Summary
Background
Findings from family and twin studies suggest that genetic contributions to psychiatric disorders do not in all cases map to present diagnostic categories. We aimed to identify specific variants underlying genetic effects shared between the five disorders in the Psychiatric Genomics Consortium: autism spectrum disorder, attention deficit-hyperactivity disorder, bipolar disorder, major depressive disorder, and schizophrenia.
Methods
We analysed genome-wide single-nucleotide polymorphism (SNP) data for the five disorders in 33 332 cases and 27 888 controls of European ancestory. To characterise allelic effects on each disorder, we applied a multinomial logistic regression procedure with model selection to identify the best-fitting model of relations between genotype and phenotype. We examined cross-disorder effects of genome-wide significant loci previously identified for bipolar disorder and schizophrenia, and used polygenic risk-score analysis to examine such effects from a broader set of common variants. We undertook pathway analyses to establish the biological associations underlying genetic overlap for the five disorders. We used enrichment analysis of expression quantitative trait loci (eQTL) data to assess whether SNPs with cross-disorder association were enriched for regulatory SNPs in post-mortem brain-tissue samples.
Findings
SNPs at four loci surpassed the cutoff for genome-wide significance (p<5×10−8) in the primary analysis: regions on chromosomes 3p21 and 10q24, and SNPs within two L-type voltage-gated calcium channel subunits, CACNA1C and CACNB2. Model selection analysis supported effects of these loci for several disorders. Loci previously associated with bipolar disorder or schizophrenia had variable diagnostic specificity. Polygenic risk scores showed cross-disorder associations, notably between adult-onset disorders. Pathway analysis supported a role for calcium channel signalling genes for all five disorders. Finally, SNPs with evidence of cross-disorder association were enriched for brain eQTL markers.
Interpretation
Our findings show that specific SNPs are associated with a range of psychiatric disorders of childhood onset or adult onset. In particular, variation in calcium-channel activity genes seems to have pleiotropic effects on psychopathology. These results provide evidence relevant to the goal of moving beyond descriptive syndromes in psychiatry, and towards a nosology informed by disease cause.
Funding
National Institute of Mental Health.
Introduction
Psychiatric nosology arose in central Europe towards the end of the 19th century, in particular with Kraepelin’s foundational distinction between dementia praecox (schizophrenia) and manic depressive insanity.1 The distinction between bipolar illness and unipolar (major) depression was first proposed in the late 1950s and became increasingly widely accepted. The major syndromes—especially schizophrenia, bipolar disorder, and major depression—were differentiated on the basis of their symptom patterns and course of illness. At the same time, clinical features such as psychosis, mood dysregulation, and cognitive impairments were known to transcend diagnostic categories. Doubt remains about the boundaries between the syndromes and the degree to which they signify entirely distinct entities, disorders that have overlapping foundations, or different variants of one underlying disease. Such debates have intensified with syndromes described subsequently, including autism spectrum disorders and attention deficit-hyperactivity disorder.
The pathogenic mechanisms of psychiatric disorders are largely unknown, so diagnostic boundaries are difficult to define. Genetic risk factors are important in the causation of all major psychiatric disorders,2 and genetic strategies are widely used to assess potential overlaps. The imminent revision of psychiatric classifications in the Diagnostic and Statistical Manual of Mental Disorders (DSM) and the International Classification of Diseases (ICD) has reinvigorated debate about the validity of diagnostic boundaries. With increasing availability of large genome-wide genotype data for several psychiatric disorders, shared cause can now be examined at a molecular level.
We formed the Psychiatric Genomics Consortium (PGC) in 2007, to undertake meta-analyses of genomewide association studies (GWAS) for psychiatric disorders and, so far, the consortium has incorporated GWAS data from more than 19 countries for schizophrenia, bipolar disorder, major depressive disorder, attention deficit-hyperactivity disorder, and autism spectrum disorders. Previous research has suggested varying degrees of overlap in familial and genetic liability for pairs of these disorders. For example, some findings3,4 from family and twin studies support diagnostic boundaries between schizophrenia and bipolar disorder and bipolar disorder and major depressive disorder, but also suggest correlations in familial and genetic liabilities.3,5 Several molecular variants confer risk of both schizophrenia and bipolar disorder.6–8 Autism was once known as childhood schizophrenia and the two disorders were not clearly differentiated until the 1970s. Findings from the past few years have emphasised phenotypic and genetic overlap between autism spec trum disorders and schizophrenia,9,10 including identification of copy number variants conferring risk of both.11 Findings from family, twin, and molecular studies12–15 suggest some genetic overlap between autism spectrum disorder and attention deficit-hyperactivity disorder.
In this first report from the PGC Cross-Disorder Group, we analyse data on genome-wide single-nucleotide polymorphism (SNP) for the five PGC disorders to answer two questions. First, what information emerges when all five disorders are examined in one GWAS? When risk is correlated across disorders, pooled analyses will be better powered than individual-disorder analyses to detect risk loci. Second, what are the cross-disorder effects of variants already identified as being associated with a specific psychiatric disorder in previous PGC analyses? We aimed to examine the genetic relation between the five psychiatric disorders with the expectation that findings will ultimately inform psychiatric nosology, identify potential neurobiological mechanisms predisposing to specific clinical presentations, and generate new models for prevention and treatment.
Methods
Samples and genotypes
The sample for these analyses consisted of cases, controls, and family-based samples assembled for previous genome-wide PGC mega-analyses of individual-level data.6,7,16,17 Cases and controls were not related. For the family-based samples, we matched alleles transmitted to affected offspring (trio cases) with untransmitted alleles (pseudo-controls). We estimated the identity-by-descent relation for all pairs of individuals to identify any duplicate individuals in the component datasets. When duplicates were detected, one member of each set was retained. We then randomly allocated these individuals, with a random number generator, to a disorder case-control dataset. Sample sizes differ from previous reports because of this allocation of overlapping individuals. All patients were of European ancestory and met criteria from the DSM third edition revised or fourth edition for the primary disorder of interest.
To ensure comparability between samples, raw genotype and phenotype data for each study were uploaded to a central server and processed through the same quality control, imputation, and analysis process (appendix).6,7 We analysed imputed SNP dosages from 1 250 922 autosomal SNPs.
Statistical analysis
In the primary analysis, we combined effects of each disease analysis by a meta-analytic approach that applied a weighted Z-score,18 in which weights equalled the inverse of the regression coefficient’s standard error. This strategy assumed a fixed-effects model, with weights indicating the sample size of the disease-specific studies. In a second analytical approach, we did a five-degree-of-freedom test by summing the χ2 values for each individual disease meta-analysis. Unlike our primary analysis, this model did not assume that all diseases had the same direction of effect and could detect allelic effects that increase risk for some diseases and decrease risk for others. The appendix describes statistical methods and results, including the handling of trios and population stratification. We also examined loci that previously achieved genome-wide significance in PGC meta-analyses of schizophrenia and bipolar disorder.6,7
To characterise the specificity of the allelic effects for our main findings, we examined the association evidence in three ways: we generated forest plots of the disorder beta coefficients with 95% CIs; we calculated a heterogeneity p value for the disorder-specific effects contributing to the overall statistics for meta-analytic association; and we undertook a multinomial logistic regression procedure with model selection19 for each main SNP for all five disorders to assess the pattern of phenotypic effects (appendix pp 8–11). To compare the fit of various models of genotype–phenotype associations, we applied established goodness-of-fit metrics (the Bayesian information criteria and the Akaike information criteria). We report the best-fitting model by Bayesian criteria and show results of both metrics for a range of models (appendix pp 38–45, 51–61).
To examine shared polygenic risk at an aggregate level between pairs of diagnoses, we used risk-score profiling as previously described.8 For each pair, we selected one disorder as a discovery dataset and the other as a target dataset and calculated the proportion of variance in the target set explained by risk scores from the discovery set with a range of statistical cutoffs for SNP inclusion in the score (appendix p 13). To assess the role of specific biological systems in the pathogenesis of the five disorders, we did pathway and eQTL analyses. Pathway analysis was by interval-based enrichment analysis (INRICH) for the full dataset consisting of linkage disequilibrium segments containing signals with association p<10−3 in the primary meta-analysis. INRICH accounts for potential genomic confounding factors, such as variable gene and pathway sizes, SNP density, linkage disequilibrium, and physical clustering of biologically related genes (appendix pp 14–16). We did eQTL enrichment analysis20 to assess whether SNPs associated with five psychiatric disorders were enriched for regulatory SNPs in post-mortem brain tissue samples compared with those with no association.21,22 To assess the specificity of this finding, we also examined eQTL datasets from three non-brain-tissue types: liver,23 skin,24 and lymphoblastoid cell lines25 (appendix pp 17–21).
Role of the funding source
The sponsor of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data and had final responsibility for the decision to submit for publication.
Results
The final dataset consisted of 33 332 cases and 27 888 controls (including pseudocontrols formed from non-transmitted alleles) distributed among the five disorder groups: autism spectrum disorders (4788 trio cases, 4788 trio pseudocontrols, 161 cases, 526 controls), attention deficit-hyperactivity disorder (1947 trio cases, 1947 trio pseudocontrols, 840 cases, 688 controls), bipolar disorder (6990 cases, 4820 controls), major depressive disorder (9227 cases, 7383 controls), and schizophrenia (9379 cases, 7736 controls). The results of the primary fixed-effects meta-analysis for all five disorders, incorporating seven multidimensional scaling components as covariates, yielded a genomic control value of λ=1·167. The λ1000 (λ rescaled to a sample of 1000 cases and 1000 controls) was 1·005 (appendix p 22). In view of evidence for substantial polygenic contributions to common psychiatric disorders, this estimate probably shows the aggregate small effect of a large number of risk variants, although a moderate degree of population stratification or technical bias cannot be excluded.
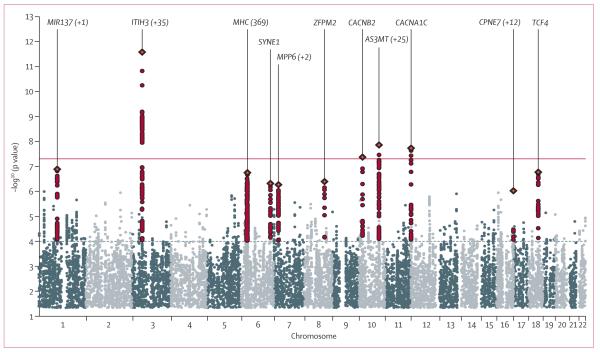
Figure 1 shows the Manhattan plot of the primary results. Four independent regions contained SNPs with p<5×10−8 (table 1; appendix pp 34–35, 25–33). The strongest association signal was on chromosome 3 at an intronic SNP within ITIH3 (table 1). This SNP is in linkage disequilibrium with SNPs encompassing several genes across a 1 Mb region (appendix p 22). The second strongest signal was in an intron of AS3MT on chromosome 10q24 (table 1). Linkage disequilibrium around this associated region encompasses several genes including CNNM2. We also recorded genome-wide significant association within CACNA1C, and finally detected significant association to a second locus on chromosome 10 in an intron of CACNB2 (table 1). We undertook conditional analyses to assess evidence for multirisk loci in a region. In these analyses, we included the most strongly associated or peak SNP plus any SNPs within 1·5 Mb of the peak SNP with association p values less than 10−4 and r2 less than 0·2 with the peak SNP based on HapMap 3 CEU data. For the chromosome 3p21 region, and regions CACNA1C and CACNB2, no additional independent association signals were detected. For the chromosome 10q24 region, an additional SNP (rs11191732), about 600 kb from the peak SNP, showed association after conditioning on the peak SNP (rs11191454) with a p value of 6·60×10−6 before conditioning and 3·88×10−5 after conditioning. Several loci previously implicated in PGC analyses of schizophrenia and bipolar disorder6,7 showed evidence for association in the cross-disorder analysis, despite not exceeding the cutoff for genome-wide significance (appendix pp 23–24). These loci include one near MIR137, TCF4, the MHC region on chromosome 6, and SYNE1 (appendix pp 23–24). The five-degree-of-freedom χ2 test did not identify any additional genome-wide significant SNPs with effects in the opposite direction among the five disorders (appendix pp 36–37).
Figure 1. Manhattan plot of primary fixed-effects meta-analysis.
Horizontal line shows threshold for genome-wide significance (p<5×10−8).
Table 1.
Five disorder meta-analysis results for regions with p<5×10−8
| Chromosome | Base-pair position* |
Nearest gene | Alleles | Frequency† | Imputation quality score (INFO) |
p value | OR (95% CI)‡ | Heterogeneity p value |
Best-fit model (BIC)§ |
|
|---|---|---|---|---|---|---|---|---|---|---|
| rs2535629 | 3 | 52808259 | ITIH3 (+ many) | G/A | 0·651 | 0·942 | 2·54×10−12 | 1·10 (1·07–1·12) | 0·27 | Five disorder¶ |
| rs11191454 | 10 | 104649994 | AS3MT (+ many) | A/G | 0·910 | 1·01 | 1·39×10−8 | 1·13 (1·08–1·18) | 0·32 | Five disorder¶ |
| rs1024582 | 12 | 2272507 | CACNA1C | A/G | 0·337 | 0·98 | 1·87×10−8 | 1·07 (1·05–1·10) | 0·0057 | BPD, schizophrenia |
| rs2799573 | 10 | 18641934 | CACNB2 | T/C | 0·715 | 0·825 | 4·29×10−8 | 1·08 (1·05–1·12) | 0·57 | Five disorder¶ |
Most strongly associated single-nucleotide polymorphisms (SNP) in associated region after clumping—ie, grouping SNPs within 250 kb of the index SNP that have r2>0·2 with the index SNP as implemented in PLINK. OR=odds ratio. BIC=Bayesian information criteria. BPD=bipolar disorder.
Detected with University of California Santa Cruz Genome Browser (version hg18).
Risk allele frequency in controls.
Estimated OR from multinomial logistic regression used in the modelling analysis.
Best-fit multinomial logistic model by BIC criteria; appendix pp 38–45 provide a comparison of BIC and Akaike information criteria across models.
Best-fit model supports an effect on all five disorders.
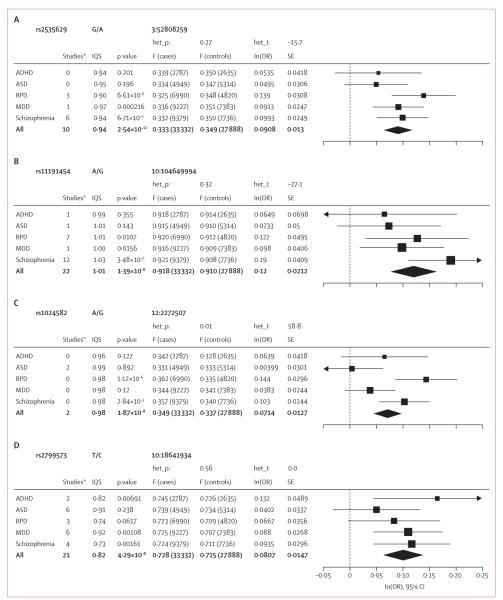
Forest plots for genome-wide significant SNPs showed the same direction of effect for most or all of the five disorders (figure 2). For three of the four associated regions, the meta-analysis heterogeneity p value was not significant and a model in which all five disorders contributed provided the best fit (table 1). The exception was rs1024582, for which the heterogeneity p value was significant and the best-fit model supported an effect limited to bipolar disorder and schizophrenia (table 1). Appendix pp 38–45 shows the profile of the Bayesian and Akaike information criteria measures for each SNP for a range of models. We examined the association between the five disorders of four SNPs showing genome-wide significant association with bipolar disorder (table 2) and ten associated with schizophrenia (table 2).6,7 Appendix pp 46–61 show forest plots and model-fitting results by disorder. The best-fitting model for seven of the 14 risk SNPs suggested disorder-specific effects for either bipolar disorder (three SNPs) or schizophrenia (four SNPs), whereas the rest were consistent with more pleiotropic models (table 2).
Figure 2. Association results and forest plots showing effect size for genome-wide significant loci by disorder.
Data in parentheses are numbers of cases or controls. Het_p=p value for the heterogeneity test. Het_I=heterogeneity test statistic. IQS=imputation quality score (INFO). ln(OR)=log of the odds ratio (OR). F=frequency. SE=standard error of the log OR. ADHD=attention deficit-hyperactivity disorder. ASD=autism spectrum disorders. BPD=bipolar disorder. MDD=major depressive disorder. *Number of studies in which the variant was directly genotyped.
Table 2.
Modelling analysis results for single-nucleotide polymorphisms (SNP) showing genome-wide significant association in previous genome-wide association studies from the Psychiatric Genome-Wide Association Study Consortium
| Chromosome | Base-pair position* | Nearest gene | Alleles | Frequency† | p value | Best-fit model (BIC)‡ | |
|---|---|---|---|---|---|---|---|
| Bipolar disorder 7 | |||||||
| rs937l601 | 6 | 152832266 | SYNE1 | T/G | 0·346 | 4·27×10−9 | BPD |
| rs10994397 | 10 | 61949130 | ANK3 | T/C | 0·065 | 7·08×10−9 | BPD |
| rs4765914§,¶ | 12 | 2290157 | CACNA1C | T/C | 0·204 | 1·52×10−8 | BPD, MDD, schizophrenia |
| rs12576775 | 11 | 78754841 | ODZ4 | G/A | 0·175 | 4·40×10−8 | BPD |
| Schizophrenia 6 | |||||||
| rs2021722 | 6 | 30282110 | MHC | C/T | 0·789 | 2·18×10−12 | Schizophrenia |
| rs1625579 | 1 | 98275522 | MIR137 | T/G | 0·801 | 1·59×10−11 | ASD, schizophrenia |
| rs12966547§ | 18 | 50903015 | CCDC68 | G/A | 0·588 | 2·60×10−10 | BPD, MDD, schizophrenia |
| rs7914558§ | 10 | 104765898 | CNNM2 | G/A | 0·587 | 1·82×10−9 | MDD, schizophrenia |
| rs11191580§ | 10 | 104896201 | NT5C2 | T/C | 0·911 | 1·11×10−8 | Five disorder |
| rs7004633 | 8 | 89829427 | MMP16 | G/A | 0·184 | 2·75×10−8 | Schizophrenia |
| rs10503253 | 8 | 4168252 | CSMD1 | A/C | 0·193 | 4·14×10−8 | Schizophrenia |
| rs17662626§ | 2 | 193692866 | PCGEM1 | A/G | 0·915 | 4·65×10−8 | ASD, schizophrenia |
| rs548181 | 11 | 124966919 | STT3A | G/A | 0·884 | 8·87×10−7 | Schizophrenia |
| rs17512836 | 18 | 51345959 | TCF4 | C/T | 0·027 | 1·05×10−6 | ASD, schizophrenia |
Peak SNPs in associated region after clumping. BIC=Bayesian information criteria. BPD=bipolar disorder. MDD=major depressive disorder. ASD=autism spectrum disorders.
Detected with University of California Santa Cruz Genome Browser (version hg18).
Risk allele frequency in controls.
Best-fit multinomial logistic model by BIC (appendix pp 46-50 [bipolar disorder] and 51-61 [schizophrenia] provide a comparison of BIC and Akaike information criteria across models).
The BIC for the best and second best models do not differ significantly (ie, greater than 2 as previously suggested26).
rs4765914 is a proxy SNP for rs4765913 based on linkage disequilibrium (481 base pairs away, r2=0·874).
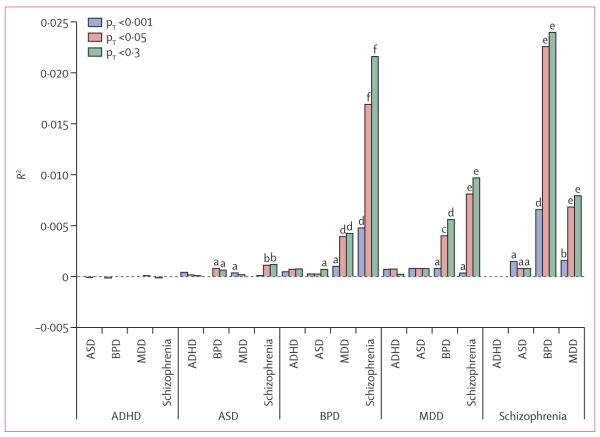
Figure 3 shows the proportion of variance explained in target sets (Nagelkerke’s pseudo R2 from logistic regression) by risk scores from the discovery sets. We noted highly significant overlap of polygenic risk between all three adult disorders (bipolar disorder, major depressive disorder, and schizophrenia), with the strongest effects noted for bipolar disorder and schizophrenia (figure 3). Overlap was reduced but still significant between aggregate genetic risk for autism spectrum disorder with schizophrenia (minimum p<10−4) and bipolar disorder (p<0·05; figure 3). No consistently significant polygenic overlap was detected between major depressive disorder and autism spectrum disorder or between attention deficit-hyperactivity disorder and any other disorder. Appendix p 63 shows additional polygene analyses combining subsets of disorders into discovery sets.
Figure 3. Pair-wise cross-disorder polygene analysis.
We derived polygene risk scores for each disorder (discovery sets) and applied them sequentially to the remaining disorders (target sets). Results are grouped by each discovery set. Each pair is shown on the x-axis and the proportion of variance explained for the target disorder (estimated via Nagelkerke’s pseudo R2) on the y-axis. For purposes of illustration, three pT cutoffs are shown, but appendix p 62 shows the proportion of variance results for a broader range of cutoffs. pT=training-set p value (used to select training set SNPs). Significance of results: a=p<0·05; b=p<10−4; c=p<10−8; d=p<1012; e=p<10−16; f=p<10−50. ADHD=attention deficit-hyperactivity disorder. ASD=autism spectrum disorders. BPD=bipolar disorder. MDD=major depressive disorder.
After correction for multiple testing (appendix pp 14–16), we noted significant enrichment for a set of calcium channel activity genes associated with catalysis of facilitated diffusion of calcium ions through a trans membrane calcium channel (appendix p 64). With a cutoff of p<10−3, 20 of 67 gene regions in this set were associated in the five-disorder meta-analysis, including voltage-gated calcium-channel subunits CACNA1C, CACNA1D, CACNA1E, CACNA1S, CACNA2D2, CACNA2D4, and CACNB2 (appendix pp 65–66). Because calcium-channel genes (mainly CACNA1C) have previously been associated with bipolar disorder, we repeated the pathway analysis with exclusion of the datasets for bipolar disorder and confirmed that enrichment was not dependent on cases with this disorder (data not shown). Appendix pp 69–70 depict functional associations between these calcium-channel activity genes on the basis of various lines of evidence.
Table 3 summarises results of the eQTL enrichment analysis for SNPs selected at varying p-value cutoffs from the primary cross-disorder meta-analysis. We noted a significantly greater proportion of brain eQTL markers in cross-disorder-selected SNPs than expected in view of their distribution of frequencies for minor alleles. We consistently noted this enrichment for two of the three studies of post-mortem brain eQTL for various p-value cutoffs (table 3) and when analyses included all SNPs or were restricted to retain only one SNP tagging (r2>0·8) an associated region. No consistent enrichment was noted in the three non-brain-tissue datasets (table 3).
Table 3.
Expression quantitative trait loci enrichment analysis for single-nucleotide polymorphisms (SNPs) from primary meta-analysis by p-value cutoffs
| Cis-regulatory eQTL datasets (enrichment p value) |
||||||
|---|---|---|---|---|---|---|
| Prefrontal cortex | Cortex | Cortex | Liver (Schadt | Skin (Ding | LPL (Stranger | |
| (Colantuoni et al27) | (Myers et al21) | (Webster et al22) | et al23) | et al24) | et al25) | |
| p<0·01 | 0·41 | 0·020 | 0·0028 | 0·37 | 0·22 | 0·11 |
| p<0·1 | 0·11 | 0·020 | 0·000027 | 0·39 | 0·15 | 0·28 |
| p<0·2 | 0·19 | 0·0030 | 0·000061 | 0·069 | 0·21 | 0·12 |
| p<0·3 | 0·012 | 0·012 | 0·00034 | 0·013 | 0·02 | 0·066 |
| p<0·4 | 0·023 | 0·0071 | 0·00030 | 0·045 | 0·044 | 0·10 |
| p<0·5 | 0·023 | 0·043 | 0·0071 | 0·22 | 0·078 | 0·0068 |
Cross-disorder risk SNPs were defined as those meeting the p-value cutoffs shown in the left column in the primary meta-analysis of five disorders, and residing within 1 M bases from human reference hg18 genes.
LPL=lymphoblastoid cell lines.
Discussion
This study is the largest genome-wide analysis of psychiatric illness so far and the first to provide evidence that specific SNPs are significantly associated with a range of childhood-onset and adult-onset psychiatric disorders. For the five disorders studied, SNPs at four loci—regions on chromosomes 3p21 and 10q24, and SNPs in two L-type voltage-gated calcium-channel subunits, CACNA1C and CACNB2—exceeded the cutoff for genome-wide significance in the primary analysis. The strongest signal was within a region on chromosome 3p21.1. Aggregate polygenic risk scores for a broad set of common variants showed cross-disorder effects for all the adult-onset disorders (bipolar and major depressive disorder, and schizophrenia) and nominally between autism spectrum disorders and both bipolar disorder and schizophrenia.
In view of extensive linkage disequilibrium in the 3p21.1 region, encompassing more than 30 genes, we could not identify the causal locus. Genome-wide significant association to the 3p21.1 region has previously been reported in GWAS of samples overlapping with ours at rs1042779 (12 kb from our peak SNP) for bipolar disorder,28 rs736408 (2 kb) for a combined bipolar disorder and schizophrenia phenotype,7 and rs2251219 (248 kb) for a combined major depressive and bipolar disorder phenotype.29 Reanalysis of this last combined dataset suggested that the signal was largely attributable to the group with bipolar disorder.30 Furthermore, the association evidence for our peak chromosome 3 SNP rs2535629 was genome-wide significant in a joint analysis of bipolar disorder and schizophrenia samples done by the PGC schizophrenia group (p=7·8×10−9).6
Our model-selection analysis was designed to characterise the range of phenotypic effects for loci that showed significant association; however, the statistical evidence and effect size for each contributing disorder can vary. Although the best-fit model for the chromosome 3p21 region included all five disorders, interpretation of these results is complicated by evidence from a PGC GWAS mega-analysis of major depressive disorder.17 In the discovery phase of that analysis, which consisted of 9240 major depressive disorder cases and 9519 controls many of whom overlap with samples reported here, the smallest association p value for this region (rs2535629) was 0·00013. However, no association was noted in a replication dataset of 6783 cases and 50 695 controls (p=0·70) for that disorder, and the combined discovery and replication phase p value was 0·0031. Thus, any association between this region and major depressive disorder is unclear.
Two of the four genome-wide significant signals in our analysis localise to introns of brain-expressed genes encoding L-type voltage-gated calcium-channel subunits (CACNA1C and CACNB2). Previous disorder-specific GWAS, overlapping with the samples included here, identified CACNA1C as a susceptibility gene for bipolar disorder,7,31 schizophrenia,6 and major depressive disorder.32 Gain-of-function mutations in CACNA1C cause Timothy syndrome, a developmental disorder in which the phenotypic range includes autism.33 Consistent with a pleiotropic role, neuroimaging studies have documented effects of CACNA1C variants on a range of structural and functional brain phenotypes, including circuitry involved in emotion processing,34 executive function,34 attention,35 and memory.36 CACNB2 encodes an auxiliary voltage-gated calcium-channel subunit that interacts with L-type calcium-channel subunits (including CACNA1C, CACNA1D, and CACNA1S) to promote their trafficking to the plasma membrane, increase their function, and regulate their modulation by other signalling proteins and molecules.3 Although previous PGC analyses (schizophrenia and bipolar disorder) did not identify CACNB2 as a risk gene, a variant in CACNB2 (52 kb from our peak SNP) was one of the main signals in an independent GWAS of bipolar disorder in Han Chinese individuals.19
The pleiotropic effects of voltage-gated calcium channels on childhood-onset and adult-onset psychiatric disorders are underscored by pathway analysis in which calcium-channel activity genes, including our main two L-type subunit genes, showed significant enrichment in the five disorder dataset. The PGC analysis of bipolar disorder reported enrichment of a pathway including CACNA1C and CACNA1D; importantly, however, we detected enrichment of these genes after exclusion of the bipolar disorder dataset. Thus, our results suggest that voltage-gated calcium signalling, and, more broadly, calcium-channel activity, could be an important biological process in psychiatric disorders. A fourth region associated with cross-disorder effects was on chromosome 10, encompassing several genes with the peak signal in an intron of AS3MT. Loci previously associated with schizophrenia and bipolar disorder6,7 had varying evidence of association for the other major psychiatric disorders. For example, a locus previously strongly associated with schizophrenia, encompassing MIR137 and DPYD on chromosome 1, showed similar evidence of association with autism spectrum disorders; this finding is consistent with reports that autism spectrum disorders are related to microdeletions of this region.38,39
Accumulating evidence, including that from clinical, epidemiological, and molecular genetic studies, suggests that some genetic risk factors are shared between neuropsychiatric disorders. Genome-wide studies have identified rare copy-number variants that confer risk of several neuropsychiatric disorders including autism, attention deficit-hyperactivity disorder, epilepsy, intellectual disability, and schizophrenia.39 Our analyses of 14 SNPs previously identified as being genome-wide significantly associated with schizophrenia and bipolar disorder suggest that some loci identified in studies of individual disorders have broader phenotypic effects. Our results suggest a diversity of findings, with some SNPs showing diagnostic specificity and others pleiotropic effects on two or more of the five disorders.
These results should be interpreted in consideration of several limitations. First, we compared models of cross-disorder effects on the basis of the most often used goodness-of-fit measures, but other criteria might yield different results. For all four of the risk loci identified in the primary meta-analysis, the selected model had a substantially better fit than any alternative models had. However, for some loci that did not reach genome-wide significance, the difference in fit between our best-fitting and alternative models was moderate (appendix pp 38–45), so more than one model could be consistent with the noted effects. Second, diagnostic misclassification (eg, reciprocal misdiagnosis of cases of schizophrenia and bipolar disorder) could produce spurious evidence of genetic overlap between disorders.40 However, a substantial degree of misdiagnosis would be needed to account for our findings of loci that affect all or subsets of five disorders whose diagnostic criteria are fairly distinct. Third, the five disorders we examined were limited to those for which large-scale GWAS datasets have been assembled by the PGC and processed through a uniform quality-control process. As further datasets become available, more comprehensive analyses of cross-disorder genetic effects on psychiatric illness should be pursued. Fourth, we restricted our analyses to individuals of European ancestry. Whether our findings apply to other populations is unknown. Finally, GWAS designs are suited to identify common variant aspects of genetic architecture; further studies (including analyses of copy-number variants and rare mutations) will be needed to account more completely for shared genetic contributions across disorders. As in almost all GWAS of complex disorders reported so far, the effect sizes of genome-wide significant loci are individually quite small and the variance they account for is insufficient for predictive or diagnostic usefulness by themselves. However, our study is the first large-scale effort to characterise allelic effects across five psychiatric disorders, incorporating single locus, multilocus, and pathway analyses.
The identification of genetic variants that confer risk of a diverse set of psychiatric disorders parallels findings from other medical specialties. Most notably, GWAS41,42 of autoimmune disorders have shown extensive overlap in genetic variants that affect a diverse range of diseases, including rheumatoid arthritis, coeliac disease, multiple sclerosis, systemic lupus erythematosus, psoriasis, Crohn’s disease, and type 1 diabetes. Our results provide insights into the shared causation of psychiatric dis orders (panel). In particular, alterations in calcium-channel signalling could represent a fundamental mechanism contributing to a broad vulnerability to psychopathology.
Supplementary Material
Panel: Research in context.
Systematic review
Psychiatric diagnoses are presently defined as descriptive syndromes on the basis of a consensus of experts. The aetiological relations among these disorders are a topic of active debate in view of the imminent revision of the Diagnostic and Statistical Manual of Mental Disorders (DSM-5) and the International Classification of Diseases (ICD-11). Many family and twin studies3–5,12–14 have documented familial and heritable overlap between subsets of the five disorders we examined. Genetic association studies6–8,32,43,44 including samples smaller than those reported here have provided some support for these associations at a molecular level. We searched the National Human Genome Research Institute (NHGRI) GWAS catalogue as of September, 2012, to identify all previous reports of genome-wide significant association for any of the five disorders. For each significant gene or gene region identified, we then searched Medline for reports of at least nominally significant association (p<0·001) with at least one of the remaining four disorders. Several such loci have been reported including CACNA1C (for schizophrenia, bipolar disorder, and major depressive disorder), ZNF804A, ANK3, DGKH, and the 3p21.1 and NCAN (schizophrenia and bipolar disorder).32,45–50 Furthermore, polygene score analyses have shown significant overlap between schizophrenia and bipolar disorder and between bipolar disorder and major depressive disorder. Finally, several rare copy-number variations associated with autism spectrum disorders have been detected in schizophrenia, attention-deficit hyperactivity disorder, bipolar disorder, and recurrent major depressive disorder.39 However, no previous studies have examined cross-disorder effects encompassing all five of the disorders examined here.
Interpretation
This analysis provides the first genome-wide evidence that individual and aggregate molecular genetic risk factors are shared between five childhood-onset or adult-onset psychiatric disorders that are treated as distinct categories in clinical practice. As such, our findings are relevant to the goal of moving beyond descriptive syndromes in psychiatry and towards a nosology informed by disease cause. The finding that genetic variants have cross-disorder effects is an empirical step towards helping clinicians understand the common co-occurrence of clinical phenotypes in individual patients. Our results implicate a specific biological pathway—voltage-gated calcium-channel signalling—as a contributor to the pathogenesis of several psychiatric disorders, and support the potential of this pathway as a therapeutic target for psychiatric disease. These results add to literature in several specialties (including autoimmune and metabolic diseases) that have begun to document widespread pleiotropy of genetic risk factors across traditional diagnostic boundaries.
Acknowledgments
We thank the study participants, and the research staffat the many study sites. This work was supported by NIMH grant U01 MH085520. Numerous grants from the National Institutes of Health (USA), and similar numbers of government grants from other countries, and substantial private and foundation support enabled this work.
Footnotes
Contributors Overall coordination: Jordan W Smoller, Kenneth Kendler, Nicholas Craddock. Writing committee: Jordan W Smoller (lead), Nicholas Craddock, Kenneth Kendler, Phil Hyoun Lee, Benjamin M Neale, John I Nurnberger, Stephan Ripke, Susan Santangelo, Patrick F Sullivan. Statistical analysis: Stephan Ripke (lead), Kenneth Kendler, Phil Hyoun Lee, Benjamin M Neale, Shaun Purcell. Editorial revisions: Richard Anney, Jan Buitelaar, Ayman Fanous, Stephen V Faraone, Witte Hoogendijk, Klaus-Peter Lesch, Douglas F Levinson, Roy H Perlis, Shaun Purcell, Marcella Rietschel, Brien Riley, Edmund Sonuga-Barke, Russell Schachar, Thomas G Schulze, Anita Thapar. PGC Cross-Disorder Group: Nicholas Craddock, Kenneth S Kendler, Jordan W Smoller (cochairs), Ayman Fanous, Benjamin Neale, Michael Neale, John I Nurnberger, Roy Perlis, Shaun Purcell, Marcella Rietschel, Susan Santangelo, Thomas G Schulze, Anita Thapar. PGC coordinating committee: Patrick F Sullivan (chair), Patrick Bender, Sven Cichon, Nicholas Craddock, Mark J Daly, Stephen V Faraone, John Kelsoe, Thomas Lehner, Douglas Levinson, Mick O’Donovan, Pablo Gejman, Jonathan Sebat, Pamela Sklar, Jordan W Smoller. See appendix for PGC Collaborators from Analysis Committee (Mark J Daly, chair), ADHD Workgroup (Stephen V Faraone, chair), Autism Workgroup (Mark Daly, Bernie Devlin, cochairs), Bipolar Disorder Workgroup (John Kelsoe, Pamela Sklar, cochairs), Major Depressive Disorder Workgroup (Patrick Sullivan, chair), Schizophrenia Workgroup (Michael O’Donovan, chair).
Conflicts of interest We declare that we have no conflicts of interest.
This online publication has been corrected. The corrected version first appeared at thelancet.com on April 19, 2013
For a full list of contributors see appendix
See Online for appendix
For INRICH see http://atgu.mgh.harvard.edu/inrich
For the NHGRI GWAS catalogue see http://www.genome.gov/26525384
References
- 1.Kraepelin E. Psychiatry. A textbook for students and physicians (translation of the 6th edition of Psychiatrie) Science History Publications; Canton, MA: 1990. [Google Scholar]
- 2.Kendler K, Eaves LJ. Psychiatric genetics (review of psychiatry) American Psychiatric Press; Washington, DC: 2005. [Google Scholar]
- 3.Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123:48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- 4.Maier W, Lichtermann D, Minges J, et al. Continuity and discontinuity of affective disorders and schizophrenia. Results of a controlled family study. Arch Gen Psychiatry. 1993;50:871–83. doi: 10.1001/archpsyc.1993.01820230041004. [DOI] [PubMed] [Google Scholar]
- 5.Lichtenstein P, Yip BH, Bjork C, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–39. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium Genome-wide association study identifies five new schizophrenia loci. Nat Gen. 2011;43:969–76. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–83. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rapoport J, Chavez A, Greenstein D, Addington A, Gogtay N. Autism spectrum disorders and childhood-onset schizophrenia: clinical and biological contributions to a relation revisited. J Am Acad Child Adolesc Psychiatry. 2009;48:10–18. doi: 10.1097/CHI.0b013e31818b1c63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.King BH, Lord C. Is schizophrenia on the autism spectrum? Brain Res. 2011;1380:34–41. doi: 10.1016/j.brainres.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 11.Levinson DF, Duan J, Oh S, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–16. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ronald A, Simonoff E, Kuntsi J, Asherson P, Plomin R. Evidence for overlapping genetic influences on autistic and ADHD behaviours in a community twin sample. J Child Psychol Psychiatry. 2008;49:535–42. doi: 10.1111/j.1469-7610.2007.01857.x. [DOI] [PubMed] [Google Scholar]
- 13.Rommelse NN, Franke B, Geurts HM, Hartman CA, Buitelaar JK. Shared heritability of attention-deficit/hyperactivity disorder and autism spectrum disorder. Eur Child Adolesc Psychiatry. 2010;19:281–95. doi: 10.1007/s00787-010-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lichtenstein P, Carlstrom E, Rastam M, Gillberg C, Anckarsater H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am J Psych. 2010;167:1357–63. doi: 10.1176/appi.ajp.2010.10020223. [DOI] [PubMed] [Google Scholar]
- 15.Williams NM, Franke B, Mick E, et al. Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13.3. Am J Psychiatry. 2012;169:195–204. doi: 10.1176/appi.ajp.2011.11060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neale BM, Medland SE, Ripke S, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010;49:884–97. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.21. published online April 3. DOI:10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–28. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MT, Chen CH, Lee CS, et al. Genome-wide association study of bipolar I disorder in the Han Chinese population. Mol Psychiatry. 2011;16:548–56. doi: 10.1038/mp.2010.43. [DOI] [PubMed] [Google Scholar]
- 20.Nicolae D, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers A, Gibbs JR, Webster JA, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–99. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 22.Webster J, Gibbs JR, Clarke J, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–58. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schadt E, Molony C, Chudin E, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding J, Gudjonsson JE, Liang L, et al. Gene expression in skin and lymphoblastoid cells: refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet. 2010;87:779–89. doi: 10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stranger B, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burnham KP, Anderson DR. Model selection and multimodel inference: a practical information-theoretic approach. 2nd edn Springer-Verlag; New York, NY: 2002. [Google Scholar]
- 27.Colantuoni C, Lipska BK, Ye T, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–23. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott LJ, Muglia P, Kong XQ, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci USA. 2009;106:7501–06. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahon FJ, Akula N, Schulze TG, et al. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21. Nat Genet. 2010;42:128–31. doi: 10.1038/ng.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breen G, Lewis CM, Vassos E, et al. Replication of association of 3p21.1 with susceptibility to bipolar disorder but not major depression. Nat Gen. 2011;43:3–5. doi: 10.1038/ng0111-3. [DOI] [PubMed] [Google Scholar]
- 31.Ferreira MA, O’Donovan MC, Meng YA, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–58. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green EK, Grozeva D, Jones I, et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry. 2010;15:1016–22. doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao P, Soong TW. CaV1.2 channelopathies: from arrhythmias to autism, bipolar disorder, and immunodeficiency. Pflugers Arch. 2010;460:353–59. doi: 10.1007/s00424-009-0753-0. [DOI] [PubMed] [Google Scholar]
- 34.Bigos KL, Mattay VS, Callicott JH, et al. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch Gen Psychiatry. 2010;67:939–45. doi: 10.1001/archgenpsychiatry.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thimm M, Kircher T, Kellermann T, et al. Effects of a CACNA1C genotype on attention networks in healthy individuals. Psychol Med. 2011;41:1551–61. doi: 10.1017/S0033291710002217. [DOI] [PubMed] [Google Scholar]
- 36.Erk S, Meyer-Lindenberg A, Schnell K, et al. Brain function in carriers of a genome-wide supported bipolar disorder variant. Arch Gen Psychiatry. 2010;67:803–11. doi: 10.1001/archgenpsychiatry.2010.94. [DOI] [PubMed] [Google Scholar]
- 37.Buraei Z, Yang J. The β subunit of voltage-gated Ca2+ channels. Physiol Rev. 2010;90:1461–506. doi: 10.1152/physrev.00057.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willemsen MH, Valles A, Kirkels LA, et al. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J Med Genet. 2011;48:810–18. doi: 10.1136/jmedgenet-2011-100294. [DOI] [PubMed] [Google Scholar]
- 39.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–41. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wray NR, Lee SH, Kendler KS. Impact of diagnostic misclassification on estimation of genetic correlations using genome-wide genotypes. Eur J Hum Genet. 2012;20:668–74. doi: 10.1038/ejhg.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cotsapas C, Voight BF, Rossin E, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhernakova A, Stahl EA, Trynka G, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J, Perlis RH, Lee PH, et al. Cross-disorder genomewide analysis of schizophrenia, bipolar disorder, and depression. Am J Psychiatry. 2010;167:1254–63. doi: 10.1176/appi.ajp.2010.09091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Blackwood DH, Caesar S, et al. Meta-analysis of genome-wide association data of bipolar disorder and major depressive disorder. Mol Psychiatry. 2011;16:2–4. doi: 10.1038/mp.2009.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Donovan MC, Craddock N, Norton N, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–55. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 46.Nyegaard M, Demontis D, Foldager L, et al. CACNA1C (rs1006737) is associated with schizophrenia. Mol Psychiatry. 2010;15:119–21. doi: 10.1038/mp.2009.69. [DOI] [PubMed] [Google Scholar]
- 47.Muhleisen TW, Mattheisen M, Strohmaier J, et al. Association between schizophrenia and common variation in neurocan (NCAN), a genetic risk factor for bipolar disorder. Schizophr Res. 2012;138:69–73. doi: 10.1016/j.schres.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Cichon S, Muhleisen TW, Degenhardt FA, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88:372–81. doi: 10.1016/j.ajhg.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng Z, Wang T, Li T, et al. Common SNPs and haplotypes in DGKH are associated with bipolar disorder and schizophrenia in the Chinese Han population. Mol Psychiatry. 2011;16:473–75. doi: 10.1038/mp.2010.86. [DOI] [PubMed] [Google Scholar]
- 50.Athanasiu L, Mattingsdal M, Kahler AK, et al. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J Psychiatr Res. 2010;44:748–53. doi: 10.1016/j.jpsychires.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.