Abstract
While studying chronic verruga peruana infections in Peru from 2003, we isolated a novel Bartonella agent, which we propose be named Candidatus Bartonella ancashi. This case reveals the inherent weakness of relying solely on clinical syndromes for diagnosis and underscores the need for a new diagnostic paradigm in developing settings.
Key words: Bartonellosis, emerging infection, Candidatus Bartonella ancashi, Peru, pathogen discovery, bacteria, verruga peruana, vector-borne infections, Bartonella
Bartonellosis is a disease caused by infection with species from the Bartonella genus. In South America, infection with B. bacilliformis, an α-2 proteobacterium, may cause a life- threatening bacterial infection (1,2). If untreated, the acute form of the illness, sometimes referred to as Oroya fever, has a high mortality rate because the bacteria invade erythrocytes, resulting in subsequent severe anemia and secondary infections. A chronic phase, termed verruga peruana, is characterized by vasculoproliferative skin lesions; some reseachers have also described an asymptomatic bacteremic phase, which may contribute to the longevity of the reservoir status of infected persons (3).
In 2007, a novel species of Bartonella (B. rochalimae) was isolated from a single traveler who had an acute febrile anemia after traveling to Peru (4). We report the identification of another novel agent of Bartonella isolated from a patient with chronic bartonellosis (collected in 2003, fully characterized in 2011–2012). We suggest that the isolate be named Candidatus Bartonella ancashi in honor of the highland region of Peru.
The Study
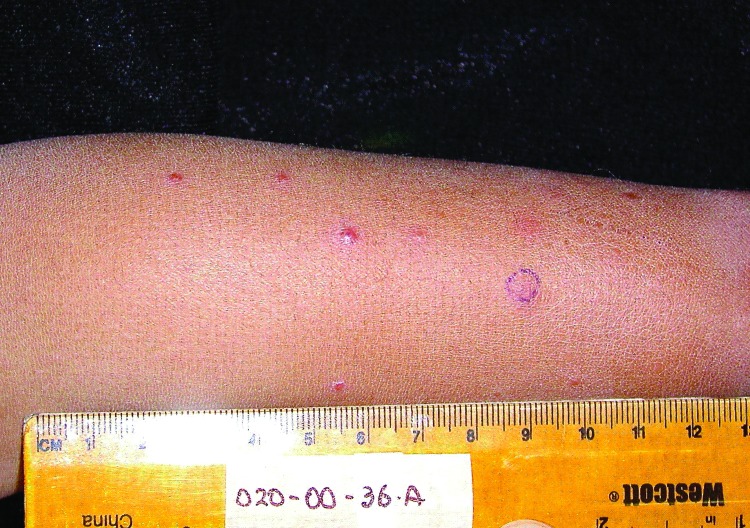
The patient was a 3-year-old boy with no known underlying medical history who was identified in 2003 as having clinical verruga peruana by the classic appearance of the eruptive nodular rash (Figure 1). He and his family lived in a rural setting near the town of Caraz, Ancash region, Peru. He lived in close proximity to numerous pets and farm animals and had experienced insect bites around the time the eruptive rash developed. His rash had been present for ≈30 days, and he had no fevers, chills, or arthralgias. Baseline laboratory studies included complete blood counts and bacterial culture for Bartonella species, using methods previously described (5,6). Briefly, the media was biphasic, consisting of Bacto agar with Proteose Peptone No. 3 (Becton, Dickinson and Co., Sparks, MD, USA), dextrose, sodium chloride and 10% defibrinated sheep’s blood, and RPMI supplemented with 10% inactivated fetal bovine serum.
Figure 1.
Clinical presentation of verruga peruana in 3-year-old boy, Peru, 2003.
The physical examination revealed that the child had 56 lesions, distributed mainly on the extremities. Laboratory values were the following: hemoglobin level 12.8 mg/dL (reference range 11–13 g/dL), hematocrit 39% (reference range 31%–43%), and platelet count of 300,000/µL (reference range 15,000–400,000); his leukocyte count was elevated at 28,000/µL (reference range 4,100–10,900) with 51% eosinophils, for which he was referred for further evaluation.
The peripheral blood smear was negative for intracellular organisms, but blood culture was positive for a Bartonella species. This species was further studied and found to be novel on the basis of genetic sequencing of the isolate obtained from the standard blood culture (7). His condition was treated with azithromycin, and the rash fully resolved (8,9). To confirm the identity of the isolate from this patient, in 2011–2012 we conducted molecular analyses (including PCR, nested PCR [nPCR], and sequencing) on the whole blood culture isolate, Bartonella species no. 20.00 (10).The isolate was characterized by sequencing 3 gene fragments.
The following conditions were used for PCR amplification. For rrs, initial denaturation was at 95°C for 1 min, followed by 45 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and elongation at 68°C for 90 s for PCR and 70 s for nPCR, and then by a final extension step at 72°C for 7 min. For gltA—95°C, denaturation was for 1 min, followed by 45 cycles of denaturation at 95°C for 30 s, annealing at 51°C for 30 s, and elongation at 68°C for 60 s, and then by the final extension step at 72°C for 7 min. For rpoB, primers were selected from the conserved regions of RNA polymerase β-subunit encoding gene (rpoB) after alignment of the rpoB from B. quintana and B. vinsonni for PCR and nPCR. PCR and nPCR were carried out by using conditions identical to those described for gltA (Table).
Table. Primers used for PCR, nested PCR, and sequencing of novel Bartonella isolate from Peru, 2011–2012*.
| Gene | Primer name | Primer sequence, 5’ → 3’ | Use | Fragment length |
|---|---|---|---|---|
| rrs | 16SU17F | AGAGTTTGATCCTGGCTCAG | PCR, nPCR, sequencing | 1,424 bp |
| 16SU1592R | AGGAGGTRATCCAGCCGCA | PCR, nPCR, sequencing | ||
| 16SU 833R | CTACCAGGGTATCTAATCCTGTT | nPCR, sequencing | ||
|
|
16S E. coli-518F |
CAGCAGCCGCGGTAATAC |
nPCR, sequencing |
|
| gltA† | BHCS 781p (F) | GGGACCAGCTCATGGTGG | PCR, sequencing | 338 bp |
|
|
BHCS 1137n (R) |
AATGCAAAAAGAACAGTAAACA |
PCR, sequencing |
|
| rpoB | BrpoB1435F | CGCATTGGTTTRCTTCGTATG | PCR | 589 bp |
| Brpo2327R | GTAGACTGATTAGAACGCTG | PCR, nPCR, sequencing | ||
| Brpo1696F | CCTACGCATTATGGTCGTATTTG | nPCR, sequencing |
*nPCR, nested PCR. †gltA primers were previously described by Eremeeva et al. (4).
PCR products were purified by using the QIAquick PCR Purification Kit (QIAGEN, Valencia, CA, USA) before sequencing. PCR products were sequenced in both directions by using the BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, CA, USA) and run on an automated 3130xl Gene Analyzer (Life Technologies). Sequences were characterized by BLAST analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequence analysis was performed by using BioEdit version 7.1.3 (Ibis Biosciences, Carlsbad, CA, USA). The multiple sequence alignments were performed with the ClustalW multiple alignment application also in BioEdit version 7.1.3. Phylogenetic trees were created with MEGA5 software using the neighbor-joining tree method with 1,000 bootstrap replicates (11).
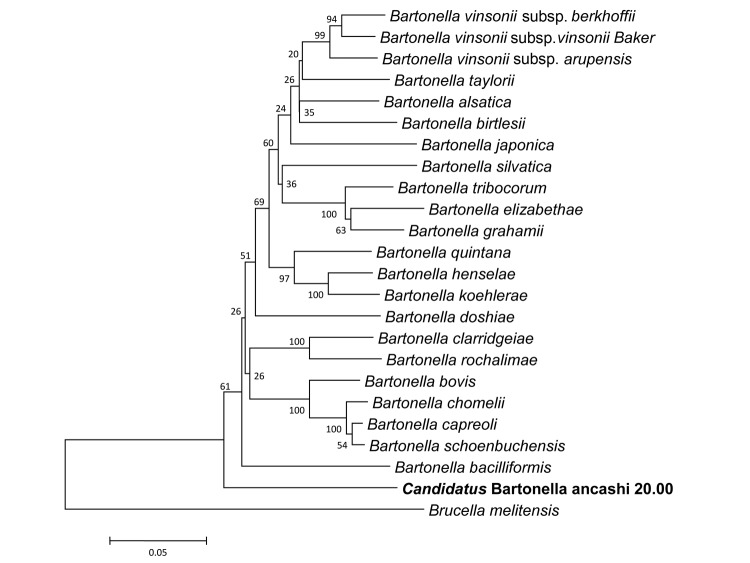
Subsequent comparison with known Bartonella species in GenBank found no identical sequences. The 1,351-bp sequence of the rrs fragment was found to be 99.0% similar to the rrs fragment of B. bacilliformis. The 312- and 589-bp fragments of gltA and rpoB, respectively, were found to be most similar to their counterparts of B. bovis at 89.4% (gltA) and 85.9% (rpoB), respectively (Technical Appendix). The sequence similarity ranges for the rrs, gltA, and rpoB for recognized Bartonella species are 97.7%–99.8%, 83.4%–96.1%, and 85.9%– 96%, respectively (12). In addition, rpoB and gltA are believed to have the best discriminating power for Bartonella species (13). La Scola et al. proposed that a new species be designated if the sequence similarities are <96% and <95.4% for a 327-bp fragment of gltA and a 825-bp fragment of rpoB, respectively (13). The sequence similarities for Candidatus Bartonella ancashi 20.00 to other known Bartonella species fall well below these suggested values, providing more evidence that this agent is unique. Phylogenetic analysis of the rrs, gltA, and rpoB, gene fragments provide additional evidence for identification of a unique Bartonella agent. The concatenated sequence of gltA and rpoB gene fragments placed the new Bartonella isolate in an exclusive clade that is most closely aligned with B. bacilliformis (Figure 2). The rrs fragment also placed the isolate in a clade with B. bacilliformis. The results from the phylogenetic analysis combined with the sequence similarity data provide evidence that this isolate, Candidatus Bartonella ancashi 20.00, is unique (12,13).
Figure 2.
Phylogeny for concatenated sequences of novel Bartonella isolate (boldface), including a 312-character fragment of gltA and a 589-character fragment of rpoB. The neighbor-joining tree method (1,000 bootstrap replicates) was employed using MEGA5 software (11), and the distances were calculated by using the Jukes-Cantor method, in which units are calculated as the number of base pair substitutions per site (10). Brucella melitensis was used as the outgroup.
Conclusions
This case underscores the inherent weakness of relying solely on clinical syndromes for diagnosis. The variety of bacteria that have been implicated in the clinical spectrum of bartonellosis is increasing as molecular methods are applied to isolates that previously were identified by using clinical criteria or biochemical testing. The novel bacterium may have similar epidemiologic, clinical, and microbiologic properties to B. bacilliformis, but without relating these data to a full molecular characterization, that assumption is precarious.
To address this public health deficiency, a new diagnostic paradigm should be deployed to developing settings such as Peru. This is particularly true for areas with high biodiversity, a point identified by other investigators who have termed these regions “hot zones” for emerging infectious diseases (14). Unfortunately, tools such as high-throughput sequencing are rare in developing settings where risk for novel pathogen emergence is highest. Investment in advanced molecular diagnostic platforms in the developing setting will be an essential tool for expanding pathogen discovery; of course, this should be accompanied by parallel investments in training in molecular laboratory techniques and analysis for resident scientists. Opportunities for grants and stable faculty positions must also be supported to encourage qualified scientists to remain in the developing setting.
Finally, evidence indicates that humans contract bartonellosis only once and that lifelong immunity results from that primary infection (15). Because of this circumstance, and the inability to identify an animal reservoir of B. bacilliformis, Peruvian scientists and others have identified bartonellosis as a disease that may be eradicated in the Andean region through development of a vaccine against B. bacilliformis, targeted treatment of patients, and vector control programs (15). This possibility may be less feasible if multiple species of Bartonella cause bartonellosis. Further molecular and immunologic studies should be undertaken if this disease is to be targeted for eradication.
Technical Appendix. Comparison of rpoB and gltA sequence similarities for Bartonella species. The 312- and 589-bp fragments of gltA and rpoB, respectively, were found to be most similar to their counterparts of Bartonella bovis at 89.4% (gltA) and 85.9% (rpoB).
Acknowledgments
We thank Jesus Gonzalez for his efforts in conducting the clinical trial in Peru.
The 3 genes have been submitted to GenBank, with the following accession numbers: rrs, KC178617, gltA, KC178618, rpoB. KC178619.
The original clinical trial was approved by the Institutional Review Boards of the Uniformed Services University of the Health Sciences, the Naval Medical Research Center, and the Universidad Peruana Cayetano Heredia in 2002, and compared the standard of care medication, rifampin, with azithromycin. The clinical trial was originally funded by Pfizer and the subsequent pathogen characterization was funded by the Department of Defense Global Emerging Infection System work unit no. 0000188M.0931.001.A0074. The funding agencies had no role in writing, editing, or approval of this manuscript.
This work was prepared as part of official duties of authors (D.L.B., T.M., B.L.S., A.L.R., L.L.) working
Biographies
in the United States Government. Title 17 U.S.C. §105 provides that “Copyright protection under this title is not available for any work of the United States Government.” The views expressed are those of the authors and do not represent the official position of the US Army, Navy or Department of Defense
Dr Blazes is an infectious diseases physician who currently directs the Military's Tropical Medicine program in Bethesda, Maryland, USA. He lived in Peru from 2004–2008, and his main interests are outbreak investigation, pathogen discovery, and novel educational methods.
Footnotes
Suggested citation for this article: Blazes DL, Mullins K, Smoak BL, Jiang J, Canal E, Solorzano N, et al. Novel Bartonella agent as cause of verruga peruana. Emerg Infect Dis [Internet]. 2013 Jul [date cited]. http://dx.doi.org/10.3201/eid1907.121718
References
- 1.Schultz MG. A history of bartonellosis (Carrion’s disease). Am J Trop Med Hyg. 1968;17:503–15 . [DOI] [PubMed] [Google Scholar]
- 2.Maguina C, Garcia PJ, Gotuzzo E, Cordero L, Spach DH. Bartonellosis (Carrion’s disease) in the modern era. Clin Infect Dis. 2001;33:772–9. 10.1086/322614 [DOI] [PubMed] [Google Scholar]
- 3.Chamberlin J, Laughlin LW, Romero S, Solórzano N, Gordon S, Andre RG, et al. Epidemiology of endemic Bartonella bacilliformis: a prospective cohort study in a Peruvian mountain valley community. J Infect Dis. 2002;186:983–90 and. 10.1086/344054 [DOI] [PubMed] [Google Scholar]
- 4.Eremeeva ME, Gerns HL, Lydy SL, Goo JS, Ryan ET, Mathew SS, et al. Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N Engl J Med. 2007;356:2381–7 and. 10.1056/NEJMoa065987 [DOI] [PubMed] [Google Scholar]
- 5.Mallqui V, Speelmon EC, Verástegui M, Maguiña-Vargas C, Pinell-Sallis P, et al. Sonicated diagnostic immunoblot for bartonellosis. Clin Diagn Lab Immunol. 2000;7:1–5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker TS, Winkler HH. Bartonella bacilliformis: colonial types and erythrocyte adherence. Infect Immun. 1981;31:480–6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maass M, Schreiber M, Knobloch J. Detection of Bartonella bacilliformis in cultures, blood, and formalin preserved skin biopsies by use of the polymerase chain reaction. Trop Med Parasitol. 1992;43:191–4 . [PubMed] [Google Scholar]
- 8.Rolain JM, Brouqui P, Koehler JE, Maguina C, Dolan MJ, Raoult D. Recommendations for treatment of human infections caused by Bartonella species. Antimicrob Agents Chemother. 2004;48:1921–33. 10.1128/AAC.48.6.1921-1933.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arroyo A. Treatment outlines for non-complicated Carrion’s disease, in Caraz, Peru. Anales de la Facultad de Medecina. 2008;69:7–11. [Google Scholar]
- 10.Maass M, Schreiber M, Knobloch J. Detection of Bartonella bacilliformis in cultures, blood, and formalin preserved skin biopsies by use of the polymerase chain reaction. Trop Med Parasitol. 1992;43:191–4 . [PubMed] [Google Scholar]
- 11.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeaiter Z, Liang Z, Raoult D. Genetic classification and differentiation of Bartonella species based on comparison of partial ftsZ gene sequence. J Clin Microbiol. 2002;40:3641–7. 10.1128/JCM.40.10.3641-3647.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 2003;11:318–21. 10.1016/S0966-842X(03)00143-4 [DOI] [PubMed] [Google Scholar]
- 14.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–3 . 10.1038/nature06536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez Clemente N, Ugarte-Gil CA, Solórzano N, Maguiña C, Pachas P, et al. Bartonella bacilliformis: a systematic review of the literature to guide the research agenda for elimination. PLoS Negl Trop Dis. 2012;6:e1819. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Technical Appendix. Comparison of rpoB and gltA sequence similarities for Bartonella species. The 312- and 589-bp fragments of gltA and rpoB, respectively, were found to be most similar to their counterparts of Bartonella bovis at 89.4% (gltA) and 85.9% (rpoB).