × 108 − 5.7 × 108 | 2 |
Fecal oxalate measurements in the study by Jiang et. al. allowed the yield of O. formigenes per mg oxalate to be estimated [34]. Total fecal oxalate recovered from stool of non-colonized and colonized subjects on the highest oxalate diet tested (750mg oxalate/1000 mg calcium) was 477 ± 60 and 93 ± 25 mg oxalate (mean ± SEM), respectively, indicating O. formigenes degraded approximately 51% of dietary oxalate ingested per day and other bacteria degraded 36% of total dietary oxalate. Total mean fecal O. formigenes number on the 750mg oxalate diet was 4.9 × 1010 ± 8.7 × 109 cells (mean ± SEM). Thus, these data suggest that approximately 1 × 108 O. formigenes cells are produced per mg of dietary oxalate degraded, assuming that degradation of intestinal oxalate by bacteria other than O. formigenes was similar in colonized and non-colonized individuals. This is in contrast to measurements of O. formigenes cell yield from oxalate in culture, where 1.1 mg dry weight of O. formigenes cells is produced per millimole of oxalate [44]. Using values from studies with E. coli, another Gram-negative bacterium, 1 mg dry weight of cells equals 4 ×109 cells [51]. Thus, 1.1 mg of O. formigenes cells would equal about 4.4 × 109 cells produced per millimole of oxalate, or about 4.9 × 107 cells / mg oxalate. Surprisingly, this cell yield from oxalate in culture medium is 2.7 fold lower than calculated in stool of colonized subjects equilibrated to the controlled high oxalate diet described above. The reasons for this are unclear. It may be due to the approximations made or to conditions in vivo producing cell yields greater than those observed with optimized culture conditions.
An important question related to O. formigenes survival and growth in the gut is its capacity to degrade crystalline as well as soluble oxalate. Ingested oxalate is present in food as the soluble anion or as insoluble, crystalline calcium oxalate. Much of the oxalate that O. formigenes encounters in the gut may be insoluble as ingested soluble oxalate may precipitate as calcium oxalate due to the concentrations of calcium and oxalate reached in intestinal fluids. The concentration of calcium in the intestinal lumen is reportedly 0.5 − 1 mM calcium [42] and is influenced by both dietary calcium and intestinal calcium secretion. The ability of O. formigenes to degrade crystalline oxalate is evident in the desert-dwelling sand rat that feeds almost exclusively on cactus, an oxalate-rich food source that contains predominantly insoluble calcium oxalate as its major calcium source [49]. How O. formigenes facilitates the release of oxalate from such crystals or in some way promotes their dissolution is unclear. Some plants have the capability of dissolving calcium oxalate crystals present in idioblasts to relocate the crystals for seed development [31, 32] or to obtain calcium when it is restricted in the environment [22]. These studies illustrate the solubility of some plant crystals and suggest that they may contain inclusions such as proteins or peptides that facilitate their dissolution.
OXALOBACTER GENOME
Genes of interest are those associated with cell to cell communication, environmental sensing, microcolony formation, host interactions and response to stress. Both O. formigenes strains contain genes involved in general stress responses, including sigma factors, heat shock proteins, universal stress protein UspA, osmotic stress, oxidative stress (including a methionine sulfoxide reductase), and cold shock (Table 3). Functional studies are needed to demonstrate the involvement, if any, of these genes in the stress response of this organism.
Table 3.
Examples of genes identified in the genome of O. formigenes strain OXCC13 and strain HOxBLS.
| Cell-cell communication, transcriptional regulators and environmental sensing | ||
|---|---|---|
| OXCC13 Gene Loci tag | HOxBLS Gene Loci tag | |
| Serine/threonine-protein kinase HipA |
OFBG_00481, OFBG_00898 | OFAG_00619 |
| two-component system sensor histidine kinase |
OFBG_01453, OFBG_00240 | OFAG_01556, OFAG_00163 |
| RNA polymerase sigma factor RpoD, RpoH and RpoN |
OFBG_00182, OFBG_02057, OFBG_01317 | OFAG_01295, OFAG_00100, OFAG_02023 |
| heat-inducible transcriptional repressor |
OFBG_02048 | OFAG_02014 |
| MarR family transcriptional regulator |
OFBG_01978, OFBG_00494, OFBG_00985 | OFAG_01943, OFAG_00564, OFAG_00562 |
| integration host factor subunit alpha and beta |
OFBG_01627, OFBG_00154 | OFAG_00071, OFAG_01588 |
| transcriptional activator MetR | OFBG_01032 | OFAG_00527 |
| Pilus genes | ||
| type I pilus protein CsuB | OFBG_00909 | |
| type I pilus protein CsuA/B | OFBG_00908 | |
| type 1 pili usher pathway chaperone CsuC |
OFBG_00910 | |
| type I pili usher protein CsuD | OFBG_00911 | |
| spore coat U domain- containing protein |
OFBG_00912 | |
| Phage genes | ||
| phage integrase | OFBG_01776 | OFAG_01357 |
| phage major capsid protein | OFBG_00323 | |
| bacteriophage tail fiber protein | OFBG_00659 | OFAG_00945 |
| Stress related genes | ||
| methionine-R-sulfoxide reductase |
OFBG_00160 | OFAG_00077 |
| cold shock domain protein CspA |
OFBG_01608 | OFAG_01569 |
| (p)ppGpp synthetase, RelA/SpoT family |
OFBG_01447, OFBG_01468, | OFAG_01420, OFAG_0.1440 |
| peroxiredoxin | OFBG_00067 | OFAG_02092 |
| hsp70-like protein | OFBG_02052 | OFAG_02018 |
| carbon starvation inducible proteins |
OFBG_00640 | OFAG_00974 |
| phosphate starvation inducible proteins | OFBG_00405 | OFAG_01173 |
| universal stress family proteins |
OFBG_00128 | OFAG_00244 |
| starvation-signaling stringent response proteins SspA and SspB |
OFBG_01881, OFBG_01880 | OFAG_01837, OFAG_01838 |
In bacteria, a major and highly conserved mechanism for sensing and responding to the outside and host environment is comprised of a two-component system. This signaling system is composed of a histidine kinase and a response regulator that enables bacteria to alter their physiological behaviour in response to changes in their immediate environment. This is accomplished by altering gene expression, enzymatic reactions or protein-protein interactions [12]. We were able to identify at least eighteen distinct open reading frames that may encode for either a histidine kinase or a response regulator protein, some of which are identified in Table 3.
The quorum sensing strategies that O. formigenes utilizes to influence group activities are not known. In Gram-negative bacteria, acylated homoserine lactones are the most common autoinducer used for cell-to-cell communication [46]. The OXCC13 and HOxBLS genomes encode several proteins with significant homology to DNA-binding response regulators in the LuxR family and may be involved in such communication (OFBG_01501, OFBG_00440, OFAG_00058, OFAG_02291). Some Gram-negative bacteria communicate using small molecules whose production depends on S-adenosylmethionine (SAM) as a substrate [62], and both O. formigenes genomic sequences encode S-adenosylmethionine synthase (OFAG_02097, OFBG_00071). Identifying the small molecules that O. formigenes secretes to influence various group behaviors may lead to strategies to improve the ability of this organism to survive in the intestine during periods of environmental stress.
The O. formigenes genomes contain numerous prophages (Table 3). The incorporation of prophages into a genome confer several advantages to an organism and assist it to cope with adverse environments, including exposure to antibiotics and osmotic, oxidative and acid stress [59]. They can also increase growth rates and enhance biofilm formation. Application of the PHAST search tool for identifying prophage sequences in bacterial genomes indicated that OXCC13 contains 4 incomplete prophages with most similarity to Enterobacteria_phage_HK620, Burholderia_phage_Bcep176, Bathycoccus sp. RCC1105 virus BpV1 and Ralstonia_phage_phiRSA1, whereas HOxBLS contains 1 intact region with similarity to Burkholderia phage KS14, and 2 incomplete regions with most similarity to Pseudomonas_phage_PAJU2 and Bacillus_phage_SPO1 [64].
Colicins [13] and microsins [50]are antimicrobial proteins and peptides, respectively, secreted by Gram-negative bacteria to inhibit the growth of similar or closely related bacterial strain(s). Genomes from both strains encode for a protein with good homology for colicin_V (OFBG_01340; OFAG_01318), and microsin B17 maturation protein (OFBG_01676; OFAG_001637). Both strains also have genes with good homology for toxin resistance, including Bacitracin resistance protein (OFAG_02041; OFBG_00014).
Clustered regularly interspaced short palindromic repeat (CRISPR) loci have been shown to protect prokaryotes from invading phages and plasmid DNA [24]. Both strains of O. formigenes encode CRISPR associated intergenic sequences, six in the genome of OXCC13 and 3 in the genome of HOxBLS.
Over fifty putative transcriptional regulators, some of which are shown in Table 3, have been identified in the genomes of O. formigenes based on the presence of conserved functional domains, indicating that O. formigenes has the machinery to modulate gene expression during stress. For example, HOxBLS and OXCC13 contain 8 and 4 genes, respectively, that contain the conserved domain Pfam01047 (MarR family), which are transcriptional regulators involved in resistance to multiple antibiotics, household disinfectants, organic solvents, and oxidative stress [21].
The genomes also contain the Hfq gene (OFBG_00195, OFAG_00111) which codes for an RNA chaperone. Hfq can bind to both small RNAs (sRNA) and mRNA [26]. The small RNA regulators have been identified in a wide range of organisms and range in length from 50 to 500 nucleotides. They play a critical role in regulating many cellular processes. Most organisms have 50 – 100 sRNAs and they remain to be identified in O. formigenes.
NUTRIENT LIMITATION
A low oxalate intake, as might occur with certain diets, such as high protein – high fat diets, fasting, and enteral and parenteral feeding, raises several questions regarding how O. formigenes responds to periods of oxalate deprivation. Is O. formigenes colonization lost on diets containing negligible oxalate? How does O. formigenes respond to starvation so that growth is arrested in a regulated manner that maximizes chances for long-term survival and persistence? Does O. formigenes develop resistance cells without dormancy [47]? Does enteric oxalate secretion from the host provide a “consistent” source of oxalate that ensures survival of O. formigenes during periods of dietary oxalate deprivation? Examining how O. formigenes survives dietary oxalate deprivation in the gut and the molecular mechanisms associated with this, may provide insight into strategies to promote O. formigenes colonization of the intestine. Searching the O. formigenes genome databases reveals several genes that are known to play an important role during periods of nutrient starvation (Table 3). The protein HipA, a member of the phosphoinositol 3/4-kinase superfamily, plays an important role in growth arrest and persistence [8], and the genomes encode a HipA domain-containing protein (OFBG_00481, OFAG_00619). In Gram-negative bacteria, the starvation response triggers the alternative sigma factor RpoS, which controls genes that make stationary cells more adaptable and resistant to challenging situations [61]. Genomes of both O. formigenes strains encode proteins with significant amino acid sequence homology to E.coli RpoS (OFAG_01295, OFBG_01317). Leucine-responsive regulatory protein (Lrp) acts as a global transcriptional regulator to coordinate cellular metabolism with the nutritional environment [40] and thus plays an important role during nutrient starvation. A blast analysis identified apparent transcriptional regulators with significant sequence homology to E.coli Lrp (OFBG_00685, OFAG_00599, OFAG_00909, OFAG_00597). The genome of both strains of O. formigenes encodes integration host factor (OFAG_00071, OFAG_01588, OFBG_00154, OFAG_01627), which has been shown to be important to cell survival during stationary phase [19]. O. formigenes genomes also encode carbon starvation inducible proteins, phosphate starvation inducible proteins, universal stress family proteins, and specific starvation-signaling stringent response proteins (Table 3), including proteins in the RelA/SpoT family (OFBG_01447, OFBG_01468, OFAG_01420, OFAG_01440), and stringent starvation proteins A (SspA) and B (SspB) (OFBG_01880, OFBG_01881, OFAG_01837, OFAG_01838). Expression of both SspA and SspB is increased during starvation-induced stress [63]. SspA is required for cell survival during acid induced stress and also functions as a transcriptional activator [27]. SspB is known to enhance the recognition of proteins marked for degradation by specific E. coli proteases including ClpA [57]. In E.coli and other bacteria the functional Clp protease is comprised of two components: a proteolytic component and one of several regulatory ATPase components. Interestingly, the genomes of both strains contain two ORFs (OFBG_00322 and OFBG_00262), that correspond to the ATP-dependent ClpP protease and its proteolytic subunit. In addition there are three ORFs (OFBG_01106, OFBG_01606, OFBG_01607), that may encode for the ClpB, ClpA and ClpS proteases, respectively.
Activation of the starvation-signaling stringent response has been shown to mediate antibiotic tolerance in Pseudomanas aeruginosa [48], and protection against osmotic stress [33], temperature stress [25], and acid stress [5]. Exploring the tolerance of O. formigenes to such stresses following nutrient limitation warrants further investigation. These studies may identify culture conditions that improve the success of recolonizing stone formers, and dietary approaches that can maintain O. formigenes colonization of the large intestine during periods of antibiotic therapy.
OXALOBACTER COLONIZATION
Little is known about how and when individuals become colonized or how it persists over time. Successful colonization of the gut presumably requires O. formigenes to occupy both mucosal and luminal niches; however, nothing is known about the biogeographic distribution of O. formigenes. The intestinal site/sites of O. formigenes colonization have only been examined in the study by Weaver and colleagues [60]. Their study, where subjects had undergone preparations for colonoscopy, showed that oxalate degradation, apparently due to O. formigenes, was detected in cecal (~90cm from anus) and/or sigmoid (~40cm from anus) brushings from 22 of 24 subjects, with concentrations of oxalate-degrading bacteria estimated to be ten-fold greater in cecal brush samples than sigmoid brushings.
The source of O. formigenes that colonizes the gut is not known. Studies to date suggest it occurs early in childhood [55], and if animal experiments provide any insight it is obtained from the environment, not directly from the mother [15]. The survival of the bacterium outside of the intestine has not been documented in any detail. It is expected that it will enter the immediate environment of human households in soil, dust, and possibly through the contamination of hand-touched objects, including pets. Interestingly, microbial 16S rRNA analysis of groundwater collected at depths of greater than 100 feet identified isolates from the Oxalobacter genus [14]. The survival of O. formigenes in various environments warrants further investigation. Improvement of techniques to increase the sensitivity of detecting low numbers of O. formigenes will facilitate answering these questions.
An understanding of both the bacterial and host genes involved in O. formigenes colonization of the large intestine will be important in identifying conditions that promote colonization. Persistent and robust colonization of the large intestine may require O. formigenes to express proteins involved in attachment to host cells. Genome analysis of both O. formigenes strains reveal a number of proteins that may be involved in this process. The OXCC13 genome contains genes that code for 4 proteins that are homologous to the type 1 pilus proteins [11], and appear to be in an operon (Table 3). Interestingly, the draft sequence of HOxBLS does not harbor genes that show good homology to these OXCC13 pilus proteins. The ability of O. formigenes to form pili could facilitate cell to cell transfer of genetic material, adhesion to host cells and the transfer of molecules to host cells.
One study has addressed the colonization/re-colonization of humans with O. formigenes. Two healthy adults became colonized following the ingestion of cultured O. formigenes [20]. It seems quite possible that O. formigenes colonization/re-colonization will prove to be an efficacious and inexpensive method for limiting calcium oxalate stone risk.
MUTUALISM: BENEFITS FOR HOST AND OXALOBACTER
A plausible hypothesis for the benefit to the host of colonization with O. formigenes is its ability to free up calcium ingested as calcium oxalate, as is most evident in studies of the sand rat and the ingestion of cactus calcium oxalate discussed above [49]. Such degradation may be an important factor in enhancing calcium bioavailability in human populations with limited access to dairy products or other rich sources of calcium and warrants further investigation.
Benefits to O. formigenes in colonizing the human gut are a secure, reasonably controlled habitat, and dietary oxalate as a food source. As oxalate consumption by humans is random, O. formigenes appears to have evolved the capability of modifying the oxalate transport properties of host cells and thus promoting its survival. This was illustrated by Freel et. al. who showed that both the colonization of rats with O. formigenes and the ingestion of O. formigenes lysates stimulated intestinal oxalate secretion [23]. Further work by Hatch et. al., showed that O. formigenes colonization of Agxt null and wild type mice resulted in normalization of plasma oxalate and urinary oxalate excretion in otherwise hyperoxalemic and hyperoxaluric animals [28]. Such changes, if confirmed and characterized, could open new avenues for therapies that increase intestinal elimination of body stores of oxalate.
OXALOBACTER AND THE RISK OF CALCIUM OXALATE STONE DISEASE
Since the discovery of O. formigenes and the recognition that it resides in the human gut and degrades oxalate, a role for the organism in stone disease has been considered. A review of colonization frequencies conducted worldwide indicated that 38 – 77% of a normal population is colonized and it was consistently observed that the colonization frequency in stone formers was about half that in normal subjects [36, 37]. Initial case-control studies with small numbers of subjects suggested colonization may be protective against stone disease [9, 43, 58]. Measurements of urinary oxalate excretion indicated that there was a lower urinary oxalate excretion in colonized compared to non-colonized individuals despite a large variability in oxalate excretion and a lack of dietary oxalate control during urine collections. The association of recurrent calcium oxalate stone disease was assessed in a study of 247 calcium oxalate stone formers and 259 matched controls [36]. The odds ratio for forming a recurrent stone when colonized was 0.3; i.e., a 70% reduction in stone risk. Surprisingly, there was no difference in urinary oxalate excretion between colonized and non-colonized individuals in either group. Although the sample size was not an issue in this study, oxalate excretion was again highly variable and dietary oxalate and calcium were not controlled. This discordance in results may be partially explained by our recently reported studies illustrating the dietary dependence of the effect [34]. A recent study involving 37 calcium oxalate kidney stone formers, of which 11 were colonized with O. formigenes, showed 24 hour urinary oxalate excretion and plasma oxalate were significantly lower in O. formigenes colonized patients compared to O. formigenes-negative patients on a controlled standardized diet [56]. Furthermore, colonization was significantly inversely associated with the number of stone episodes. A lower plasma oxalate levels in colonized patients supported findings in rodents that O. formigenes enhances or induces enteric oxalate secretion and warrants further examination.
ANTIBIOTIC EXPOSURE
Several studies have indicated that the intake of antibiotics can result in the loss of colonization [37, 38, 43], and this is supported by lower prevalence of O. formigenes in both cystic fibrosis patients [53], and calcium oxalate stone formers who are frequently prescribed antibiotics. [43, 54]. Stone patients frequently receive antibiotic therapy during surgical procedures to remove stones, placing colonized patients at risk of losing O. formigenes colonization. In a test of the sensitivity of 4 different strains of O. formigenes to commonly used antibiotics we observed that all of them were resistant to amoxicillin, amoxicillin/clavulanic acid (augmentin), ceftriaxone (rocephin), and vancomycin [41]. Resistance to amoxicillin and clavulanic acid is in keeping with both the OXCC13 and HOxBLS strain having the gene that encodes for beta lactamase (OFAG_00902; OFBG_00689). Strain OXCC13 contains the 5-nitroimidazole antibiotic resistance gene (OFBG_00777) and is resistant to nitrofuratonin, whereas strain HOxBLS lacks the gene and is sensitive to this antibiotic [41]. O. formigenes is also likely to be resistant to chloramphenicol, as both OXCC13 and HOxBLS contain a gene with good homology to chloramphenicol acetyltransferase (OFAG_00880; OFBG_00696). These antibiotic resistance patterns suggest that it may be prudent to tailor antibiotic therapy and not treat stone patients with antibiotics which will eradicate O. formigenes.
CONCLUSIONS
The genome of O. formigenes encodes traits important for its survival and retention in the hostile environment of the gastrointestinal tract. However, the mechanisms by which O. formigenes establishes itself as a resilient member of the fecal microbiome are not known. As more features of the intestinal microbiome are defined, the characteristics of O. formigenes that have enabled it to establish its niche should become apparent. Unraveling these mechanisms is especially important with respect to the colonization/recolonization of non-colonized stone formers and how this impacts stone risk. Further studies on the factors involved in re-colonization and its stabilization are warranted in light of this. The range of conditions where O. formigenes lowers stone risk remains to be clearly defined.
ACKNOWLEDGEMENTS
This research was supported in part by NIH grants DK087967 and DK062284.
The “Oxalobacter formigenes Sequencing Project, Broad Institute of Harvard and MIT (http://www.broadinstitute.org/)” and the Integrated Microbial Genomes (IMG) system, supported by the NIH Human Microbiome Project, were used for genomic data analysis.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
- 1.Allison MJ, Cook HM. Oxalate degradation by microbes of the large bowel of herbivores: the effect of dietary oxalate. Science. 1981;212:675–676. doi: 10.1126/science.7221555. [DOI] [PubMed] [Google Scholar]
- 2.Allison MJ, Littledike ET, James LF. Changes in ruminal oxalate degradation rates associated with adaptation to oxalate ingestion. J Anim Sci. 1977;45:1173–1179. doi: 10.2527/jas1977.4551173x. [DOI] [PubMed] [Google Scholar]
- 3.Anantharam V, Allison MJ, Maloney PC. Oxalate:formate exchange. The basis for energy coupling in Oxalobacter. J Biol Chem. 1989;264:7244–7250. [PubMed] [Google Scholar]
- 4.Argenzio RA, Liacos JA, Allison MJ. Intestinal oxalate-degrading bacteria reduce oxalate absorption and toxicity in guinea pigs. J Nutr. 1988;118:787–792. doi: 10.1093/jn/118.6.787. [DOI] [PubMed] [Google Scholar]
- 5.Arnold KW, Kaspar CW. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61:2037–2039. doi: 10.1128/aem.61.5.2037-2039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baetz AL, Allison MJ. Purification and characterization of oxalyl-coenzyme A decarboxylase from Oxalobacter formigenes. J Bacteriol. 1989;171:2605–2608. doi: 10.1128/jb.171.5.2605-2608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baetz AL, Allison MJ. Purification and characterization of formyl-coenzyme A transferase from Oxalobacter formigenes. J Bacteriol. 1990;172:3537–3540. doi: 10.1128/jb.172.7.3537-3540.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 9.Batislam E, Yilmaz E, Yuvanc E, Kisa O, Kisa U. Quantitative analysis of colonization with real-time PCR to identify the role of Oxalobacter formigenes in calcium oxalate urolithiasis. Urol Res. 2012;40:455–460. doi: 10.1007/s00240-011-0449-8. [DOI] [PubMed] [Google Scholar]
- 10.Bentley SD, Parkhill J. Comparative genomic structure of prokaryotes. Annu Rev Genet. 2004;38:771–792. doi: 10.1146/annurev.genet.38.072902.094318. [DOI] [PubMed] [Google Scholar]
- 11.Capitani G, Eidam O, Glockshuber R, Grutter MG. Structural and functional insights into the assembly of type 1 pili from Escherichia coli. Microbes Infect: 2284-2290. 2006 doi: 10.1016/j.micinf.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connon SA, Tovanabootr A, Dolan M, Vergin K, Giovannoni SJ, Semprini L. Bacterial community composition determined by culture-independent and -dependent methods during propane-stimulated bioremediation in trichloroethene-contaminated groundwater. Environ Microbiol. 2005;7:165–178. doi: 10.1111/j.1462-2920.2004.00680.x. [DOI] [PubMed] [Google Scholar]
- 15.Cornelius JG, Peck AB. Colonization of the neonatal rat intestinal tract from environmental exposure to the anaerobic bacterium Oxalobacter formigenes. J Med Microbiol. 2004;53:249–254. doi: 10.1099/jmm.0.05418-0. [DOI] [PubMed] [Google Scholar]
- 16.Cornick NA, Allison MJ. Anabolic Incorporation of Oxalate by Oxalobacter formigenes. Appl Environ Microbiol. 1996;62:3011–3013. doi: 10.1128/aem.62.8.3011-3013.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornick NA, Allison MJ. Assimilation of oxalate, acetate, and CO2 by Oxalobacter formigenes. Can J Microbiol. 1996;42:1081–1086. doi: 10.1139/m96-138. [DOI] [PubMed] [Google Scholar]
- 18.Dawson KA, Allison MJ, Hartman PA. Isolation and some characteristics of anaerobic oxalate-degrading bacteria from the rumen. Appl Environ Microbiol. 1980;40:833–839. doi: 10.1128/aem.40.4.833-839.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ditto MD, Roberts D, Weisberg RA. Growth phase variation of integration host factor level in Escherichia coli. J Bacteriol. 1994;176:3738–3748. doi: 10.1128/jb.176.12.3738-3748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan SH, Richardson AJ, Kaul P, Holmes RP, Allison MJ, Stewart CS. Oxalobacter formigenes and its potential role in human health. Appl Environ Microbiol. 2002;68:3841–3847. doi: 10.1128/AEM.68.8.3841-3847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellison DW, Miller VL. Regulation of virulence by members of the MarR/SlyA family. Curr Opin Microbiol. 2006;9:153–159. doi: 10.1016/j.mib.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Franceschi VR. Calcium oxalate formation is a rapid and reversible process in Lemna minor L. Protoplasma. 1989;148:130–137. [Google Scholar]
- 23.Freel RW, Hatch M, Green M, Soleimani M. Ileal oxalate absorption and urinary oxalate excretion are enhanced in Slc26a6 null mice. Am J Physiol Gastrointest Liver Physiol. 2006;290:G719–728. doi: 10.1152/ajpgi.00481.2005. [DOI] [PubMed] [Google Scholar]
- 24.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 25.Givskov M, Eberl L, Moller S, Poulsen LK, Molin S. Responses to nutrient starvation in Pseudomonas putida KT2442: analysis of general cross-protection, cell shape, and macromolecular content. J Bacteriol. 1994;176:7–14. doi: 10.1128/jb.176.1.7-14.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 3. 2011 doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen AM, Gu Y, Li M, Andrykovitch M, Waugh DS, Jin DJ, Ji X. Structural basis for the function of stringent starvation protein a as a transcription factor. J Biol Chem. 2005;280:17380–17391. doi: 10.1074/jbc.M501444200. [DOI] [PubMed] [Google Scholar]
- 28.Hatch M, Gjymishka A, Salido EC, Allison MJ, Freel RW. Enteric oxalate elimination is induced and oxalate is normalized in a mouse model of primary hyperoxaluria following intestinal colonization with Oxalobacter. Am J Physiol Gastrointest Liver Physiol. 2011;300:G461–469. doi: 10.1152/ajpgi.00434.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmes RP, Assimos DG. The impact of dietary oxalate on kidney stone formation. Urol Res. 2004;32:311–316. doi: 10.1007/s00240-004-0437-3. [DOI] [PubMed] [Google Scholar]
- 30.Hoppe B, Groothoff JW, Hulton SA, Cochat P, Niaudet P, Kemper MJ, Deschenes G, Unwin R, Milliner D. Efficacy and safety of Oxalobacter formigenes to reduce urinary oxalate in primary hyperoxaluria. Nephrol Dial Transplant. 2011;26:3609–3615. doi: 10.1093/ndt/gfr107. [DOI] [PubMed] [Google Scholar]
- 31.Ilarslan H, Palmer RG, Horner HT. Calcium oxalate crystals in developing seeds of soybean. Ann Botany. 2001;88:243–257. [PubMed] [Google Scholar]
- 32.Ilarslan H, Palmer RG, Imsande J, Horner HT. Quantitative determination of calcium oxalate and oxalate in developing seeds of soybean (Leguminosae) Am J Botany. 1997;84:1042–1046. [PubMed] [Google Scholar]
- 33.Jenkins DE, Chaisson SA, Matin A. Starvation-induced cross protection against osmotic challenge in Escherichia coli. J Bacteriol. 1990;172:2779–2781. doi: 10.1128/jb.172.5.2779-2781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang J, Knight J, Easter LH, Neiberg R, Holmes RP, Assimos DG. Impact of dietary calcium and oxalate, and Oxalobacter formigenes colonization on urinary oxalate excretion. J Urol. 2011;186:135–139. doi: 10.1016/j.juro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang DC, Venkataraman PA, Dumont ME, Maloney PC. Oligomeric state of the oxalate transporter, OxlT. Biochemistry. 2011;50:8445–8453. doi: 10.1021/bi201175y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaufman DW, Kelly JP, Curhan GC, Anderson TE, Dretler SP, Preminger GM, Cave DR. Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J Am Soc Nephrol. 2008;19:1197–1203. doi: 10.1681/ASN.2007101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly JP, Curhan GC, Cave DR, Anderson TE, Kaufman DW. Factors related to colonization with Oxalobacter formigenes in U.S. adults. J Endourol. 2011;25:673–679. doi: 10.1089/end.2010.0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kharlamb V, Schelker J, Francois F, Jiang J, Holmes RP, Goldfarb DS. Oral antibiotic treatment of Helicobacter pylori leads to persistently reduced intestinal colonization rates with Oxalobacter formigenes. J Endourol. 2011;25:1781–1785. doi: 10.1089/end.2011.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhner CH, Hartman PA, Allison MJ. Generation of a proton motive force by the anaerobic oxalate-degrading bacterium Oxalobacter formigenes . Appl Environ Microbiol. 1996;62:2494–2500. doi: 10.1128/aem.62.7.2494-2500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landgraf JR, Wu J, Calvo JM. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J Bacteriol. 1996;178:6930–6936. doi: 10.1128/jb.178.23.6930-6936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lange JN, Wood KD, Wong H, Otto R, Mufarrij PW, Knight J, Akpinar H, Holmes RP, Assimos DG. Sensitivity of human strains of Oxalobacter formigenes to commonly prescribed antibiotics. Urology. 2012;79:1286–1289. doi: 10.1016/j.urology.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindahl A, Ungell AL, Knutson L, Lennernas H. Characterization of fluids from the stomach and proximal jejunum in men and women. Pharm Res. 1997;14:497–502. doi: 10.1023/a:1012107801889. [DOI] [PubMed] [Google Scholar]
- 43.Mittal RD, Kumar R, Bid HK, Mittal B. Effect of antibiotics on Oxalobacter formigenes colonization of human gastrointestinal tract. J Endourol. 2005;19:102–106. doi: 10.1089/end.2005.19.102. [DOI] [PubMed] [Google Scholar]
- 44.MJ A, KA D, WR M, JG F. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch Microbiol. 1985;141:1–7. doi: 10.1007/BF00446731. [DOI] [PubMed] [Google Scholar]
- 45.Muller JA, Ross RP, Sybesma WF, Fitzgerald GF, Stanton C. Modification of the technical properties of Lactobacillus johnsonii NCC 533 by supplementing the growth medium with unsaturated fatty acids. Appl Environ Microbiol. 2011;77:6889–6898. doi: 10.1128/AEM.05213-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nasser W, Reverchon S. New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators. Anal Bioanal Chem. 2007;387:381–390. doi: 10.1007/s00216-006-0702-0. [DOI] [PubMed] [Google Scholar]
- 47.Navarro Llorens JM, Tormo A, Martinez-Garcia E. Stationary phase in gram-negative bacteria. FEMS Microbiol Rev. 2010;34:476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palgi N, Ronen Z, Pinshow B. Oxalate balance in fat sand rats feeding on high and low calcium diets. J Comp Physiol B. 2008;178:617–622. doi: 10.1007/s00360-008-0252-1. [DOI] [PubMed] [Google Scholar]
- 50.Rebuffat S. Microcins in action: amazing defence strategies of Enterobacteria. Biochem Soc Trans. 2012;40:1456–1462. doi: 10.1042/BST20120183. [DOI] [PubMed] [Google Scholar]
- 51.Roberts RB, P. H. Abelson, D. Cowie, E. T. Bolton, RJ Britten. Studies of Biosynthesis in Escherichia coli. Carnegie Institution of Washington Publication 607, Washington, D.C. 1955 [Google Scholar]
- 52.Ruan ZS, Anantharam V, Crawford IT, Ambudkar SV, Rhee SY, Allison MJ, Maloney PC. Identification, purification, and reconstitution of OxlT, the oxalate: formate antiport protein of Oxalobacter formigenes. J Biol Chem. 1992;267:10537–10543. [PubMed] [Google Scholar]
- 53.Sidhu H, Hoppe B, Hesse A, Tenbrock K, Bromme S, Rietschel E, Peck AB. Absence of Oxalobacter formigenes in cystic fibrosis patients: a risk factor for hyperoxaluria. Lancet. 1998;352:1026–1029. doi: 10.1016/S0140-6736(98)03038-4. [DOI] [PubMed] [Google Scholar]
- 54.Sidhu H, Schmidt ME, CorneliusT JG, Thamiselvam S, Khan SR, Hesse A, Peck AB. Direct correlation between hyperoxaluria/oxalate stone disease and the absence of the gastrointestinal tract dwelling bacterium Oxalobacter formigenes: Possible prevention by gut recolonization or enzyme replacement therapy. J Am Soc Nephrol. 1999;10:S334–S340. [PubMed] [Google Scholar]
- 55.Sidhu H, Yenatska L, Ogden SD, Allison MJ, Peck AB. Natural colonization of children in the Ukraine with the intestinal bacterium, Oxalobacter formigenes, using a PCR-based detection system. Mol Diagnosis. 1997;2:89–97. doi: 10.1054/MODI00200089. [DOI] [PubMed] [Google Scholar]
- 56.Siener R, Bangen U, Sidhu H, Honow R, von Unruh G, Hesse A. The role of Oxalobacter formigenes colonization in calcium oxalate stone disease. Kid Int. 2013 doi: 10.1038/ki.2013.104. [DOI] [PubMed] [Google Scholar]
- 57.Song HK, Eck MJ. Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine. Mol Cell. 2003;12:75–86. doi: 10.1016/s1097-2765(03)00271-5. [DOI] [PubMed] [Google Scholar]
- 58.Troxel SA, Sidhu H, Kaul P, Low RK. Intestinal Oxalobacter formigenes colonization in calcium oxalate stone formers and its relation to urinary oxalate. J Endourol. 2003 doi: 10.1089/089277903321618743. [DOI] [PubMed] [Google Scholar]
- 59.Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM, Wood TK. Cryptic prophages help bacteria cope with adverse environments. Nat Commun 1: 147. 2010 doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weaver GA, Krause JA, Allison MJ, Lindenbaum J. Distribution of Digoxin-reducing, Oxalate-degrading, and Total Anaerobic Bacteria in the Human Colon. Microb Ecol Health D. 1992;5:227–234. [Google Scholar]
- 61.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei Y, Perez LJ, Ng WL, Semmelhack MF, Bassler BL. Mechanism of Vibrio cholerae autoinducer-1 biosynthesis. ACS Chem Biol. 2011;6:356–365. doi: 10.1021/cb1003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams MD, Ouyang TX, Flickinger MC. Starvation-induced expression of SspA and SspB: the effects of a null mutation in sspA on Escherichia coli protein synthesis and survival during growth and prolonged starvation. Mol Microbiol. 1994;11:1029–1043. doi: 10.1111/j.1365-2958.1994.tb00381.x. [DOI] [PubMed] [Google Scholar]
- 64.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]