SUMMARY
Nutrient amino acid transporters (NATs) of solute carrier family 6 (SLC6) mediate uptake of essential amino acids in mammals and insects. Phylogenomic analysis of the Caenorhabditis elegans (Ce) SLC6 family identifies five genes paralogous to an insect-specific NAT subfamily. Here we cloned and characterized the first representative of the identified nematode-specific transporters, SNF-5. SNF-5 mediates broad spectrum cation-coupled transport of neutral amino acids with submillimolar affinities and stoichiometry of 1 AA:1 Na+, except for 1 l-Pro:2 Na+. Unexpectedly, it transports acidic l-Glu− and l-Asp− (1 AA−:3 Na+), revealing it to be the first member of a new B0,− system among characterized SLC6 transporters. This activity correlates with a unique positively charged His+ 377 in the substrate-binding pocket. snf-5 promoter-driven enhanced green fluorescent protein labels intestinal cells INT1-9 and three pairs of amphid sensory neurons: ASI, ADF and ASK. These cells are intimately involved in control of dauer diapause, development, metabolism and longevity. The snf-5 deletion mutants do not show apparent morphological disorders, but increase dauer formation while reducing dauer maintenance upon starvation. Overall, the present study characterized the first nematode-specific NAT and revealed important structural and functional aspects of this transporter. In addition to the predictable role in alimentary amino acid absorption, our results indicate possible neuronal roles of SNF-5 as an amino acid provider to specific neuronal functions, including sensing of amino acid availability.
KEY WORDS: SLC6; glutamate; B0,− system; dauer; embryonic arrest; amphid sensory neurons; ASI; ASK; ADF; snf-5; transceptor
INTRODUCTION
The set of essential amino acids is very similar across animals. Except for some specific additions, it includes ~50% of the proteinogenic amino acids that utilize complex synthesis cascades that consume the lion's share of the total energy budget in autotrophic bacteria and plants (Payne and Loomis, 2006; Boudko, 2010). The heterotrophy of these amino acids accelerates development and reproduction in animals by reducing the energy and time necessary to synthesize amino acids, peptides and proteins. However, this adaptation demands mechanisms for environmental acquisition and systemic balance of the essential substrates in agreement with the metabolic quorum of the organism, specific tissues and individual cells. To avoid a collapse of essential amino acid metabolism, organisms must monitor the availability and tightly control the systemic balance of these substrates. Mechanisms for sensing and acquiring essential amino acids are especially critical for animal survival, development, reproduction and coordination of metabolic diapauses in nutrient-limited environments. However, essential amino acid sensing mechanisms are unknown and the existing knowledge of transport systems for these substrates is very limited.
A few transporters with the capacity to transport essential amino acids were identified and functionally characterized in model mammalian organisms, as well as dipteran and lepidopteran arthropods [summarized in Boudko (Boudko, 2012)]. Published results suggest that trafficking of essential amino acids requires synergistic action of secondary transporters from two gene families, namely solute carrier family (SLC) 6 (Chen et al., 2004; Bröer, 2006) and SLC7 (Verrey et al., 2003). Members of these families play prominent roles in apical and basolateral translocation of essential amino acids through alimentary epithelia and consecutive absorption through plasma membranes of different tissues and cells. Specifically, SLC6 members mediate Na+ or K+ ion motive force-coupled transport, providing an active mechanism for systemic and cellular accumulation of essential amino acids against large chemical gradients. In contrast, SLC7 members facilitate transmembrane diffusion and/or aid in reciprocal exchange of essential and metabolic amino acids across a cellular membrane, providing optimal transport mechanisms for favorable or small unfavorable substrate gradients, respectively (Hansen et al., 2011). Several other secondary transporters aiding in the bulk absorption of nutrient amino acids include: proton-coupled small oligopeptide transporters SLC15 (Daniel and Kottra, 2003), proton-coupled amino acid transporters SLC36 (Boll et al., 2004), sodium-dependent SLC38 (Mackenzie and Erickson, 2003) and sodium-independent SLC43 transporters. However, based on their substrate specificities and limited conservation across different animal organisms, their contributions toward the balance of the essential amino acid pool may be secondary or dispensable (Boudko, 2012).
The present study focuses on a population of amino acid transporters of the SLC6 family. Several members of this family were initially identified as sodium-dependent transporters for monoamine and GABA neurotransmitters. However, a subsequent study revealed broader substrate specificities for some of these transporters, demonstrating that SLC6 family members actually form several subfamilies specialized in the absorption of amino acids and various derivatives of amino acid metabolism, including the aforementioned neurotransmitters (Chen et al., 2004). More recently, several orphan transporters of this family were cloned and characterized as Na+-coupled transporters for large aliphatic and aromatic amino acids that are essential in Metazoa. Comparative phylogenomic analysis showed that these transporters form a large polyphyletic subfamily, representing several branches of the SLC6 family, which we have named the nutrient amino acid transporter subfamily (NAT-SLC6) (Boudko et al., 2005a). The mammalian NAT-SLC6 paralogs, in general, share properties with the canonical broad spectra neutral (0 charged) amino acid transport system (B0), except for a more specialized proline-selective IMINO system transporter (Kowalczuk et al., 2005). In addition to B0-like transporters, the insect NAT-SLC6 subfamily includes several selective transporters with increased selectivity and affinity for phenylalanine (Meleshkevitch et al., 2006), tryptophan (Meleshkevitch et al., 2009a) or methionine (Meleshkevitch et al., 2009b). Mammalian and insect NATs are highly expressed in the apical membranes of absorptive regions of the alimentary canal, as well as in other organs with elevated requirements for essential amino acids, including reproductive and neuronal tissues (Boudko, 2012). The homology between insect and mammalian NAT-SLC6 members, combined with the apparent absence of alternative mechanisms with similar functionality, suggests a principal role for the identified subfamily in active absorption and distribution of essential amino acids.
Here we explore the emerging NAT-SLC6 paradigm further by identifying and analyzing a group of Caenorhabditis elegans transporters that are closely related to the characterized insect NATs. To corroborate the role and compare the properties of the identified nematode transporters against known NATs, we have cloned, heterologously expressed and functionally characterized the first representative of the nematode-specific SLC6 cluster, a transporter named SNF-5 (after the snf-5 gene of WormBase nomenclature). The putative contributions and integration of SNF-5 to neuronal and alimentary functions, as well as overall nematode biology, are analyzed and discussed.
MATERIALS AND METHODS
Bioinformatic analysis
Selected nematode genomes were analyzed for annotated SLC6 members using the NCBI's BLASTp. The identified sequences were aligned on top of a comprehensive reference alignment of human and insect transporters of the SLC6 family. Different alignment algorithms were employed to determine the optimal alignment pattern. Subsequently, manual editing using local automatic and manual realignments was used. Finally, the sequence of bacterial SLC6-NAT LeuT (NSBI, NP_214423) and a representative 2D map (PDB, 2A65) of its annotated 3D structure (Yamashita et al., 2005) were aligned to explore sequence–structure correlation of selected NATs. The sequence assembling, alignment, editing, and preparation of final figures were completed using Geneious V5.5 Pro (www.geneious.com). The phylogenetic analysis and tree reconstruction were performed using MEGA5 (Tamura et al., 2011). The phylogenetic tree was visualized with FigTree (http://tree.bio.ed.ac.uk) and Inkscape (http://inkscape.org) software.
Cloning and heterologous expression
The open reading frame (ORF) of sodium neurotransmitter family member 5 (snf-5) was cloned from an adult C. elegans cDNA library using exact primers with HindIII (5′-AAG CTT ATG GCC GAT TCG GGC AGC AAT GA-3′) and NotI (5′-GCG GCC GCT TAA TAA ACT TGA TGA TAT GTA-3′) restriction site flanking sequences (underlined). The selected PCR products, representing the complete snf-5 ORF, were sequenced and inserted into the HindIII/NotI-digested expression vector pXOON (Jespersen et al., 2002). cRNA for oocyte injections was obtained by in vitro transcription of PmeI-linearized SNF-5-pXOON plasmids using mMessage mMachine, a high-yield, capped RNA transcription kit (Ambion, Austin, TX, USA). Surgically isolated and collagenase-treated stage V–VI Xenopus laevis oocytes (Nasco, Fort Atkinson, WI, USA) were injected with ~40 ng of SNF-5 cRNA and incubated for 2–4 days at 17°C in sterile oocyte incubation medium. SNF-5-injected oocytes were analyzed in a small volume perfusion chamber using two-electrode voltage clamp techniques identical to those used to characterize previously identified NATs (Meleshkevitch et al., 2006; Meleshkevitch et al., 2009a).
Electrophysiological analysis
Assays were performed in a 50 μl constant-flow chamber. KCl-filled (1.5 mol l−1) sharp glass microelectrodes with 0.5–1 MΩ resistance were connected via AgCl wire to a two-electrode OC-725C Oocyte Clamp Amplifier (Warner Instruments, Hamden, CT, USA). Two AgCl/1 mmol l−1 KCl/agar bridges were used to connect the chamber with a ‘virtual ground’ head-stage for precise current recording. Current–voltage signals were acquired and analyzed using parallel inputs of MiniDigi and DigiData1322A analog/digital converters and pCLAMP software (Molecular Devices, Sunnyvale, CA, USA). SigmaPlot 11 software (Systat Software, San Jose, CA, USA) was used for reconstruction of current–voltage plots and Hill approximation. The compositions of solutions used for ion substitution assays are as follows: 98N (98 mmol l−1 NaCl, 2 mmol l−1 KCl, 0.5 mmol l−1 MgCl2 6H2O, 0.5 mmol l−1 CaCl2 2H2O, 10 mmol l−1 HEPES), 98K (98 mmol l−1 KCl, 2 mmol l−1 NaCl, 0.5 mmol l−1 MgCl2 6H2O, 0.5 mmol l−1 CaCl2 2H2O, 10 mmol l−1 HEPES) and 98N-Cl (98 mmol l−1 NaGluconate, 2 mmol l−1 KGluconate, 0.5 mmol l−1 MgSO4 7H2O, 0.5 mmol l−1 CaGluconate, 10 mmol l−1 HEPES). Kinetic profiles and constants were derived by curve fitting of normalized data sets with a three-parameter sigmoidal Hill function: y = a×xb/(cb+xb) upon 200 iterative steps, where a=max(y)=derived normalized mean maximum current; b=η=order of the transport process; and c=K0.5(x,y), the substrate concentration at half-maximal current. Saturation kinetics data were acquired via staircase concentration change of substrates under the specified voltage clamp parameters. Substrates were applied stepwise until the induced current was nearly saturated.
Isotope uptake assays
Uptake assays were performed ~4 days after RNA injection on oocytes using a previously described protocol (Meleshkevitch et al., 2006). Briefly, distilled water- and SNF-5 RNA-injected oocytes were exposed, in triplicate, to a 98N solution supplemented with 0.02 μCi ml−1 radioactively labeled amino acid (repeated for all utilized amino acids). Uptake was terminated after 15 min by washing oocytes three times in 3 ml of ice-cold 98N solution. The three identically treated oocytes were then placed into scintillation vials. After addition of 200 μl of 10% sodium dodecyl sulfate (SDS), each scintillation vial was vortexed to dissolve the oocytes within the fluid. Subsequently, 2 ml of scintillation fluid was added, vortexed, and radioactivity was counted using a Beckman-Coulter LS 6500 scintillation counter (Beckman-Coulter, Fullerton, CA, USA). The statistical evaluation and quantitative conversion of radioactivity to molar amount substrates was calculated as described previously (Meleshkevitch et al., 2006). The radiolabeled amino acids were acquired from Moravek (Brea, CA, USA) and have specific radioactivity in Ci mmol−1 as follows: l-Ala[2.3-3H], 49.4; l-Glu[2,4-3H], 40.0; Gly[2-3H], 6.5; l-Leu[4,5-3H], 144.0; Hys[ring2,5-3H], 55.4; l-Met[1-14C], 0.054; l-l-Phe[ring2,4-3H], 23.0; Pro[2,3,4,5-3H], 71.3; and l-Tyr[ring3,5-3H], 48.0.
Nematode preparation and maintenance
Nematodes were cultured on standard nematode growth medium (NGM) plates seeded with 0.05 ml of OP50 strain Escherichia coli bacteria at 20°C. Strains were maintained via transfer of three appropriately staged nematodes to freshly seeded plates prior to the exhaustion of the OP50 food supply (~every 4 days). The snf-5 knockout strain (RB687) underwent two separate outcrossings with wild-type N2 nematodes to eliminate contribution of background mutations to observed phenomena. The final strain, which we named HKK479, was confirmed homozygous for the deletion via PCR with specific primers.
Analysis of snf-5 promoter-driven EGFP expression
Confocal microscopy was performed on individuals of the BC14858 strain of nematodes (Caenorhabditis Genetics Center, Minneapolis, MN, USA), as well as a strain generated in our laboratory via previously described techniques (Mello et al., 1991), and selected for a more robust signal, both of which express enhanced green fluorescent protein (EGFP) under the snf-5 specific promoter. Due to the mosaic nature of C. elegans chromosomal arrays, we analyzed and summarized expression patterns from different animals within the same transgenic line. Control (N2) and EGFP-labeled nematodes were immobilized with 4 mmol l−1 sodium azide on a flattened drop of agar placed on a slide. An Olympus FluoView FV10i scanning laser confocal microscope (Olympus America, Center Valley, PA, USA) was used to acquire 30–60 frame long Z-stack scans. GFP-specific settings of excitation/emission=489/510 nm were used to obtain images of EGFP expression in the nematode body. Representative images were assembled as maximum projection of confocal Z-stacks using the FIJI implementation of ImageJ (Schindelin et al., 2012). The images of control and labeled nematodes acquired under identical excitation/emission and recording gain settings exhibited no overlapping background florescence.
Starvation assays
The effects of starvation on nematode proliferation, growth and dauer formation were assessed. Briefly, single wild-type and snf-5 mutant animals were picked and placed on seeded NGM plates at 20°C. Animals were allowed to feed and reproduce freely. Plates were imaged at days 1, 5, 7 and 15 post-plating to assess the number of eggs, L1, L2 and L3/L4 nematodes present on each plate. Additional plates, started in parallel, were checked for exhaustion of the food lawn. Four days after exhaustion of the food lawn, these plates were imaged and assessed for worm density and the presence of advanced stage (L4/adult) nematodes, and subsequently flooded with 1% SDS and scored after 15 min for the presence of dauers (live thrashing animals).
Diapause recovery assays
The ability of nematodes to recover from L1 and dauer diapause states was assessed. For L1 recovery assays, wild-type and snf-5 mutant animals were treated with hypochlorite to obtain eggshell protected embryos; embryos were allowed to hatch in M9 medium overnight and were age-synchronized at L1 stage. After 24 h, animals were placed to OP50-seeded plates and assessed for the ability to exit L1 diapause and return to a normal life cycle. To assess dauer recovery, wild type and snf-5 mutant animals were allowed to feed freely and exhaust their food supplies. Four days after exhaustion of the food lawn, plates were flooded with 1% SDS and after 15 min, individual nematodes were transferred to OP50-seeded plates and assessed for the ability to exit dauer diapause and return to a normal life cycle.
Statistical analysis
Values depicted in this study represent the means ± s.d. or s.e.m. for at least three experimental repeats performed on no less than three different, yet identically treated, experimental subjects, e.g. oocytes/nematodes/nematode plates. Electrophysiological and uptake data were obtained for both control (water-injected or non-injected) and experimental (SNF-5-injected) oocytes. Assuming unequal expression of the transporter in the different oocyte batches, the electrophysiological values were normalized to the maximum value obtained in individual oocytes. To obtain an accurate representation of the effects of SNF-5 expression on amino acid transport uptake, values from control non-injected oocytes were pairwise subtracted from values recorded from SNF-5-injected oocytes. Data from time-course studies of nematode strain egg laying and hatching were analyzed via a two-way repeated-measures ANOVA. Significance and data fidelity tests were used and included in figure legends as required.
RESULTS
Phylogenetic analysis of SNF-5 and related transporters
To identify nematode relatives of the NAT-SLC6 subfamily, we completed phylogenomic analysis of SLC6 members from selected metazoan genomes. BLASTp of the mosquito AeAAT1 (Boudko et al., 2005a) versus the protein database of C. elegans yields 17 SLC6 members, allowing a maximum cutoff value of E=0.008 for the most distant predicted protein sequence, C09E8.1. An optimized alignment of identified nematode versus human and mosquito SLC6 transporters showed 18.8% pairwise sequence identity (PSI) and 226 overall identical sites. Using a similar approach, we identified sets of SLC6 members in other nematodes, including: 15 Caenorhabditis briggsae (Cb, PSI 21.3%), 19 Caenorhabditis brenneri (Cbre, PSI 21.3%), 15 Caenorhabditis remanei (Cre, PSI 20.6%), seven Ascaris suum (As, PSI 20.4%), four Brugia malayi (Bmal, PSI 20.4%) and nine Trichinella spiralis (Tspir, PSI 18.8%) members (Table 1). A phylogenomic tree of identified CeSLC6 transporters versus mammalian and insect transporters revealed a five-gene branch located below the insect NAT cluster (iNAT; Fig. 1A). Y43D4A.1, a duplicated short fragment resembling the 102 AA N terminus of snf-9, was removed from subsequent analysis. Based on the phylogenetic proximity to insect NATs (iNAT cluster in Fig. 1), we may anticipate similar roles for the paralogous group of nematode transporters labeled in Fig. 1 as SNFs 2, 4, 5, 7 and 9, which we have named the nematode NATs. Despite clear identification of SLC6 family transporters, no apparent NAT representatives were found in the current drafts of genomes from parasitic nematodes.
Table 1.
Numbers of SLC6 and NAT-SLC6 paralogs in genomes of selected species
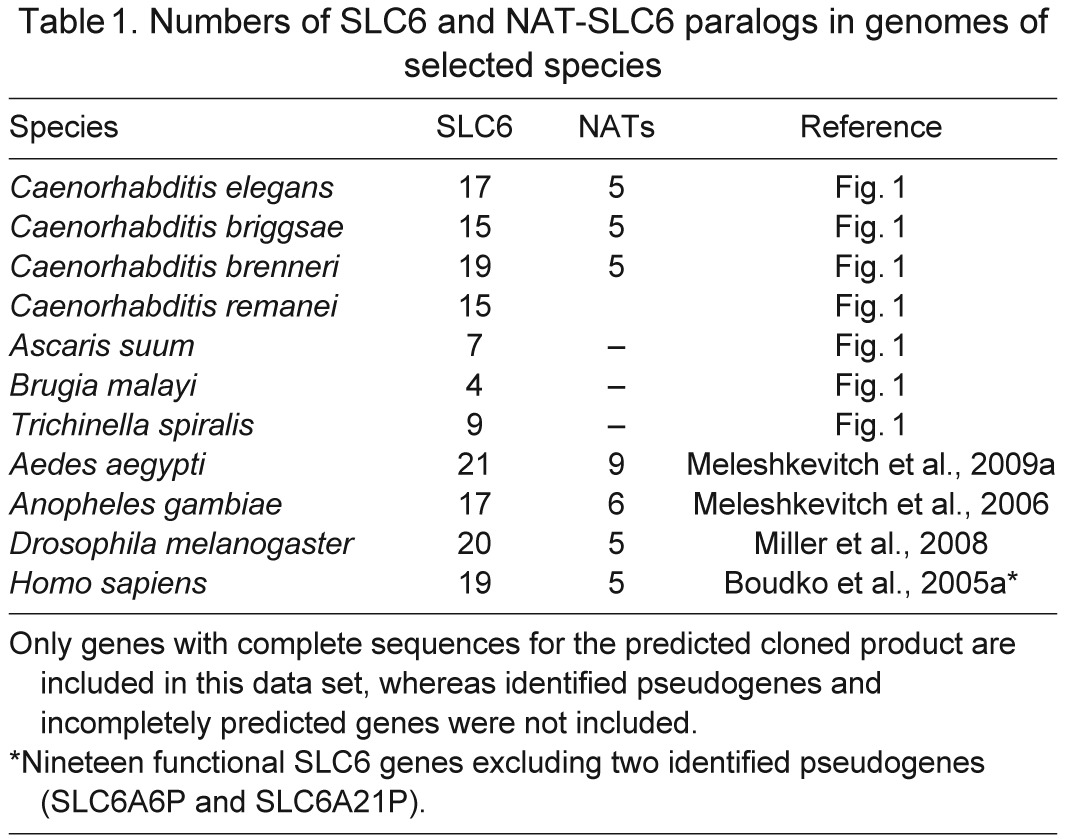
Fig. 1.
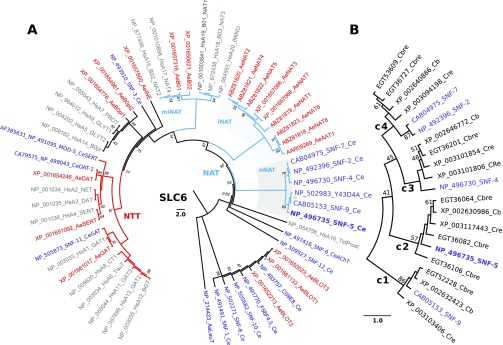
Relationships of nematode nutrient amino acid transporters (NATs) in a phylogenomic tree of the SLC6 family. (A) The evolutionary history was inferred using the UPGMA method (Sneath and Sokal, 1973) based on complete sets of SLC6 members from human (Hs), mosquito Aedes aegypti (Ae) and nematode Caenorhabditis elegans (Ce) genomes (also indicated by different colors). The NCBI protein accession numbers, followed by the name of the transporters from HUGO or WormBase (Version WS234) nomenclature, are shown. For human transporter names, the redundant SLC6 string was removed. The bootstrap consensus tree inferred from 2000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches, except for support values >95%, which were deleted (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter=1). The analysis involved 57 amino acid sequences. The tree was rooted with a sequence of bacterial transporter LeuT (Yamashita et al., 2005). All ambiguous positions were removed for each sequence pair. There were a total of 455 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011). (B) The phylogenetic branch of NATs from selected Caenorhabditis genomes: Cb, C. briggsae; Cbre, C. brenneri; Cre, C. remanei. Four paralogous NAT clusters are marked with large numbers 1–4. NAT, nutrient amino acid transporters resembling (n) nematode, (i) insect, and (mi) mammal and insect clusters, also highlighted by gray background; NTT, neurotransmitter transporter.
In contrast, our analysis of SLC6 transporters in free-living Caenorhabditis species showed expansion of the NAT subfamily to four orthologous groups, numbered in Fig. 1B as c1–c4 considering the apparent order of gene duplication. SNF-2 and SNF-7 are products of the recent SLC6 duplication in C. elegans. Another duplication resulted in the pair of CreNATs in cluster 3. These transporters are orthologous to SNF-4 (Fig. 1B), which is annotated as a pseudogene in WormBase (snf-4). The predicted snf-4 transcript has significant loss of structural fragments, including two sites in transmembrane domain 3 (TMD3) that are involved in substrate coordination (Fig. 2A). In contrast, theoretical products of two SNF-4 coding genes in C. remanei (XP_003101806 and XP_003101854) maintain typical SLC6 structure without such deletions, and could code for functional transporters. Three orthologous fragments of SNF-5, found in the draft annotations of the C. brenneri genome (EGT36064, EGT36082 and EGT36106), are theoretically predicted short products of alternative splicing of one gene. Therefore, despite long-term divergence >20 Mya and noticeable differences in ecological and physiological adaptations (Cutter, 2008; Thomas, 2008), the cluster of putative NATs in free-living nematode species represents a conserved subset of transporter-coding genes with minor species-specific gene duplications. SNF-5, with a single ortholog in each analyzed Caenorhabditis species, was selected for initial functional expression and characterization.
Fig. 2.
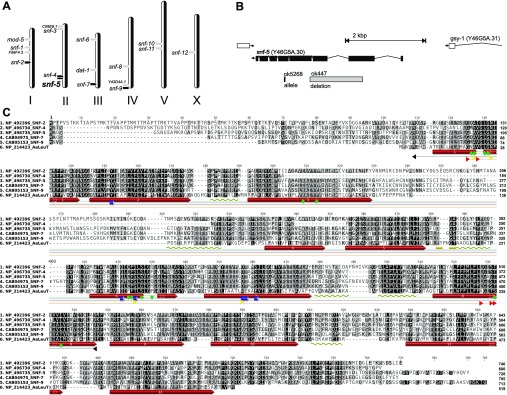
(A) Relative distribution of SLC6 transporters (gray arrowheads) and putative nematode NATs (black arrowheads) in C. elegans genome. (B) Localization and splicing diagram of snf-5 (SNF-5) in chromosome II. Gray bar indicates the deletion fragment in the mutant nematode that was analyzed in the present work. (C) Alignment of nematode NATs relative to the sequence and experimentally determined 2D structure of the bacterial transporter LeuT. Color shapes indicate relative positions of sites that are respectively implicated in: green, amino acid coordination in occlusion state of the transporters; yellow, first Na+ binding; red, second Na+ binding; blue, Cl− binding; light gray, outside gate; black, inside gates. Transmembrane domain numbers inside red bars indicate specific transmembrane domains. Gold arrows and coils indicate relative positions of conserved β-strands and sub-membrane α-helices.
Molecular cloning and structural analysis of SNF-5
The predicted splicing of the snf-5 gene, and the sequence of the SNF-5 protein, were confirmed by molecular cloning from a normalized cDNA collection of adult nematodes. The longest identified ORF has a 2187-base sequence, encoding a 728 amino acid protein, with an apparent molecular mass of 81.7 kDa. It is the eight-exon splicing product of the snf-5 (Y46G5A.30) gene. This gene is located at the 12,861,948–12,865,821 bp region in the negative strand of chromosome II, in proximity to its closest relative, pseudogene snf-4 (Fig. 2A).
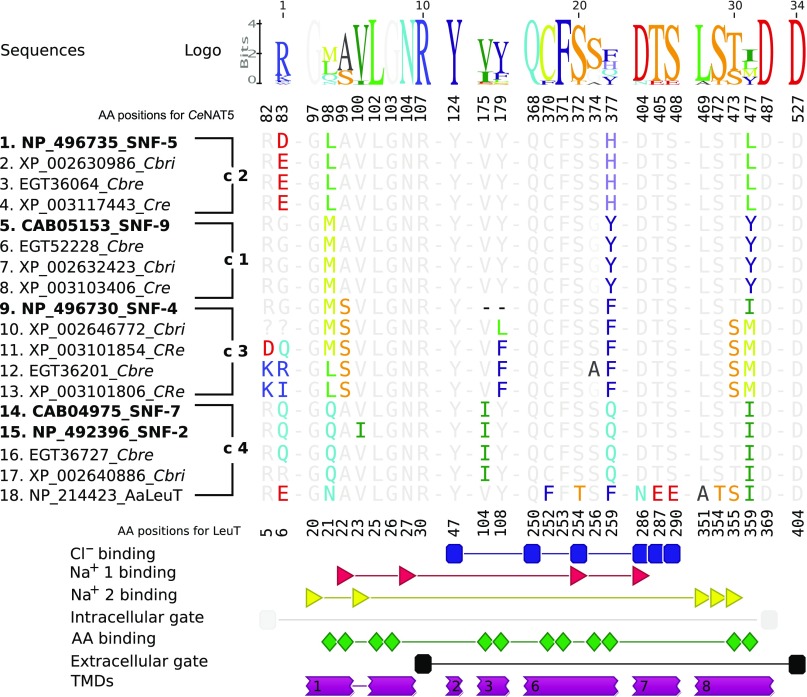
A comprehensive alignment of CeNATs with selected characterized SLC6 transporters and a bacterial member of the SLC6 family with determined crystal structure, LeuTAa of Aquifex aeolicus (Yamashita et al., 2005), provides insight into structural aspects of nematode transporters (Fig. 2C). It identifies a strong correlation between the theoretically predicted 12 TMD structure of CeNATs, based on protein sequence hydropathy (analysis of SNF-5 sequence using TopPred at http://mobyle.pasteur.fr/), and an alignment with the experimentally determined 2D structure and TMDs of the bacterial transporter (Fig. 2C). Despite only moderate PSI (~20%) of LeuT with nematode NATs, the alignment reveals more substantial conservation of TMD regions, ranging from 35 to 65% PSI. Conservation is especially high for TMDs 1, 2, 3, 6, 7 and 8, those that assemble to form the substrate-binding pocket (SBP) of the protein, a key determinant of its transport function. To examine the structural specificity of the CeNATs, we aligned selected nematode transporters with characterized insect and mammalian NATs and extracted and compared SBP-forming fragments (Fig. 3). None of the previously characterized transporters match the SBP profile of any of the putative nematode transporters. In contrast, the SBP profiles within orthologous clusters of nematode transporters, shown in Fig. 1B, were virtually identical (Fig. 3). This finding suggests two important features. First, there are specific structural adaptations of nematode transporters versus mammalian and insect NATs. Second, there is strong conservation of these adaptations amongst free-living nematode species. Of 12 organic substrate-coordinating residues (Fig. 3, green shapes), nine are relatively conserved, whereas three change between phylogenetic groups, introducing substantial changes in the volumes of the SBPs and substrate interaction frames of the transporters (Fig. 3, Logo). Relative to the SNF-5 sequence, the non-conserved residues are 98L-M/Q in TMD1, 377H-Y/F/Q in TMD6, and 477L-Y/M/I. In particular, Histidine 377 is interesting, because from currently available data this is the first basic amino acid found within the overall neutral composition of organic substrate-binding residues of SLC6 transporters. The presence of the relatively small and positively charged histidine is expected to broaden the substrate spectra and increase the ability to bind acidic amino acids in SNF-5 orthologs. To test this hypothesis and determine the exact transport properties of SNF-5, we functionally expressed and characterized its activity in Xenopus laevis oocytes.
Fig. 3.
Alignment of substrate-binding pockets (SBPs) of selected nematode transporters versus the SBP of LeuT. The residues that form SLC6 SBPs were identified upon alignment of selected SLC6 members versus LeuT sequence and secondary structure and extracted from the complete alignment (Fig. 2C). Structural motifs interacting with particular substrates are shown on the graphical diagram below the alignment as annotated on the right. Caenorhabditis elegans (Ce) NATs are indicated in bold and followed by four groups of orthologs transporters marked as c1–c4, as in the phylogenetic tree (Fig. 1B). The abbreviation strings used for representative species are: Cbri, C. briggsae; Cbre, C. brenneri; Cre, C. remanei.
Electro-physiological characterization of SNF-5
Amino acid-induced current became detectable on day 3 after injection of SNF-5 cRNA into X. laevis oocytes, demonstrating successful functional expression of this transporter in the selected heterologous system. Nineteen canonical l-amino acids and achiral glycine induce inward currents of different amplitudes from 5 to 125 nA in SNF-5-injected oocytes (Fig. 4A). These currents were statistically verified as significantly different from currents induced by the same substrates in control deionized-water-injected or uninjected oocytes, except for l-Lys- and l-Tyr-induced currents, which insignificantly differ from controls at concentrations ≤1 mmol l−1 (Fig. 4C, t-test for P<0.05 and N>2). The largest currents were generated by application of l-Pro and l-Met. Decreased currents were seen for the set of aromatic amino acids, with values for l-Trp at ~40% of the response seen for l-Pro (Fig. 4C). The profile of substrate-induced currents was comparable, but not identical, to broad substrate spectra transporters of the B0 system previously reported in mammals (Bröer et al., 2004) and insects (Miller et al., 2008). Specifically, SNF-5 exhibits a distinct order of amino acid/current amplitudes that may suggest distinct substrate preferences and substrate transport efficacy for SNF-5 versus previously characterized NATs. Contrary to B0-like transporters, SNF-5 expression generates significant current upon application of the acidic amino acids l-Glu (~75% of l-Pro) and l-Asp (~55% of l-Pro). We tested the capacity of SNF-5 to couple transport of other SLC6 substrates, including amino acid-derived neurotransmitters and their precursors. In addition to proteinogenic amino acids, SNF-5 generates significant currents in response to application of betaine and glutathione (Fig. 4B). In contrast, application of other potential SLC6 substrates, such as serotonin, dopamine, octopamine, GABA, l-DOPA, 5-hydroxytryptophan (5-HTP), taurine, creatine or any of the d-enantiomers of the 19 proteinogenic amino acids, did not induce considerable current in SNF-5-expressing oocytes (data not shown).
Fig. 4.
Specificity of SNF-5-coupled amino acid-induced currents. (A) Typical recording of amino acid-induced currents in an SNF-5-expressing oocyte. The oocyte was placed in a small volume constant-flow chamber on the fifth day after cRNA injection and maintained at −50 mV holding potential using a two-electrode voltage clamp. Amino acids were applied via a multiplexing perfusion system switching from 98N basal medium to 1 mmol l−1 amino acid solution in the same medium for the indicated intervals (black bars). The responses to similar concentrations of amino acid in control deionized-water-injected oocytes were <2 nA or undetectable at the presented scale (N>10, data not shown). (B) Response of an SNF-5-expressing oocyte to metabolic amino acids: betaine (Bet), taurine (Tau), and glutathione (GSSG). (C) Statistically validated responses of SNF-5-injected oocytes to 1 mmol l−1 l-amino acids in Na+ (98N) and K+ (98K) media, black and gray bars, respectively. Bars are relative amplitudes of substrate-induced currents normalized versus l-Pro, mean ± s.d., N>2 different oocytes for each data point. #Values that are not significantly different between Na+ and K+ specific samples; ##values that were not significantly different between control and SNF-5 injected oocytes (t-test, P>0.05, N>2). (D) Superposed current traces generated in response to 1 mmol l−1 solutions of l-Met and l-Glu in 98N medium and after complete substitution of Na+ with a larger NMDG+ ion.
Ion selectivity of SNF-5
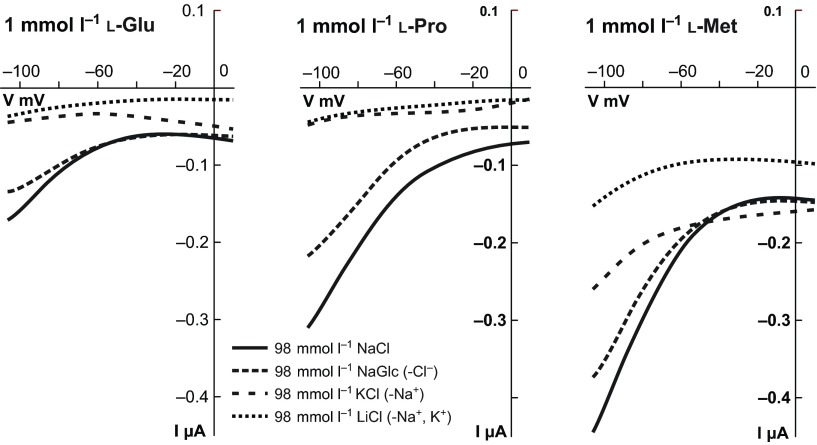
To determine the ion preferences of the SNF-5 transport mechanism, we analyzed amino acid-induced currents in solutions with different compositions of Na+ and K+ ions. The substitution of Na+ with K+ substantially, but not entirely, reduces substrate-induced currents across all tested substrates (Fig. 4C,D). Interestingly, SNF-5 displays an apparent shift of relative substrate preferences from l-Pro and l-Met in Na+ solution toward l-Gly and l-Ala in K+ solution. Substitution of canonical alkali cations with equimolar concentrations of smaller Li+ or larger N-methyl-d-glucamine+ (NMDG) (Fig. 4D, Fig. 5 dotted lines) and choline+ (data not shown) reversibly abolished substrate-induced currents. To test the requirement for Cl− in SNF-5 substrate transport, we administered selected substrates in a modified 98 Na+ medium, in which Cl− is substituted with gluconate−. This substitution has little to no effect on substrate-induced currents (Fig. 5, 98 NaGlc saline).
Fig. 5.
Electrophoretic and ion-dependent characteristics of SNF-5. A collection of current–voltage (I–V) plots recorded from SNF-5-expressing oocytes. The plots were built from reciprocal current and voltage amplitudes recorded during 2 s voltage ramp stimulus, e.g. initial holding potential −50 mV, drop to −110 mV, 2 s ramp increase up to +10 mV, and final drop to −50 mV. The three different panels represent IV collections for selected amino acids: 1 mmol l−1 l-Glu, l-Pro and l-Met in four different solutions: normal medium (98 mmol l−1 NaCl); Cl−-free medium with Cl− substituted by gluconate− (98 mmol l−1 NaGlc); Na+-free medium with Na+ substituted by K+ (98 mmol l−1 KCl); and Na+/K+-free medium with major alkali cations substituted by Li+ (98 mmol l−1 LiCl). The ion-specific I–V plots are indicated by distinct line patterns as shown in the inset. Each I–V plot represents the amino acid-specific component acquired as a current response during application of amino acid (~90% saturation point), with point by point subtraction of leak current recorded immediately before the amino acid application. Values were acquired from different oocytes and were subsequently filtered (0.1 Hz) and normalized under the assumption that amplitudes of leak and amino acid-induced currents are linear functions of SNF-5 expression.
Radiolabelled substrate uptake by SNF-5
Relative and competitive assays of radioactive isotope-labeled substrate uptake were employed to validate coupling of SNF-5 mediated currents to amino acid absorption. SNF-5-injected oocytes showed significant increases in the accumulation of labeled amino acids compared with uninjected oocytes (Fig. 6). The comparative uptake assay of selected amino acids revealed the highest accumulation rate for l-Pro and l-Met, corroborating data from the electrophysiological study (Fig. 4). Non-polar amino acids also strongly competed with l-Pro for uptake, reducing the accumulation of l-Pro to the greatest degree when compared with basic, acidic or aromatic amino acids (Fig. 4B).
Fig. 6.
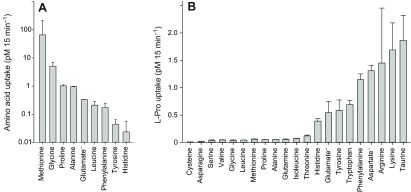
SNF-5-coupled uptake of radiolabeled amino acids. (A) Comparative uptake: 4 days after injection, SNF-5-expressing and uninjected control oocytes from the same batch were bathed in 0.02 μCi ml−1 radiolabeled substrate diluted in 98 mmol l−1 NaCl solution. After 15 min exposure, oocytes were washed in several changes of ice-cold NaCl solution to terminate uptake and were subsequently processed for scintillation counting. Bars represent the mean ± s.e.m. substrate accumulation after pairwise subtraction of disintegrations per minute counted in uninjected oocytes (N>2). (B) Competitive uptake: SNF-5-expressing and uninjected oocytes were exposed to 0.02 μCi ml−1 3H-l-Pro in 98 mmol l−1 NaCl solution containing 5 mmol l−1 unlabeled competing amino acid (2.5 mmol l−1 for Tyr). After 15 min exposure, oocytes were washed and processed for scintillation counting. Bars represent the mean ± s.e.m. uptake of radiolabeled l-Pro under competition with the listed substrates (N>2).
Kinetic properties of SNF-5
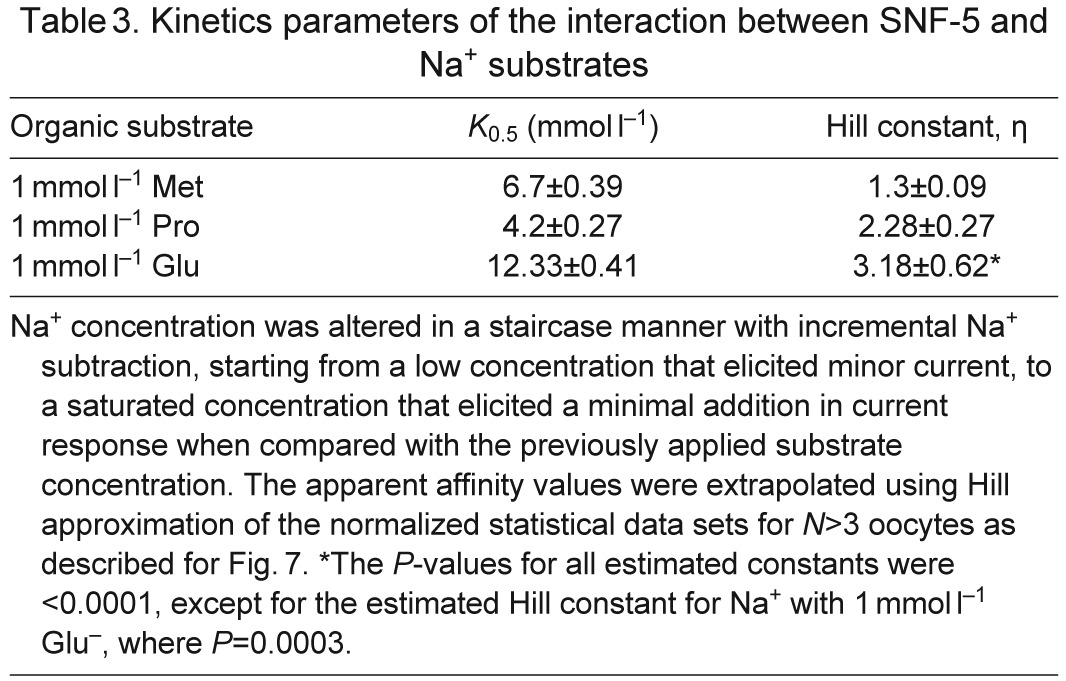
SNF-5 exhibits saturable kinetics upon interaction with organic and inorganic substrates (Fig. 7A). It shows submillimolar apparent affinity to all tested amino acids, with the highest affinity for l-Ser (21 μmol l−1), which is comparable to the affinity for l-Met (27 μmol l−1) and approximately four times higher than the affinity for l-Pro (95 μmol l−1; Fig. 7A, Table 2). This affinity profile generally corroborates both the electrophysiological and substrate uptake data, showing high apparent affinity for non-polar and polar amino acids, those which induced larger currents, and low affinities for aromatic and basic substrates, those which induced smaller currents. However, despite high substrate-induced currents, the K0.5 for the acidic amino acids were in the range of hundreds of μmol l−1, 10–20 times higher than the most preferable substrates. The estimated Hill constants for all tested amino acid were close to 1, suggesting that SNF-5 transports one amino acid molecule per transport event. The apparent K0.5 of SNF-5 for Na+ varies slightly upon combination with different organic substrates. For example, the estimated K0.5 Na+ was ~4.2 mmol l−1 when the transporter was exposed to 1 mmol l−1 l-Pro, and ~6.7 mmol l−1 when tested with 1 mmol l−1 l-Met (Fig. 7B, Table 3). Moreover, SNF-5 mechanisms showed different estimated stoichiometry for Na+ ions in combination with different amino acids. For example, the estimated Hill constant for Na+ was ~1 during transport of l-Met, ~2 during transport of l-Pro and ~3 during transport of l-Glu, resulting in 1:1, 1:2 and 1:3 apparent transport stoichiometry, respectively.
Fig. 7.
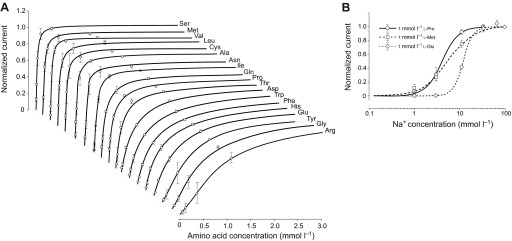
Kinetic properties of SNF-5 interaction with substrates. (A) Amino acid saturation profiles. The specified amino acids were applied in staircase concentration increments: 0.011<0.033<0.11<0.33<1.1<3.3<10 mmol l−1 prepared in 7.2 pH buffered 98 mmol l−1 Na+ medium. Graphs show only up to the 3 mmol l−1 concentrations of the specified amino acids. (B) Na+ saturation profiles with linear and logarithmic scale, respectively. The specified amino acids were applied at 1 mmol l−1 concentrations in media containing variable concentrations of Na+. Stepwise, these concentrations were: 1<3.3<10<33<98 mmol l−1, where Na+ was substituted with equimolar concentrations of N-methyl-d-glucamine (NMDG+). Data points are mean ± s.e.m. current responses normalized to maximum current in individual oocytes, for N>3 different oocytes. The data were extrapolated to fit a three-parameter sigmoidal Hill approximation f=ImaxIη/(K0.5η +Iη) without parametric constraints of Michaelis–Menten (η=1). Estimated affinities (K0.5) and apparent order of translocation events (η, Hill constant) for amino acid and Na+ substrates are summarized in Tables 2 and 3, respectively.
Table 2.
Kinetics parameters of the interaction between SNF-5 and amino acid substrates
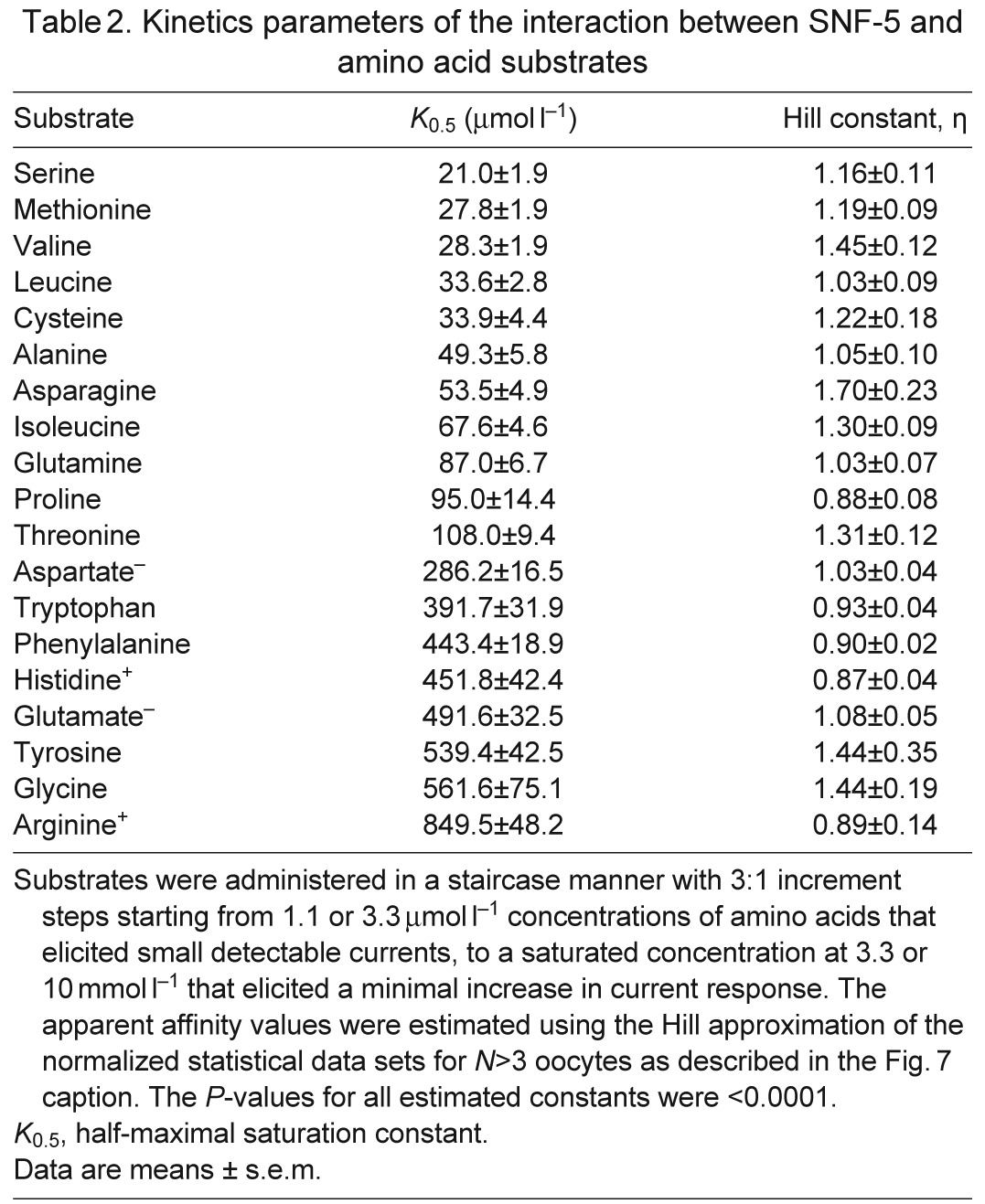
Table 3.
Kinetics parameters of the interaction between SNF-5 and Na+ substrates
Analysis of snf-5 expression in nematode tissues
To determine the tissue-specific activity of snf-5, we analyzed expression of a transcriptional EGFP reporter under an snf-5-specific promoter using previously generated strain (BC14858) and a strain newly generated in our laboratory. Both strains showed a particularly intense EGFP signal in intestinal cells INT1-9 of the alimentary canal, a principal organ for nutrient amino acid absorption (Fig. 8A–C for INT1-3 and C for INT8 and 9). EGFP expression exhibits a more intensive and aggregated pattern in the epithelial cells of the mature gut (Fig. 8B), but more even and diffuse expression in the alimentary canal of younger worms (Fig. 8C). This fact suggests specificity of the labeling and gradual aggregation of the reporter protein. EGFP signal was detected in the pharynx in some, but not all, analyzed preparations. EGFP expression was also detected in the posterior cells of the alimentary canal, two cells of the rectal gland (Fig. 8A,B), and several cells that approximately co-localize with the positions of the DVA, DVB or DVC neurons. Further identification of these cells will require additional analysis.
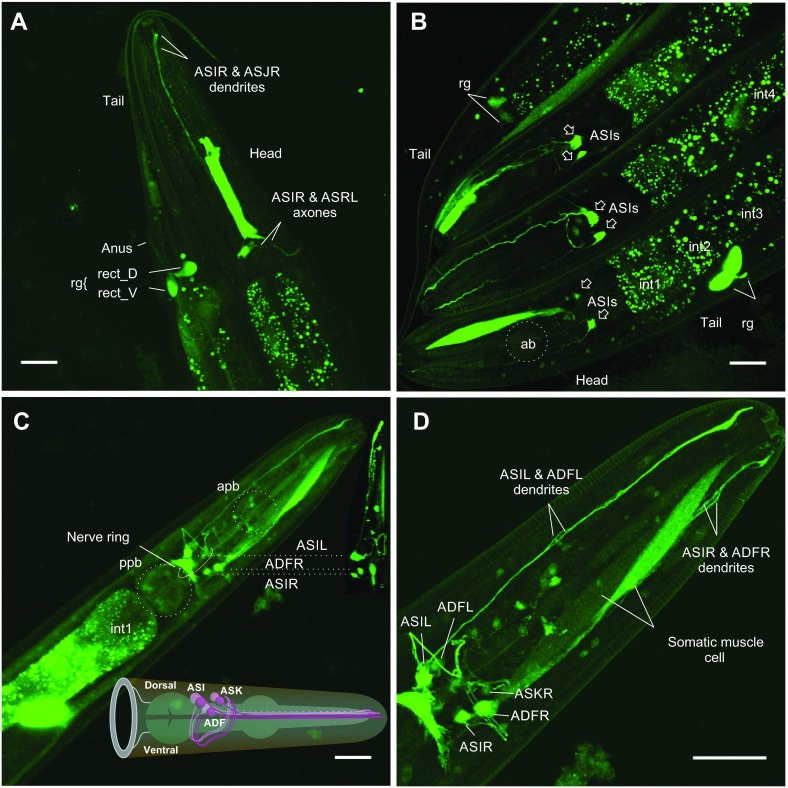
Fig. 8.
Patterns of snf-5 expression in the alimentary canal and neuronal system. (A) Right lateral view of EGFP expression under snf-5 promoter in anterior and posterior fragments of the nematode body. (B) Simultaneous view of three nematode heads and two nematode tails, demonstrating typical variations of enhanced green fluorescent protein (EGFP) expression across BC14858 worms. (C,D) Ventral maximum projection of the anterior portion of the nematodes from preparations of the snf-5 promoter-driven EGFP strain generated in our laboratory. Both strains showed similar pattern of EGFP expression. In addition, our strain showed brighter labeling and allows for better reconstruction of the neuronal labeling pattern. Images were assembled from 40–68 scanned frames using 20× and 60× objectives on an LM10 laser scanning confocal microscope (Olympus). Individual cells were identified using information and neuronal maps available at http://www.wormatlas.org and labeled according to WormBase nomenclature. ab, anterior bulb; apb, anterior pharyngeal bulb; ppb, posterior pharyngeal bulb; AS, identified anterior sensory neurons I, J and K pairs, both left (L) and right (R); AD, identified amphid dual ciliated sensory neuron pair F, both left (L) and right (R); int#, specific intestinal cells; rg, rectal gland with labeled rect_D and rect_V cells. Scale bars, 20 μm.
Expression of EGFP reporter was also identified in three pairs of amphid sensory neurons that are localized near the dorsal-anterior surface of the posterior pharyngeal bulb (Fig. 8). Considering the localization to the lateral ganglia of the head, the relative somatic positions identified via 3D reconstruction of confocal sections, the axonal projections, and the projections and shapes of dendrites visualized in a maximum projection of confocal stack (Fig. 8C, inset; Fig. 8D), these cells represent the ASI, ADF and ASK neurons (Fig. 8C, inset). The identification of ASI and ASK was supported by visual inspection of the relative distribution of EGFP signal and DiI labeling in amphid neurons. The ASI and ADF neurons exhibited intense labeling in the somata and along the axonal and dendritic projections. In contrast, ASK labeling was weak in the somatic regions and neuronal branches. EGFP expression was also detected in a population of small neurons at the dorsal surface of the anterior pharyngeal bulb (Fig. 8D). These cells, as well as ASK and ADF, were labeled only in the strain generated in our laboratory. The BC14858 strain produces less intense labeling of the ASI neurons, and other cells were difficult to visualize at a fluorescence gain optimized for ASI (Fig. 8A,B). The constitutive expression of SNF-5 in the alimentary canal corresponds with a universal expression profile expected for NATs. In contrast, strong expression of a NAT in the identifiable amphid sensory neurons, which are involved in sensing of nutrient amino acids and other environmental signals, as well as efferent coordination of chemotaxis, metabolism and developmental strategy, is an important new finding. It was particularly intriguing as, until now, there have been no clear data as to whether, and how, animals determine the availability and regulate the metabolism of essential amino acids at a systemic level. As no phenotype has yet been described for snf-5, we analyzed the impact of snf-5 deletion on nematode biology and developmental strategy under different nutrient conditions.
Analysis of snf-5 deletion mutants
To reveal the role of SNF-5, we compared wild-type N2 animals and snf-5(ok447) mutants with a large deletion in the second, third and fourth exons (Fig. 2B). In the initial assay, we monitored and analyzed worm populations of both strains originating from individual animals plated to three separate OP50-seeded plates. Regional images were captured every 3 h during the initial egg laying phase of nematode reproduction and at specific stages of nematode development, including days 1, 5, 7 and 15 (see Fig. 9 for representative image sequence). Day 15 corresponds to ~10 days after the bacterial lawn was exhausted. From the image analysis, we assessed the number of eggs, L1, L2 and L3/L4 nematodes present on the plate. We detected no significant effect of strain over time in the reproductive capacity as measured by egg count (F9,36=1.2586, P=0.2924) or early stage larval density (F7,28=0.4954, P=0.8297) between wild-type and snf-5 mutant groups. Likewise, we observed no apparent difference in the overall densities of L1 nematodes at any point throughout the specified time course (Fig. 9A). In addition, no apparent differences were seen for L2 larvae, which were not distinguished from dauer individuals in the initial assay (Fig. 9A). The differences between wild-type and snf-5 mutant groups became prominent at time points beyond the exhaustion of bacterial food on the plates. Specifically, L3, L4 and adult stage nematodes (identified by the presence of developed eggs within the individual) were present in snf-5 mutants, while these stages were significantly limited, and mostly absent, at ~7 days in N2 samples (Fig. 9A). These results may suggest a reduced ability of snf-5 mutants to enter or/and maintain dauer. To solve the dilemma, and characterize snf-5 mutants further, we examined L1-specific metabolic arrest and L2-specific dauer formation using synchronized larvae groups obtained via egg synchronization, and constrained experimental conditions in which no nutrients were initially provided. No differences were detected between wild-type and snf-5 mutant groups in the ability of L1 larvae to undergo and maintain metabolic arrest (N=3 plates for each sample/control pair), as well as complete recovery after 24 h of starvation (N=10 individuals for each sample/control pair; all individuals recovered). In contrast, formation of dauer larvae upon starvation, assessed via SDS treatment of food-exhausted plates, was significantly increased in the snf-5 mutant versus N2 control (Fig. 9C; mean increase 213% for N=3; P=0.029). The starvation-induced dauers from both strains completely recovered upon reintroduction to food, with no apparent difference (N=10 individuals for each sample/control pair; all individuals recovered). Together, our data showed no detectable contribution of SNF-5 to nematode biology without nutrient deprivation, and a significant contribution of SNF-5 in dauer regulation upon nematode starvation.
Fig. 9.
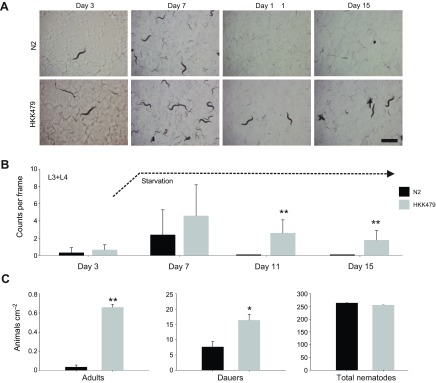
Analysis of the snf-5 deletion phenotype. (A) A sequence of representative snapshot images on days 3, 7, 11 and 15 during monitoring of nematode population in control N2 and snf-5 knockout (HKK479) strains of C. elegans. Scale bar, 1 mm. (B) Graphical representation of nematode density. Bars are mean + s.e.m. nematode count for N≥9 samples; three randomly distributed frames (4.2×5.6 mm; 23.5 mm2) in at least three different plates. Graph represents the overall trend (day #) in the quantity of L3 and older nematodes in experimental nematode populations from control (N2 strain; black bars) and snf-5 knockout (HKK479 strain; gray bars) nematodes over the 15-day period. Counting was completed considering approximate L1, L2 and L3 stage nematodes (L2 was not distinguished from dauer larvae, and L3 size were combined with L4 and adult nematodes). Day 7 represents the point of starvation, ~2 days after the bacterial food lawn was exhausted (dashed line). No differences were seen between L1 and L2 larval stages (N=12 plates). (C) Graphical representations of adult nematode survival, dauer formation and total population densities. Bars are mean ± s.e.m. nematode density for N≥3 replicates. Graphs represent the density of the indicated nematode population ~4 days after the exhaustion of food supply. * and ** indicate statistical significance at P<0.05 and P<0.001, respectively.
DISCUSSION
SLC6 is one of the largest families of secondary transporters with diverse roles in metazoan metabolism and neurochemical signaling (Chen et al., 2004). It includes two phylogenetically and functionally specialized subfamilies: the animal-specific subfamily of neurotransmitter transporters (NTTs), with several orthologous branches of monoamine and amino acid neurotransmitter transporters (Kristensen et al., 2011); and a more polyphyletic subfamily of NATs (Boudko et al., 2005a). NATs, paralogous to those present in Bacteria and Archaea, tend to form species-specific clusters in different animal clades. Moreover, NATs appear to be much more diverse in substrate specificity compared with NTTs (Boudko, 2010). Previous studies have shown that NATs are broadly and universally involved in the alimentary absorption and distribution of essential amino acids across different metazoan organisms and tissues (Boudko et al., 2005a; Boudko et al., 2005b). Moreover, several characterized NATs are expressed in specific neurons and possess substrate preferences for essential precursors of neurotransmitter synthesis (Meleshkevitch et al., 2006; Meleshkevitch et al., 2009a; Boudko, 2012). Hence, comparative analysis of NAT functions, descending through the history of life, is crucial for compiling the origin, expansion and biological roles of the SLC6 family members. As systemic and neuronal transporters of essential amino acids, NATs are considerable targets for pharmacological correction of metabolic and neuronal disorders, as well as for selective intervention in pest control. The cloning and functional analysis of new NATs, in combination with the emerging potential to model functional domains of SLC6 transporters (Yamashita et al., 2005), provides unique opportunities for unraveling structure–function relationships and elucidating the molecular evolution of substrate selectivity in these transporters. Here we describe a NAT cluster and summarize characteristics of the first NAT representative from an important biological model, C. elegans.
Phylogeny
The identification and characterization of new nematode NATs is an important milestone of SLC6 study. Our analysis revealed 17 SLC6 members in the C. elegans genome. Five of these genes had been previously analyzed. These include: snf-6 (Kim et al., 2004), snf-11 (Mullen et al., 2006), mod-5 (Ranganathan et al., 2001) and dat-1 (Jayanthi et al., 1998), which encode putative acetylcholine, GABA, serotonin and dopamine transporters, respectively. An essential role in immune signaling has been also been demonstrated for snf-12 (Dierking et al., 2011). However, the substrates of this transporter remain unknown. Phylogenomic trees of the SLC6 family including nematode representatives have been published previously (Boudko et al., 2005a; Meleshkevitch et al., 2009b; Boudko, 2010; Dierking et al., 2011). The phylogenomic analysis presented in this work focuses on identification of NAT homologs and includes additional nematode species. Our results are generally consistent with previous studies, aiding in the identification of four orthologous clusters of NATs in free-living nematodes (Fig. 1A,B).
Five of 17 identified SLC6 genes form a cluster at a phylogenetic position anticipated for the nematode NATs that includes the CeNAT subfamily (Fig. 1, nNAT). The phylogenomic analysis, coupled with functional characterization, identifies at least one transporter among the putative nematode NATs that exhibits typical NAT properties and tissue distribution. The identification of a NAT cluster in basal Ecdysozoa, represented by nematodes, supports a NAT paradigm that was previously limited to mammalian and insect members (Boudko et al., 2005b; Boudko, 2012). Based on the data, we may now conclude that NATs exist in free-living nematode species and form a paralogous NAT cluster (Fig. 1), as was the case for the formation of insect and mammalian branches of the NAT subfamily. Nematode-specific NATs form four orthologous groups that are expected to comprise transporters with homologous functions (Fig. 1B). This expansion pattern is consistent with gene duplication-driven expansion and rapid stabilization of new transport phenotypes upon acquisition of new ecological and nutrient niches. The combined phylogenetic and structural analysis demonstrates explicit correlation of phylogenetic clustering (Fig. 1) with conservation in the amino acid profile of the SBP (Fig. 3), a principal determinant of transporter function. This finding is important for further analysis of structure–function relationships in that it exposes the structural components involved in evolution of substrate selectivity. The phylogenomic analyses of the current framework of nematode genomes also lead to an unexpected finding: an apparent lack of NATs and a marked reduction of the SLC6 family in the parasitic nematode species (Table 1). Putative NAT homologs form the entire SLC6 family in bacteria, archaea, fungi and some protozoan organisms, and comprise a significant portion of the metazoan SLC6 family. SLC6 members have not currently been identified in plants. To our knowledge, the absence of the NAT-SLC6 subfamily in parasitic nematodes represents the first case of complete extinction of these transporters in animals. We anticipate that such a tradeoff is the result of parasitic adaptations to nutrient-rich environments, where NAT function can be compensated for by other transport mechanisms, such as passive uniporters and exchangers, or proton-dependent transporters. We remark here that the present analysis of parasitic nematode transporters is based on draft annotations of the genomes of these organisms that require substantial improvement.
Transport properties
Only snf-11 has been previously heterologously expressed and characterized with direct transport assays (Mullen et al., 2006). Thus, SNF-5 represents the second transporter in this category. From the electrophysiological assays, SNF-5 appears to be a Na+- or K+-driven, Cl−-independent, l-isomer-specific, broad-spectrum neutral amino acid transporter with an unusual additional capacity to transport acidic amino acids. These properties identify a new transport system, which we named here as B0,−, following the generally accepted convention for amino acid transport systems (Bannai et al., 1984; Christensen et al., 1994). Recent cloning of several orphan transporters from the mammalian SLC6 family revealed the molecular identity of the B0 system, which currently incorporates four characterized orthologous groups identified as B0AT1 (SLC6A19) (Romeo et al., 2005), B0AT2 (SLC6A15) (Takanaga et al., 2005b; Bröer et al., 2006), B0AT3 (SLC6A18) (Singer et al., 2009) and NTT4 (SLC6A17) (Zaia and Reimer, 2009) members. Combined, these transporters represent 80% of the NAT subfamily in mammals. They are similar, but not identical, in their preference for large aliphatic and aromatic amino acid substrates. However, these transporters are notably different in expression profile, suggesting that duplication of mammalian NATs is driven mainly by tissue and subcellular membrane specialization. Some substrate specialization is also evident among mammalian NATs. For example, the closest relatives of the B0 system, SLC6A20 transporters, represent the IMINO system, which specializes in the transport of conditionally essential l-Pro (Kowalczuk et al., 2005; Takanaga et al., 2005a). The degree of selectivity of SLC6A20 orthologs to l-Pro versus other neutral amino acids is varied in different mammalian models (Kowalczuk et al., 2005; Takanaga et al., 2005a; Ristic et al., 2006). The substrate narrowing specializations are far more pronounced for some insect-specific NATs. For example, NAT expansion in mosquitoes is larger than that of mammalian NATs and, in addition to B0-like transporters (Boudko et al., 2005a; Miller et al., 2008), includes currently identified Phe-, Trp- and Met-selective members (Meleshkevitch et al., 2006; Meleshkevitch et al., 2009a; Meleshkevitch et al., 2009b). The functional characterization of SNF-5 showed that B0-like transporters exist, and are structurally conserved, in free-living nematodes, making them one of the most universal and phylogenetically ancient types of metazoan NATs. One of the striking differences of SNF-5 versus the traditional B0 system is its capacity to transport acidic amino acids, which was identified in electrophysiological and radiolabeled uptake experiments (Figs 4, 5 and 6). This property is unique, given that no previously characterized SLC6 transporter has been reported with the potential to generate substantial currents or mediate uptake with acidic amino acids. The acidic amino acids are substrates of phylogenetically and structurally distinct transporters of the SLC1 family (Kanai and Hediger, 2004). From the structural comparison (Fig. 3), we anticipate that interaction of SNF-5 with acidic substrates results from the basic His 377 in the fragment of the SNF-5 SBP that is associated with TMD6, and appears to be conserved across SNF-5 orthologs from other nematodes. However, this hypothesis, as well as the biological significance of such an unusual expansion of substrate spectra, requires additional study. Another interesting property of SNF-5 is its ability to use the ion K+ in addition to Na+ as a motive force for substrate transport. Reported mammalian B0ATs are Na+ dependent. However, it appears that some insect NATs have evolved the capacity to use the energy of the inverted gradient of K+ ions in addition to (Meleshkevitch et al., 2006), or even in favor of (Castagna et al., 1998), Na+ electrochemical gradients. These types of ion selectivity correlate with insect adaptations to unstable or limited availability of nutrient Na+. Similarly, free-living nematodes may experience variable and limited availability of Na+ and K+ in their habitats. In particular, CeNAT5 ion dependency is consistent with the nutrient adaptations of nematodes feeding on soil microorganisms.
The reported apparent stoichiometry of substrate translocation events in mammalian SLC6 members A15 (Takanaga et al., 2005b), A19 (Camargo et al., 2005) and A20 (Takanaga et al., 2005a) is ~1. It may be different for A18 (Romeo et al., 2006) and A17 members (Zaia and Reimer, 2009), for which it was technically difficult to determine. It is generally accepted that coupling stoichiometry is a constrained parameter, ranging from 1 to 3 Na+ ions per substrate molecule between different members of the SLC6 family (Albers and Radzik, 2004). However, increasing electrophysiological evidence suggests a more flexible concept, e.g. a single Na+ is necessary, and may be sufficient, to trigger the conformation cycle of SLC6 transporters with distinct preferable stoichiometry, while additional Na+ merely increases the ion-generated force. Moreover, several Na+ ions may cooperate in changes of SLC6 conformation via allosteric binding sites with different affinities and, therefore, produce complexes with different stoichiometry dependent on substrate concentration. For example, a Trp-selective NAT-SLC6 transporter from Anopheles mosquitoes showed a preferred 1 Na+:1 AA stoichiometry at sub-millimolar concentrations of l-Trp, and 2:1 stoichiometry at inferior substrate concentrations. Such a ‘gear shift’ has physiological rationale, as it doubles the motive force of the transporter's conformational shift at low substrate concentrations and enables accumulation against concentration gradients a full order of magnitude higher. Our analysis showed that SNF-5 physiological and kinetic parameters generally overlap with those of insect NATs, indicating a preferred stoichiometry of 1:1. This is not surprising considering similar parameters are shared between mammalian and insect B0-like transporters that act with submillimolar apparent affinities for amino acids. One surprising finding was the identification of acidic amino acid-induced currents that cannot be generated upon 1 Na+:1 Glu− stoichiometry. Hence, we more carefully tested the stoichiometry for all analyzed substrates and established that in addition to 1:1 stoichiometry for the majority of neutral amino acids, SNF-5 prefers to transport 3 Na+ with Glu− or Asp− and 2 Na+ with the small achiral Gly zwitterion. A finding of differing substrate stoichiometry is novel, but fits well with the theory that the SBP of transporters can accommodate additional Na+ during interaction with small amino acids, as well as attract additional Na+ upon interaction with small anionic amino acids. The increased Na+ coordination may be obligatory for transport of acidic amino acids and glycine, and unique to SNF-5's mechanism. Alternatively, and more conceivably, substrate-dependent stoichiometry is a more common phenomenon that can be identified in other SLC6 members.
Physiological role of SNF-5
Here we took advantage of a relatively simple transgenic nematode technique for monitoring and disrupting gene expression. As expected, we found strong constitutive expression of a transcriptional snf-5-specific EGFP reporter in the absorptive epithelial cells of the alimentary canal. This finding is consistent with the anticipated NAT role of SNF-5 in the nutrient absorption of amino acids, as was similarly identified for insect NATs via in situ hybridization and immunolabeling (Meleshkevitch et al., 2006; Meleshkevitch et al., 2009a; Okech et al., 2008). The transcriptional marker cannot address membrane localization of the transporter. However, we expect apical docking of this transporter, as was reported from immunolabeling of insect and mammalian NATs in the insect gut and mammalian intestinal and renal epithelia (Okech et al., 2008; Romeo et al., 2006; Vanslambrouck et al., 2010). In contrast, SNF-5 expression in rectal gland cells may indicate basolateral localization, supporting the secretory function of the organ, as was previously identified for insect NATs in the salivary gland and other secretory tissues (Okech et al., 2008). Previous studies in insects showed unique patterns of NAT expression in neurons of the central ganglia and sensory system, which, together with the aromatic amino acid specificity of these transporters, suggests their role as substrate providers for the synthesis of monoamine neurotransmitters (Meleshkevitch et al., 2006; Meleshkevitch et al., 2009a). A similar function has been suggested for B0 transporters in GABA-ergic components of the mammalian brain (Takanaga et al., 2005b). In our study, cell-specific expression of snf-5 promoter-driven EGFP was identified in the amphid sensory neurons ASI, ASK and ADF. All of these neurons are intimately involved in worm chemoreception (Bargmann and Horvitz, 1991a). Moreover, ASK and ASI represent gustatory neurons responsible for sensing the availability of nutrient amino acids and triggering local or dispersal searching (Gray et al., 2005), whereas ADFs couple food availability to serotoninergic signaling (Jafari et al., 2011). The cellular mechanism for sensing external amino acids is unknown. Nonetheless, considering the high relative expression of SNF-5 and the associated amino acid-dependent sodium current it transmits, we may postulate a significant contribution of this transporter to the modulation of the transmembrane potentials and electrical activities of the identified amphid sensory neurons. As a result, SNF-5 is a good candidate to be the long-elusive sensor for monitoring the availability of nutrient amino acids in the environment and systemic circulation. SNF-5 also may supply the synthesis of signaling peptides whose sequences are enriched in essential amino acids. Specifically, ASI synthesizes a diversity of neuropeptide-like proteins, including NLP-1, NLP-5, NLP-6, NLP-7, NLP-9, NLP-14, NLP-18, NLP-24, NLP-27 (Nathoo et al., 2001) and insulin-like peptide INS-9 (Pierce et al., 2001); ASK releases glutamate and synthesizes NLP-8, NLP-10 and NLP-14 (Nathoo et al., 2001); and serotonergic ADF also synthesizes FRMF-amide-like peptide FLP-6, insulin-like peptide INS-1 and neuropeptide-like protein NLP-3. As a provider of essential tryptophan, SNF-5 may support synthesis of serotonin in ADF neurons.
To reveal the biological contributions of snf-5, we tested the effect of its knockout in worms developing under different nutrient conditions. In agreement with concurrent snf-5 RNAi reports (Kamath et al., 2003; Rual et al., 2004), our analysis showed no apparent differences in nematode morphology or reproduction between wild-type and snf-5 mutants grown under unlimited food supply. Similarly, our assay did not indicate notable morphological changes in starved snf-5 mutants. Hence, despite high SNF-5 expression in the alimentary canal, its contribution in the alimentary absorption of essential amino acids can be effectively compensated by auxiliary transport mechanisms.
In contrast, our assay showed involvement of snf-5 in a neuronal mechanism that coordinates worm metabolism and developmental strategy with environmental conditions. Specifically, starved snf-5 mutants showed significant increases of L3, L4 and adult worms compared with wild-type worms, in which all individuals either underwent dauer formation or died. This result was surprising, as we may expect more dauers in subjects that might experience more rapid nutrient deprivation due to the lack of a nutrient amino acid absorbing mechanism. Indeed, in additional assays we found a significant increase in dauer formation in the snf-5 mutant. Because the metabolic contribution of SNF-5 was initially ruled out, we must consider the neuronal functions of SNF-5 to explain the apparent conflict of observed snf-5 phenotypes. The most plausible, if not only, possible explanation is that the snf-5 mutant has a disorder in the sensory mechanisms that signal nutrient amino acid availability to a dauer control mechanism. We can address this subject more precisely through the specific roles of the identified amphid sensory neurons that express SNF-5. In addition to their roles in the sensing of amino acids and chemotaxis (Bargmann and Horvitz, 1991a), the SNF-5 expressing sensory neurons are known to control worm metabolism and development (Bargmann and Horvitz, 1991b), social feeding (de Bono et al., 2002), longevity (Alcedo and Kenyon, 2004; Bishop and Guarente, 2007), diet-restriction-induced life span extension (Bishop and Guarente, 2007) and dauer formation (Albert et al., 1981; Fielenbach and Antebi, 2008). Specifically, ablation of the ASI neurons is known to reduce chemotaxis to amino acids (Bargmann and Horvitz, 1991a), as well as trigger dauer formation (Bargmann and Horvitz, 1991b) by removing an ASI-coupled inhibition (Schackwitz, et al., 1996). ADF has the same modality as ASI, but relays via a different set of signaling molecules and cascades. Hence, SNF-5 coupled depolarization or amino acid influx may potentially contribute to ASI/ADF mediated dauer inhibition, while SNF-5 disruption will mimic reduction of amino acid availability and promote dauer formation. In contrast, ablating the ASK neurons reduces dauer entry under inducing conditions and disables the recovery of dauer larvae (Bargmann and Horvitz, 1991b; Schackwitz et al., 1996). Thus, the significant increase of worms that ignore dauer-inducing conditions among snf-5 mutants is consistent with an ASK ablation phenotype (Schackwitz et al., 1996). Therefore, SNF-5-assisted signaling in specific amphid sensory neurons fits well with the known dauer control functions, and may explain the increased, but incomplete, dauer formation in the snf-5 mutant as a disorder of amino acid sensing in ASI/ADF and ASK neurons. Based on the experimental data and literature, we hypothesize that some NATs may regulate metabolism and development, acting as neuronal sensors of amino acid availability. However, these conclusions remain speculative, as we have not yet formally demonstrated that the snf-5 mutant allele causes the observed phenotypes.
ACKNOWLEDGEMENTS
We thank Dr H. J. Kelly Oh (Dr Kim's laboratory) for her valuable help and technical assistance on the experimental work with nematodes, and Drs Hui Li (Dr Sackin's laboratory) and Junjie Tong (Dr Ebihara's laboratory) for providing the Xenopus laevis oocytes used in this study. In addition, we would like to thank Drs Peterson, Hawkins and Wolf for their very helpful discussions, critiques and insights.
LIST OF ABBREVIATIONS
- EGFP
enhanced green fluorescent protein
- K0.5
half-maximal saturation constant
- NAT
nutrient amino acid transporter
- NGM
nematode growth medium
- NTT
neurotransmitter transporter
- ORF
open reading frame
- PSI
pairwise sequence identity
- SBP
substrate-binding pocket
- SDS
sodium dodecyl sulfate
- SLC6
solute carrier family 6
- TMD
transmembrane domain
FOOTNOTES
COMPETING INTERESTS
No competing interests declared.
FUNDING
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). This work was supported in part by faculty startup funds from the Rosalind Franklin University for Medicine and Science; the Graduate Student Fund from the Department of Physiology and Biophysics, Rosalind Franklin University of Medicine and Science; National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID) grant AI030464 to D.Y.B.; and National Institutes of Health/National Institute of Neurological Disorders and Stroke (NIH/NINDS) grant NS077018 to H.K. Deposited in PMC for release after 12 months.
REFERENCES
- Albers S., Radzik T. (2004). Algorithms – ESA 2004. In Proceedings of the 12th Annual European Symposium, Bergen, Norway, September 14-17, 2004. Berlin: Springer; [Google Scholar]
- Albert P. S., Brown S. J., Riddle D. L. (1981). Sensory control of dauer larva formation in Caenorhabditis elegans. J. Comp. Neurol. 198, 435-451 [DOI] [PubMed] [Google Scholar]
- Alcedo J., Kenyon C. (2004). Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron 41, 45-55 [DOI] [PubMed] [Google Scholar]
- Bannai S., Christensen H. N., Vadgama J. V., Ellory J. C., Englesberg E., Guidotti G. G., Gazzola G. C., Kilberg M. S., Lajtha A., Sacktor B., et al. (1984). Amino acid transport systems. Nature 311, 308 [DOI] [PubMed] [Google Scholar]
- Bargmann C. I., Horvitz H. R. (1991a). Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7, 729-742 [DOI] [PubMed] [Google Scholar]
- Bargmann C. I., Horvitz H. R. (1991b). Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251, 1243-1246 [DOI] [PubMed] [Google Scholar]
- Bishop N. A., Guarente L. (2007). Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447, 545-549 [DOI] [PubMed] [Google Scholar]
- Boll M., Daniel H., Gasnier B. (2004). The SLC36 family: proton-coupled transporters for the absorption of selected amino acids from extracellular and intracellular proteolysis. Pflugers Arch. 447, 776-779 [DOI] [PubMed] [Google Scholar]
- Boudko D. Y. (2010). Molecular ontology of amino acid transport. In Epithelial Transport Physiology (ed. Gerencser G.), pp. 379-472 Springer, NY: Humana-Springer Verlag; [Google Scholar]
- Boudko D. Y. (2012). Molecular basis of essential amino acid transport from studies of insect nutrient amino acid transporters of the SLC6 family (NAT-SLC6). J. Insect Physiol. 58, 433-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudko D. Y., Kohn A. B., Meleshkevitch E. A., Dasher M. K., Seron T. J., Stevens B. R., Harvey W. R. (2005a). Ancestry and progeny of nutrient amino acid transporters. Proc. Natl. Acad. Sci. USA 102, 1360-1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudko D. Y., Stevens B. R., Donly B. C., Harvey W. R. (2005b). Nutrient amino acid and neurotransmitter transporters. In Comprehensive Molecular Insect Science, Vol. 4 (ed. Gilbert L. I., Iatrou K., Gill S. S.), pp. 255-309 Amsterdam: Elsevier; [Google Scholar]
- Bröer S. (2006). The SLC6 orphans are forming a family of amino acid transporters. Neurochem. Int. 48, 559-567 [DOI] [PubMed] [Google Scholar]
- Bröer A., Klingel K., Kowalczuk S., Rasko J. E., Cavanaugh J., Bröer S. (2004). Molecular cloning of mouse amino acid transport system B0, a neutral amino acid transporter related to Hartnup disorder. J. Biol. Chem. 279, 24467-24476 [DOI] [PubMed] [Google Scholar]
- Bröer A., Tietze N., Kowalczuk S., Chubb S., Munzinger M., Bak L. K., Bröer S. (2006). The orphan transporter v7-3 (slc6a15) is a Na+-dependent neutral amino acid transporter (B0AT2). Biochem. J. 393, 421-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo S. M., Makrides V., Virkki L. V., Forster I. C., Verrey F. (2005). Steady-state kinetic characterization of the mouse B0AT1 sodium-dependent neutral amino acid transporter. Pflugers Arch. 451, 338-348 [DOI] [PubMed] [Google Scholar]
- Castagna M., Shayakul C., Trotti D., Sacchi V. F., Harvey W. R., Hediger M. A. (1998). Cloning and characterization of a potassium-coupled amino acid transporter. Proc. Natl. Acad. Sci. USA 95, 5395-5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. H., Reith M. E., Quick M. W. (2004). Synaptic uptake and beyond: the sodium- and chloride-dependent neurotransmitter transporter family SLC6. Pflugers Arch. 447, 519-531 [DOI] [PubMed] [Google Scholar]
- Christensen H. N., Albritton L. M., Kakuda D. K., MacLeod C. L. (1994). Gene-product designations for amino acid transporters. J. Exp. Biol. 196, 51-57 [DOI] [PubMed] [Google Scholar]
- Cutter A. D. (2008). Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 25, 778-786 [DOI] [PubMed] [Google Scholar]
- Daniel H., Kottra G. (2003). The proton oligopeptide cotransporter family SLC15 in physiology and pharmacology. Pflugers Arch. 447, 610-618 [DOI] [PubMed] [Google Scholar]
- de Bono M., Tobin D. M., Davis M. W., Avery L., Bargmann C. I. (2002). Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature 419, 899-903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierking K., Polanowska J., Omi S., Engelmann I., Gut M., Lembo F., Ewbank J. J., Pujol N. (2011). Unusual regulation of a STAT protein by an SLC6 family transporter in C. elegans epidermal innate immunity. Cell Host Microbe 9, 425-435 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783-791 [DOI] [PubMed] [Google Scholar]
- Fielenbach N., Antebi A. (2008). C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22, 2149-2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. M., Hill J. J., Bargmann C. I. (2005). A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 102, 3184-3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen I. A., Boudko D. Y., Shiao S. H., Voronov D. A., Meleshkevitch E. A., Drake L. L., Aguirre S. E., Fox J. M., Attardo G. M., Raikhel A. S. (2011). AaCAT1 of the yellow fever mosquito, Aedes aegypti: a novel histidine-specific amino acid transporter from the SLC7 family. J. Biol. Chem. 286, 10803-10813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari G., Xie Y., Kullyev A., Liang B., Sze J. Y. (2011). Regulation of extrasynaptic 5-HT by serotonin reuptake transporter function in 5-HT-absorbing neurons underscores adaptation behavior in Caenorhabditis elegans. J. Neurosci. 31, 8948-8957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayanthi L. D., Apparsundaram S., Malone M. D., Ward E., Miller D. M., Eppler M., Blakely R. D. (1998). The Caenorhabditis elegans gene T23G5.5 encodes an antidepressant- and cocaine-sensitive dopamine transporter. Mol. Pharmacol. 54, 601-609 [PubMed] [Google Scholar]
- Jespersen T., Grunnet M., Angelo K., Klaerke D. A., Olesen S. P. (2002). Dual-function vector for protein expression in both mammalian cells and Xenopus laevis oocytes. Biotechniques 32, 536-538, 540 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231-237 [DOI] [PubMed] [Google Scholar]
- Kanai Y., Hediger M. A. (2004). The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflugers Arch. 447, 469-479 [DOI] [PubMed] [Google Scholar]
- Kim H., Rogers M. J., Richmond J. E., McIntire S. L. (2004). SNF-6 is an acetylcholine transporter interacting with the dystrophin complex in Caenorhabditis elegans. Nature 430, 891-896 [DOI] [PubMed] [Google Scholar]
- Kowalczuk S., Bröer A., Munzinger M., Tietze N., Klingel K., Bröer S. (2005). Molecular cloning of the mouse IMINO system: an Na+- and Cl-dependent proline transporter. Biochem. J. 386, 417-422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen A. S., Andersen J., Jørgensen T. N., Sørensen L., Eriksen J., Loland C. J., Strømgaard K., Gether U. (2011). SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacol. Rev. 63, 585-640 [DOI] [PubMed] [Google Scholar]
- Mackenzie B., Erickson J. D. (2003). Sodium-coupled neutral amino acid (System N/A) transporters of the SLC38 gene family. Pflugers Arch. 447, 784-795 [DOI] [PubMed] [Google Scholar]
- Meleshkevitch E. A., Assis-Nascimento P., Popova L. B., Miller M. M., Kohn A. B., Phung E. N., Mandal A., Harvey W. R., Boudko D. Y. (2006). Molecular characterization of the first aromatic nutrient transporter from the sodium neurotransmitter symporter family. J. Exp. Biol. 209, 3183-3198 [DOI] [PubMed] [Google Scholar]
- Meleshkevitch E. A., Robinson M., Popova L. B., Miller M. M., Harvey W. R., Boudko D. Y. (2009a). Cloning and functional expression of the first eukaryotic Na+-tryptophan symporter, AgNAT6. J. Exp. Biol. 212, 1559-1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meleshkevitch E. A., Voronov D., Miller M., Fox J., Popova L., Boudko D. (2009b). Novel methionine-selective transporters from the neurotransmitter sodium symporter family. Amino Acids 37, 84 [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. M., Popova L. B., Meleshkevitch E. A., Tran P. V., Boudko D. Y. (2008). The invertebrate B0 system transporter, D. melanogaster NAT1, has unique d-amino acid affinity and mediates gut and brain functions. Insect Biochem. Mol. Biol. 38, 923-931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen G. P., Mathews E. A., Saxena P., Fields S. D., McManus J. R., Moulder G., Barstead R. J., Quick M. W., Rand J. B. (2006). The Caenorhabditis elegans snf-11 gene encodes a sodium-dependent GABA transporter required for clearance of synaptic GABA. Mol. Biol. Cell 17, 3021-3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathoo A. N., Moeller R. A., Westlund B. A., Hart A. C. (2001). Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc. Natl. Acad. Sci. USA 98, 14000-14005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okech B. A., Meleshkevitch E. A., Miller M. M., Popova L. B., Harvey W. R., Boudko D. Y. (2008). Synergy and specificity of two Na+-aromatic amino acid symporters in the model alimentary canal of mosquito larvae. J. Exp. Biol. 211, 1594-1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne S. H., Loomis W. F. (2006). Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryot. Cell 5, 272-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce S. B., Costa M., Wisotzkey R., Devadhar S., Homburger S. A., Buchman A. R., Ferguson K. C., Heller J., Platt D. M., Pasquinelli A. A., et al. (2001). Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 15, 672-686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan R., Sawin E. R., Trent C., Horvitz H. R. (2001). Mutations in the Caenorhabditis elegans serotonin reuptake transporter MOD-5 reveal serotonin-dependent and -independent activities of fluoxetine. J. Neurosci. 21, 5871-5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristic Z., Camargo S. M., Romeo E., Bodoy S., Bertran J., Palacin M., Makrides V., Furrer E. M., Verrey F. (2006). Neutral amino acid transport mediated by ortholog of imino acid transporter SIT1/SLC6A20 in opossum kidney cells. Am. J. Physiol. 290, F880-F887 [DOI] [PubMed] [Google Scholar]
- Romeo E., Dave M. H., Kleta R., Ristic Z., Camargo S. M. R., Makrides V., Wagner C. A., Verrey F. (2005). B(0)AT1 (SLC6A19) and other SLC6 orphan transporters: Hartnup disorder and beyond. FASEB J. 19, A747 [Google Scholar]
- Romeo E., Dave M. H., Bacic D., Ristic Z., Camargo S. M., Loffing J., Wagner C. A., Verrey F. (2006). Luminal kidney and intestine SLC6 amino acid transporters of B0AT-cluster and their tissue distribution in Mus musculus. Am. J. Physiol. 290, F376-F383 [DOI] [PubMed] [Google Scholar]
- Rual J. F., Ceron J., Koreth J., Hao T., Nicot A. S., Hirozane-Kishikawa T., Vandenhaute J., Orkin S. H., Hill D. E., van den Heuvel S., et al. (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14 10B, 2162-2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackwitz W. S., Inoue T., Thomas J. H. (1996). Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17, 719-728 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. (2012). FIJI: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer D., Camargo S. M., Huggel K., Romeo E., Danilczyk U., Kuba K., Chesnov S., Caron M. G., Penninger J. M., Verrey F. (2009). Orphan transporter SLC6A18 is renal neutral amino acid transporter B0AT3. J. Biol. Chem. 284, 19953-19960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath P. H. A., Sokal R. R. (1973). Numerical Taxonomy. San Francisco, CA: Freeman; [Google Scholar]
- Takanaga H., Mackenzie B., Suzuki Y., Hediger M. A. (2005a). Identification of mammalian proline transporter SIT1 (SLC6A20) with characteristics of classical system imino. J. Biol. Chem. 280, 8974-8984 [DOI] [PubMed] [Google Scholar]
- Takanaga H., Mackenzie B., Peng J. B., Hediger M. A. (2005b). Characterization of a branched-chain amino-acid transporter SBAT1 (SLC6A15) that is expressed in human brain. Biochem. Biophys. Res. Commun. 337, 892-900 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28, 2731-2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. H. (2008). Genome evolution in Caenorhabditis. Brief Funct. Genomic Proteomic 7, 211-216 [DOI] [PubMed] [Google Scholar]
- Vanslambrouck J. M., Bröer A., Thavyogarajah T., Holst J., Bailey C. G., Bröer S., Rasko J. E. (2010). Renal imino acid and glycine transport system ontogeny and involvement in developmental iminoglycinuria. Biochem. J. 428, 397-407 [DOI] [PubMed] [Google Scholar]
- Verrey F., Closs E. I., Wagner C. A., Palacin M., Endou H., Kanai Y. (2003). CATs and HATs: the SLC7 family of amino acid transporters. Pflugers Arch. 447, 532-542 [DOI] [PubMed] [Google Scholar]
- Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. (2005). Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215-223 [DOI] [PubMed] [Google Scholar]
- Zaia K. A., Reimer R. J. (2009). Synaptic vesicle protein NTT4/XT1 (SLC6A17) catalyzes Na+-coupled neutral amino acid transport. J. Biol. Chem. 284, 8439-8448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. (1965). Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins (ed. Bryson V., Vogel H. J.), pp. 97-166 New York, NY: Academic Press; [Google Scholar]