Abstract
Identification and validation of protein targets of bioactive small molecules is an important problem in chemical biology and drug discovery. Currently, no single method is satisfactory for this task. Here, we provide an overview of common methods for target identification and validation that historically were most successful. We have classified for the first time the existing methods into two distinct and complementary types, the “top-down” and “bottom-up” approaches. In a typical top-down approach, the cellular phenotype is used as a starting point and the molecular target is approached through systematic narrowing down of possibilities by taking advantage of the detailed existing knowledge of cellular pathways and processes. In contrast, the bottom-up approach entails the direct detection and identification of the molecular targets using affinity-based or genetic methods. A special emphasis is placed on target validation, including correlation analysis and genetic methods, as this area is often ignored despite its importance.
1. Introduction
Small molecules are widely used as tools to study and perturb biological systems. Bioactive small molecules provide an opportunity to quickly switch on or off the activity of proteins in both temporally and spatially controlled manner, which is unmatched by any other method. The main obstacle to a wider use of small molecules for studying biology is the difficulty of identifying compounds that can specifically alter the function of a given protein or protein family. Historically, many drugs and bioactive small molecules, particularly natural products, were discovered based on their desired or undesired physiological effects at the cellular or organismal level. Using ad hoc approaches, researchers have painstakingly identified the protein targets of many widely used drugs and bioactive natural products. This has led to a deeper understanding of the physiological functions of the newly identified protein targets and sometimes of the pathophysiology of diseases that the drugs were used to intervene. With the advent of contemporary chemical biology, the availability of high-throughput phenotypic screening platforms coupled with the significant increase in the number of chemical compounds in both public and private sectors has led to an emergence of a growing collection of chemical compounds that are known to interfere with various biological processes. In contrast to the discovery of biologically active compounds, however, target identification and validation has remained a major bottleneck that has prevented the wider use of small molecules in biological research and impeded to certain extent drug development.
In this review, we attempt to provide an overview of some of the most widely used and effective methods for identification and validation of protein targets of bioactive small molecules. Due to space limitations, we will not be able to cover all the available methods. This review is divided into two parts. In the first part, we will discuss the common methods used to identify candidate target proteins. In the second part, we will cover methods commonly applied to validate physiological relevance of newly identified protein targets.
2. Target Identification
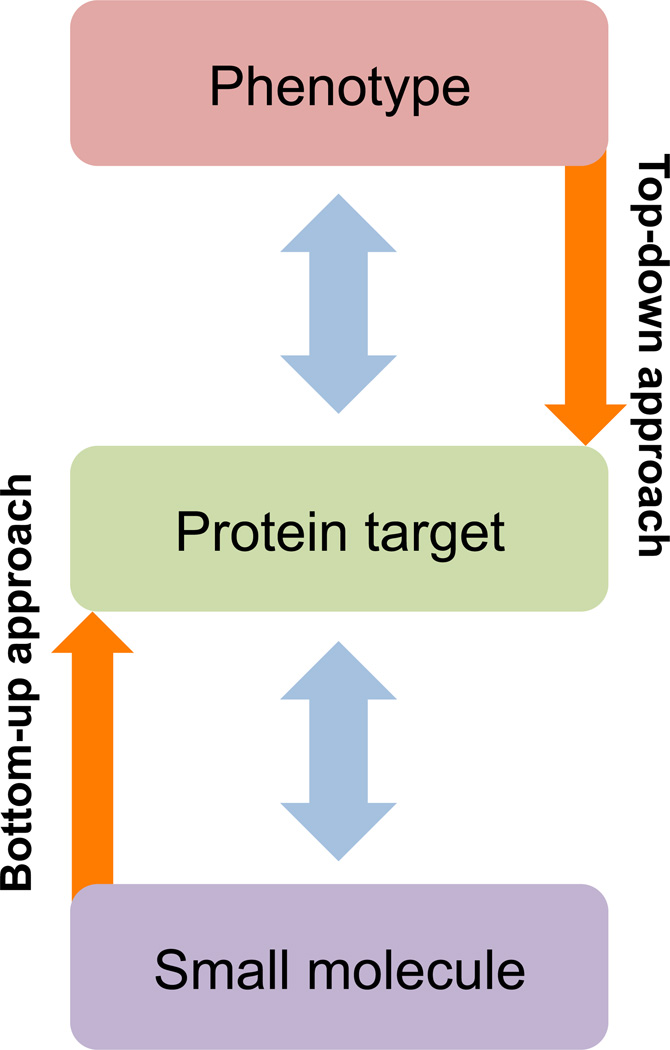
We have broadly divided target identification methods into two general classes, the bottom-up and the top-down approaches. For definition, we place the small molecule, its protein target and its cellular phenotype on a vertical line with the protein target at the bottom and the global cellular phenotype at the top (Figure 1). The first approach consists of methods that allow for the direct identification of the protein target using genetics or affinity-based methods. We call this the “bottom-up” approach because it starts at the “bottom” with identification of the target protein before one goes “up” to the next level to explain the phenotype through perturbation of the function of the target protein. The second approach consists of methods that allow identification of the protein target by exploiting the existing knowledge of a given cellular process that is perturbed by the small molecule. We call this the “top-down approach” as it allows one to narrow “down” the possible targets based on a general understanding of what part of cellular and/or organismal physiology the small molecule affects and the proteins known to be involved in the relevant process.
Figure 1.
Schematic representation of top-down and bottom-up approaches to target identification
2.1. Bottom-up Approach
Up to now, the two most successful methods used for target identification are affinity purification and genetics. Genetics is the method of choice for target identification of antibiotics and antifungal drugs. Affinity purification has been successfully applied to drugs and small molecules that are active only in mammalian cells or whole organisms. Each method has its advantages and disadvantages. Affinity purification requires chemical modification of the compound under study that in turn requires prior knowledge on the structure/activity relationship of the compound. In contrast, genetic approaches employ unmodified compound. But genetic approaches require an appropriate cellular readout that is suitable for selection. Thus, not all biologically active small molecules can be readily studied using genetic methods (e.g. compounds that do not inhibit cell proliferation). In addition, genetic approaches may not lead to the identification of the direct target for small molecules, even though it may shed light on the pathways in which the target lie or with which the target protein may genetically interact. A high degree of uncertainty exists for both approaches and it is best to use both methods whenever possible to increase the chances of success.
2.1.1. Affinity-based methods
Affinity purification is one of the most widely used methods for target identification. It was successfully applied to a variety of small molecules and drugs. The basic premise of this approach is that a protein that binds a small molecule with the highest affinity is likely to mediate the biological activity of the compound. This approach comprises at least two distinct methods. In one method, a small molecule of interest is attached to a resin and then affinity chromatography is performed using an appropriate cell and tissue lysate in the presence or absence of excess free small molecule. Here, the goal is to identify proteins that bind to the affinity resin and are competed away in the presence of excess free compound. In the other method, a radioactively or fluorescently labeled small molecule is used to measure the binding activity of cell or tissue lysates to facilitate the purification of a fraction that contains the majority of drug binding activity.
Classically, affinity chromatography approach is used with a small molecule immobilized on a resin such as agarose or sepharose or other types of beads (Figure 2). Affinity resin is incubated with a cell lysate of choice, washed several times with a lysis buffer and proteins are eluted by either a solution of high concentrations of free small molecule or simply by boiling in Laemmli sample buffer. The eluted proteins are analyzed on an SDS-PAGE gel using silver staining. The identity of specific binding proteins that can be competed away by the free drug is determined by mass spectrometry. Success is dependent on a number of factors, including what cell or tissue type and what lysis buffer to use, how long to incubate the beads with the lysate and how stringent the washes should be. Unfortunately, there are no rules for arriving at the most optimal conditions and they are usually determined by trial and error. A prerequisite for a successful use of affinity chromatography approach is to attach the compound to the resin in such a way that it does not significantly abrogate its binding activity. Another important factor is that the target protein is abundant in the tissue or cell type that one uses. More recently, a variation of this method has emerged that takes advantage of the high affinity between biotin and streptavidin or avidin. Instead of directly immobilizing small molecules to solid phase, they are coupled to biotin. Such biotin-small molecule conjugates can be fully verified chemically and evaluated in relevant phenotypic assays to ensure that the chemical modifications allowed for retention of the biological activity of the small molecules. Some of the known examples of small molecules whose targets were identified using this approach include norepinephrine (- adrenergic receptor),1 FK506 (FKBP),2 trapoxin (histone deacetylase),3 TNP-470 (Methionine aminopeptidase 2),4, 5 thalidomide (cereblon or CRBN),6 pateamine A (eukaryotic initiation factor or eIF4A),7, 8 methyl-gerfelin (glyoxalase I),9 and spliceostatin A (SF3b involved in splicing).10
Figure 2.
Schematic representation of protein target identification using affinity chromatography
In conventional affinity chromatography experiments, the putative binding protein(s) is first detected on an SDS-PAGE gel upon silver staining before the protein band is excised for identification by mass spectrometry. The recent development of quantitative proteomics is changing the way target proteins are identified. Over the last decade, much effort has been made to develop methods for identification of protein-protein interaction on a genome wide scale.11, 12 A major goal was to better understand the functions of proteins present in various genomes. As a result, new methods for identifying specific interacting proteins that are cumulatively called quantitative proteomics were developed.13–15 It started with efforts to identify protein interaction partners using affinity chromatography. But instead of small molecules, antibodies were attached to the resin to immunoprecipitate a specific protein along with its interacting partners that would be identified using mass spectrometry. With the development of more sensitive and more accurate mass spectrometers, it became possible to identify hundreds of proteins in a given experiment. Many of these proteins are not even visible on the gel. The problem is that the majority of those proteins bound non-specifically to the resin and since mass spectrometry is inherently non-quantitative it was difficult to determine which protein(s) bound specifically to the protein of interest. In order to solve this specificity problem, several groups developed methods to quantify the relative amounts of a particular protein bound to loaded and control resins. Two of the most popular quantitative mass spectrometry methods are iTRAQ and SILAC.16, 17 The theory behind both of those methods is that samples are labeled with different isotopes. The samples are mixed in a 1:1 ratio before the mixture is subject to MS analysis. The enrichment of any given protein can be detected by the ratio between peptides that have heavy vs light isotopes. After repeating the pull-down experiment, it is possible to obtain a list of specific binding proteins based on that ratio for follow-up studies. Recently these techniques have been applied to target identification for small molecules and drugs.18–21
The second type of affinity-based methods relies on drug binding assays to guide target purification. The goal of this approach is to purify the binding protein (receptor) of a small molecule to homogeneity using drug binding assays to track the binding activity during protein chromatography and fractionation. A radioactively or fluorescently labeled small molecule such as a drug is usually incubated with a selected cell or tissue lysate and the protein-drug complex is separated from the free drug by various techniques including centrifugation, gel filtration, or absorption to charcoal.22, 23 Scintillation counting, spectrophotometry or other methods can then be applied to follow the protein fraction that binds the drug. This method is similar to that used for enzyme purification guided by enzymatic activity of a given protein or protein complex. When the putative binding protein is purified to near homogeneity, it can be subjected to identification by mass spectrometry. Binding specificity can be assessed by competition with excess unlabeled drug. Kd can also be determined by measuring the amount of drug bound to the protein at different drug and lysate/protein concentrations.24 In fact, receptors for the majority of known drugs were identified using this method, including adrenergic receptors25–27, opioid receptors,28, 29 cyclophilins,30 and FKBP12.31 In many of these studies, affinity chromatography was used side by side with binding assays as one of the purification steps. Binding assays are essential for identification of appropriate lysis methods for optimal receptor solubilization and for identification of buffer conditions for maximal binding of the drug to its receptor. It is apparent that the optimal approach for target identification using these methods should consist of preparation of the extract enriched with target protein using binding assays followed by affinity chromatography in conjunction with quantitative mass spectrometry.
2.1.2. Genetic methods
The basic concept of genetic methods for target identification is to genetically perturb cells or whole organisms by mutagenesis, protein overexpression or knockdown and select for clones that are resistant to a particular small molecule. Identification of the gene(s) that caused the resistance can often lead to the target of a small molecule, though one needs to be cautious when analyzing genetic resistance since there are multiple ways resistance can arise. The most common phenotype that is exploited in genetic methods is cell viability. In this section we will first describe target identification using yeast genetics and then discuss examples of genetic methods applied to mammalian cells.
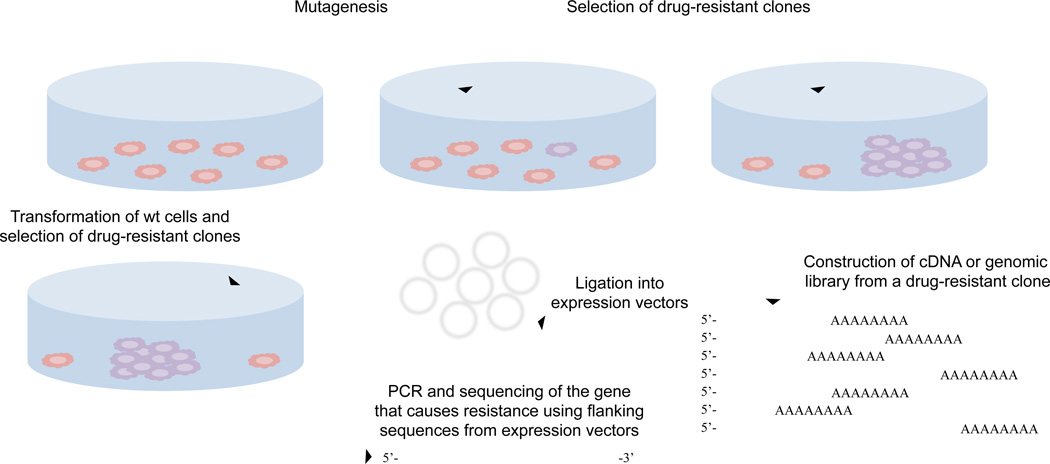
In the classic genetic method of target identification, bacterial or yeast cells are mutagenized and clones that are resistant to a small molecule of interest are selected by screening (Figure 3). For yeast and bacterial cells it is often possible to select resistant clones that arise from spontaneous mutations. Once resistant clones are isolated, genomic DNA can be isolated and used to prepare a library. The genomic DNA library is used to transform wild type cells followed by selection on the small molecule. The genomic DNA that confers resistance of wild type cells to the drug can be isolated and sequenced, which will lead to the identification of the mutation on a specific gene that gave rise to resistance. Further studies will be required to assess whether the newly identified mutant gene product is the direct protein target for the small molecule. Occasionally, this approach can also be used in reverse whereby the DNA from wild type strain that confers sensitivity to the resistant clone is identified.
Figure 3.
Schematic representation of protein target identification using genetic methods in yeast, bacterial or mammalian cells
An illuminating example of target identification using yeast genetics is the identification of the target of the immunosuppressive and antiproliferative natural product rapamycin (TOR)32, whose mammaliam counterpart (mTOR) was later identified using affinity chromatography.33, 34 The ability of rapamycin to arrest yeast cell cycle in G1 phase made it possible to apply yeast genetics to identify its target. Rapamycin, like another immunosuppressant FK506, works by first binding to its receptor FK506 binding protein (FKBP or FKBP proline rotamase 1 (FPR1) in yeast).2, 35, 36 This binding is necessary but not sufficient for its antiproliferative activity since cells without FPR1 are viable and completely resistant to rapamycin. Yeast cells can spontaneously become resistant to rapamycin with a frequency of 10−7 so it was unnecessary to mutagenize cells in this case.35 Heitman et al. isolated 18 independent spontaneous rapamycin-resistant strains. Using complementation analysis, they showed that 15 of 18 mutations were in FKBP gene and the other 3 mutations were in 2 genes that were named TOR1 and TOR2. Genomic libraries were prepared from rapamycin resistant strains as described above and the sequence of TOR1 and 2 genes was identified from the genomic fragments that caused rapamycin resistance in wild type strains.37 Ensuing studies validated TOR1 and TOR2 as the target of rapamycin.
In the last few years, genome-wide yeast knockout collections have become available where each gene is deleted in one strain and replaced by a DNA bar code to facilitate identification of the deletion mutants.38, 39 These knockout collections have been exploited to facilitate target identification as well. Several groups have used those collections to identify targets of some known and unknown drugs. There are two different approaches used. The first approach is to use a collection of haploinsufficiency diploid yeast strains in which one copy of a gene is deleted. The underlying principle for this approach is that cells that express half the amount of the target protein for a given small molecule will become hypersensitive to the molecule in comparison to the wild type strain. This approach was validated on a number of small molecules with known targets.40 The key to success is to choose a concentration of the drug around or slightly lower than the IC50 so that the difference in sensitivity between wild type and deletion strains could be easily observed. Deletion strains can either be assayed separately using OD600 measurements in a plate reader41 or they can be pooled and then the sensitive strains can be identified by the decrease of the bar code amount for a particular strain using microarrays.42, 43 Sphingolipid biosynthesis was identified as the target of a potential anticancer agent dihydromotuporamine C using these methods.41 The second complementary approach is to use a haploid yeast deletion collection to screen for drug sensitivity.44 The downside to this approach is that it is unlikely to yield a direct target of the drug since the complete deletion of the target protein is expected to produce non-viable yeast cells and will be absent from the collection. Nevertheless, this approach may yield information on genes that are functionally connected to the target of the small molecule under scrutiny. Similarly, gene overexpression screens in yeast can be used to identify targets of small molecules based on the expectation that cells with overexpressed target will become partially resistant to the small molecules.45, 46
Given that many essential biological pathways are conserved between yeast and mammals, it is likely that many biologically active small molecules will be active in both. The homologue of the target that is identified in yeast can then be confirmed as a target in the corresponding organism where the small molecule of interest is also active. However, the use of yeast genetics for target identification does have its limitations. A number of mammalian cellular pathways are absent in yeast, which is evident from the much larger genome and proteome of mammals than those of yeast. Thus, yeast genetics will not be applicable to those small molecules that affect processes unique to mammalian cells. In addition, some small molecules cannot be used in yeast because of their low uptake and/or high efflux in comparison to mammalian cells.
Although less common, similar genetic approaches have been used with mammalian cells (Figure 3). Mammalian cells can be readily mutagenized to select for clones that are resistant to a particular small molecule.47–49 The advantage of using chemical mutagenesis is that dominant mutants can often be obtained using this method. Usually cells are treated with ethyl methanesulfonate (EMS), washed and allowed to proliferate for one cell cycle, as replication is required for mutations to incorporate into the genome. Then the cells are treated with a high concentration (10–1000 times higher then IC50) of the small molecule of interest for days or even weeks to allow resistant cells to expand to form small colonies. Such drug-resistant colonies from different plates are then isolated and expanded in the presence of the drug. The most difficult part of this method is to isolate the gene that causes the resistance. One method is functional expression cloning.50–52 First, a cDNA library is prepared from the resistant mutant cell line and cloned into a retroviral expression vector. Then the wild type cell line is transduced with the retroviral library and colonies that become resistant to the drug are isolated as described above. The gene that causes the resistance is incorporated into the genome of the wild type cell and can be amplified using PCR with primers complimentary to retrovirus-derived flanking regions and then sequenced to identify the mutant protein that confers resistance. Further development of sequencing technologies and wider availability of genome-wide mammalian ORF libraries will make genetic methods of target identification in mammalian cells even more efficient in the years to come.
A “neo-genetic” approach for identifying novel protein-protein interactions, the yeast two-hybrid system,53 was adapted to develop the yeast three-hybrid system for target identification.54 This method is based on the ability of a heterodimer between a small molecule of interest and another small molecule with a known high-affinity protein receptor to induce the formation of a ternary complex between two hybrid proteins to activate gene transcription. The original system was developed using the steroid hormone dexamethasone and its receptor, the rat glucocorticoid receptor, as the existing pair of small molecule/receptor. A limitation of the original version of the system is the requirement for high affinity between small molecule of interest and the target. Subsequently, a new ligand-receptor pair, methotrexate-dihydrofolate reductase, was developed that showed higher sensitivity.55 That system was successfully used to discover targets for inhibitors of protein kinases.56 Most recently, an even more sensitive SNAP-tag-based yeast three-hybrid system was developed that allowed for the identification of an enzyme involved in tetrahydrobiopterin biosynthesis as the target for sulfasalazine and its metabolites.57
2.2. Top-down approach
In contrast to the bottom-up approach discussed above, the top-down approach entails a path of going from a phenotype to specific protein target by exploiting the existing knowledge of a given cellular process. In this section we will describe a framework for the top-down approach along with some useful methods and examples from the literature where this approach was successfully applied. Top-down approach always starts with a phenotype that a compound of interest causes in cells or in organisms. All bioactive small molecules of course have some phenotype (e.g., cell proliferation inhibition). The goal is to go from the global phenotype in a stepwise fashion to eventually arrive at the protein target. A number of successful examples of the top-down target identification have been reported in the literature, including the identification of tubulin as the target of the anticancer drug taxol,58, 59 Sec61 complex as the target of a cyclodepsipeptide natural product that inhibits protein secretion,60 and kinesin Eg5 as the target of a mitosis inhibitor named monastrol61, to name a few. Described in more details below are three examples in this category. As our knowledge of biology increases, the top-down approach will become more and more effective.
The top-down approach was used successfully for the elucidation of the mechanism of action of a transcription inhibitor 5,6-dichloro- -D-ribofuranosylbenzimidazole (DRB). In the late 1970s, DRB was shown to be a selective inhibitor of hnRNA and mRNA synthesis.62–64 It inhibited hnRNA synthesis only by 60–70% but completely (>95%) inhibited cytoplasmic poly(A) containing RNA. After only several minutes of exposure to the drug, cells preferentially incorporated radioactively labeled nucleotides into long hnRNAs compared to untreated cells.63 This was taken as an important clue that DRB is an inhibitor of transcription initiation rather than elongation. These in vivo observations were later confirmed using in vitro transcription assays where DRB was found to inhibit RNA polymerase II-driven transcription at concentrations similar to those used in vivo without affecting purified RNA polymerase II activity.65–67 A more detailed investigation of the effects of DRB on in vitro transcription using pulse-chase assays showed that DRB inhibited the synthesis of full-length transcription only when added during the synthesis of the first 500 nucleotides or less and has no effect on elongating RNA polymerase II complex after it synthesized more then 500 nucleotides.68, 69 Furthermore, DRB had no effect on synthesis of RNA less than 100 nucleotides. This effect was explained by the observation that RNA polymerase II undergoes pausing soon after transcription initiation and the activity of positive transcription elongation factor (p-TEF) is required for the formation of productive elongation complex. This activity is inhibited by DRB. Now that the activity that is inhibited by DRB was identified, it was possible to fractionate cell extracts using this activity as readout. pTEFb was purified this way and was shown to contain 2 subunits – Cdk9 and cyclin T.70–72 DRB inhibited Cdk9 kinase activity at concentrations similar to those required for its effects on in vitro and in vivo transcription. Cdk9 was later shown to promote productive elongation by phosphorylating RNA polymerase II C-terminal domain and several negative regulators of elongation.71, 73 Thus, the top-down approach not only revealed the molecular target of DRB as Cdk9, but also led to identification of a new mechanism for transcription regulation.
Another bioactive small molecule whose mechanism was elucidated using the top-down approach is brefeldin A (BFA). In late 1980s, BFA was identified as a potent inhibitor of protein secretion.74 Using careful pulse-chase and electron microscopy experiments, BFA was shown to inhibit transport of proteins from ER to Golgi and cause the disassembly of the Golgi complex.74, 75 Time course experiments showed that Golgi complex with its resident proteins is absorbed into the ER within 10 min after BFA treatment.76, 77 Disassembly of the Golgi complex proceeds through a dramatic intermediate state with long tubular projections spread throughout cytoplasm that is visible as soon as 2 min after BFA treatment. The absorption of Golgi complex into ER happens because BFA only inhibits transport of proteins from ER to Golgi but does not affect microtubule-dependent retrograde transport of proteins from Golgi to ER. Remarkably, all these effects are completely reversed within 20 min after BFA is washed out. BFA also causes the fusion of early endosomes and trans-Golgi network through mechanism similar to fusion of Golgi complex and ER.78, 79 The first clue to the biochemical mechanism of action of BFA came from a serendipitous observation that BFA promoted dissociation of a 110-kDa peripheral membrane protein from the Golgi apparatus within 30 s after treatment.80 This process was inhibited by pre-incubation with GTPγS suggesting the participation of GTP binding proteins.81 The 110-kDa protein was later found to be one of the major coat protein subunits of coated vesicles in Golgi and renamed β-COP.82 BFA was shown to block the formation of non-clathrin coated vesicle from Golgi using a cell-free assay with purified Golgi.83 This observation provided the first molecular understanding of BFA action and further studies focused on explaining the mechanism of vesicle formation inhibition by BFA and its dependence on GTP. At about the same time, several groups reported identification of ADP-ribosylation factor (ARF) small GTPase as a component of the coat of Golgi-derived vesicles that was required for β-COP binding to Golgi memberanes in GTP dependent manner.84–86 Investigation of an effect of BFA on ARF showed that BFA inhibited ARF binding to Golgi membrane through inhibition of GDP-GTP exchange catalyzed by an unknown ARF GTP exchange factor (ARF GEF) present in Golgi fractions.87, 88 ARF GEF that is inhibited by BFA was purified using protein chromatography coupled with assay for BFA-inhibited ARF GEF activity.89–91 It turned out that BFA is able to inhibit a number of ARF GEFs (BIG1, BIG2 and GBF1) that contain a common Sec7 domain. Independently, Sec7-containing yeast proteins GEA1p, GEA2p and Sec7p were later identified as targets of BFA using yeast genetics,92, 93 supporting the observations made in mammalian cells.
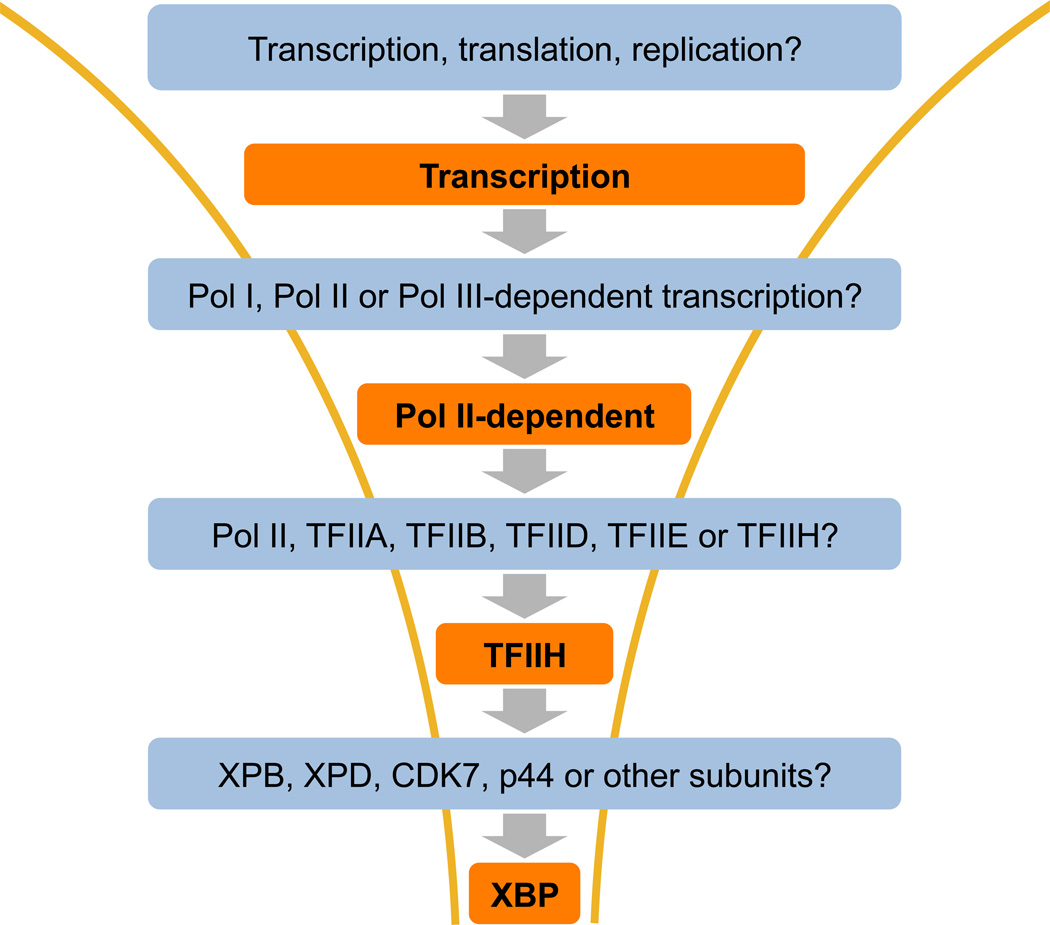
Most recently, the molecular target for triptolide, an anti-proliferative and immunosuppressive natural product isolated from a Chinese medicinal plant, was identified using the top-down approach as well (Figure 4).94 Since its initial isolation and identification in the early 1970s,95 the mechanism of action of triptolide has attracted the attention of chemists and biologists. This interest was intensified when it was reported to possess potent anticancer activity through screening of the NCI 60 cancer cell lines. Using the “bottom-up” approach with a [3H]-labeled triptolide, one group discovered a calcium channel polycystin-2 as a possible target96 and another group detected the covalent binding of a 90-kD nuclear protein that bound to triptolide.97 Unfortunately, polycystin-2 cannot account for the general anti-proliferative activity of triptolide. Having detected the covalent binding of the 90-kD protein from cell lysate, it is theoretically possible to purify the putative triptolide target using [3H]-triptolide binding as an assay. But that did not happen. On a different front, the extensive synthetic efforts led to a large number of triptolide analogs and suggested that the C14 hydroxyl group is suitable for tethering an affinity probe. Affinity probes attached to the C14 OH group led to the isolation of dCTP pyrophosphatase, which does not seem to be relevant to the anti-proliferative activity of triptolide either.98
Figure 4.
Top-down approach to identify XPB as the molecular target for triptolide
We took a top-down approach by beginning with a clue that triptolide may affect transcription.99, 100 We began by systematically examining the effects of triptolide on the synthesis of DNA, RNA and proteins in cancer cell lines. We quickly found that triptolide preferentially affected cellular RNA synthesis, corroborating with findings of earlier reports. Fortunately, the mechanism and the key regulatory proteins of transcription initiation have been well characterized and there are a number of reconstituted in vitro transcription assays with defined protein components. Using those assays, we found that triptolide appeared to specifically affect TFIIH-mediated transcriptional initiation and nucleotide excision repair (NER). Four of the ten subunits of TFIIH possess defined enzymatic activities. A systematic examination of those activities narrowed the target to XPB, a DNA-dependent ATPase that is required for unwinding the double stranded DNA in the promoter region and around damaged nucleotides to allow RNAPII-dependent transcription initiation and NER to proceed. Subsequent experiments verified XPB as a physiologically relevant target for triptolide. It is interesting to note that for reasons that remain unknown, we are still unable to detect binding of affinity probes of triptolide by modifying triptolide at the C14 OH group to XPB both in vitro and in cell culture, highlighting the limitation of any given approach for target identification.
3. Target Validation
Target validation is a crucial step in validating the physiological relevance of newly identified small molecule targets. It is also a process during which new biological insights can be gained on the function of the target protein. Just because a small molecule binds to a particular protein or a mutation in a particular gene causes resistance does not guarantee that the protein is the physiological target of the small molecule. Necessary and sufficient criteria for target validation include demonstration that a small molecule binds directly to the target protein and that expression of the functional target protein that is deficient in binding to the small molecule leads to the resistance of cells or organisms to this small molecule. In reality, very few targets meet these criteria, underscoring the difficulty of target validation. We will briefly review two commonly used methods for target validation here.
3.1. Correlation between small molecule-target interaction and cellular activity
Correlation analysis is a most versatile and general method of target validation. It can be applied to virtually any target. The goal of the correlation analysis is to generate analogs of small molecule of interest that will have different affinity for the target protein and then determine if there is a good correlation between the affinity for the target protein and the potency of causing the corresponding phenotype. For example, if one analog is 10 times more potent at binding to a target protein then it should be 10 times more potent at causing the corresponding phenotype. If there is such a correlation, then it is strong evidence that this phenotype is caused by binding of this small molecule to the target protein. There are examples in the literature where correlation method is used for target validation but the correlation that is observed is 10 to 1 instead of 1 to 1, which in fact is the evidence against the putative protein as a relevant target of the small molecule. The key to a successful application of this method is to synthesize analogs of the bioactive small molecule that will have wide range of IC50 values spanning at least 3 orders of magnitude. It is also important to make compounds with IC50 values that are more or less equally distributed over those 3 orders of magnitude of activity and not concentrated around one particular value. Correlation analysis was successfully used for target validation of a number of bioactive small molecules including triptolide (XPB)94 and fumagilin (MetAP2).4 This method was used in 1970s to show that specific binding of 15 morphine analogs to homogenates of guinea pig intestine longitudinal muscle perfectly correlated (slope 0.98, r = 0.97) with their ability to inhibit electrically induced contraction of the guinea pig intestine.101 This observation helped to solidify the proposition that binding to opioid receptors led to biological activity of morphine analogs and paved the way to the eventual isolation of these receptors.
3.2. Genetic approaches
If a bioactive small molecule causes a cellular phenotype by binding to a specific protein and (often) causes loss of its function, it is expected that (1) Overexpression of the target protein will confer resistance to the small molecule; (2) Deletion of the target protein will cause the same cellular phenotype as the treatment of cells with the small molecule and reduction of its expression will cause hypersensitivity to the small molecules; (3) Mutants of the target protein that no longer bind to the small molecule but retain cellular activity of wild type protein will confer resistance to the small molecule. The genetic approaches of target validation entail experiments to address all three aforementioned aspects. Ideally, a complete validation of target should meet all three expectations.
The easiest and most widely used method of target validation is to overexpress, knock down or less often, knock out the target protein. It is sometimes possible to observe resistance or sensitization by several orders of magnitude for the IC50 values for small molecules using those perturbations. Often, resistance or sensitization cannot be achieved because it is impossible to efficiently overexpress (e.g. protein is part of multi-subunit complex) or knock out (e.g. protein is essential) the target protein. Another caveat of this approach is that overexpression or knockdown of any drug binding protein (not necessarily the physiological target) will lead to partial resistance or sensitivity to that drug, making it difficult to draw conclusions. Therefore, other methods of target validation should be used unless dramatic resistance or sensitization to the small molecule of interest is achieved.
Another genetic approach to target validation is to generate a mutant protein that is resistant to small molecule binding but retains its normal function. This method can lead to a very convincing validation of a physiological target and is comparable to correlation analysis in that regard. However, it is often difficult to generate a mutant target protein that is defective in small molecule binding while remaining active. This approach can be significantly facilitated if genetic methods of target identification were used and the mutations that perturb binding are already known.
A good example of the use of a genetic method for target validation is the identification of cereblon (CRBN) as the target of thalidomide’s teratogenic activity.6 Thalidomide was widely used as a sedative in the 1960s and was often prescribed to pregnant women. Before its teratogenic effect was discovered, more than 10000 affected children were born from mothers taking thalidomide.102, 103 CRBN is a substrate receptor of E3 ubiquitin ligase complex that also contains DDB1, Cul4A and Roc1. Researchers generated Y384A and W386A double mutant of CRBN (CRBNYW/AA) that was deficient for thalidomide binding by systematically mutating conserved residues in the C-terminal 104 amino acid long thalidomide-binding region of CRBN. To prove that CRBN is the physiological target of thalidomide’s teratogenicity, chicks and zebrafish were used as models to assess thalidomide’s teratogenicity. Overexpression of CRBNYW/AA but not wild type CRBN in chicks and zebrafish led to a complete resistance to teratogenic effect of thalidomide in those organisms. In addition, it was shown that knockdown of CRBN in zebrafish also led to similar defects as thalidomide treatment further validating it as a physiological target.
4. Conclusion
We described a number of approaches for identification and validation of protein targets of bioactive small molecules. Each method has its own strengths and weaknesses. The choice of an appropriate subset of methods for target identification and validation will depend on the chemistry and the cellular phenotype associated with the small molecule. With the advent and improvement of target identification and validation methods, the bottle-neck of target identification in chemical biology will continue to be widened and eventually cease to exist, paving the way for small molecules to become more widely used in biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lefkowitz RJ, Haber E, O'Hara D. Proc Natl Acad Sci U S A. 1972;69:2828. doi: 10.1073/pnas.69.10.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harding MW, Galat A, Uehling DE, Schreiber SL. Nature. 1989;341:758. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- 3.Taunton J, Hassig CA, Schreiber SL. Science. 1996;272:408. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 4.Griffith EC, Su Z, Turk BE, Chen S, Chang YH, Wu Z, Biemann K, Liu JO. Chem Biol. 1997;4:461. doi: 10.1016/s1074-5521(97)90198-8. [DOI] [PubMed] [Google Scholar]
- 5.Sin N, Meng L, Wang MQ, Wen JJ, Bornmann WG, Crews CM. Proc Natl Acad Sci U S A. 1997;94:6099. doi: 10.1073/pnas.94.12.6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Science. 2010;327:1345. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 7.Low WK, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO. Mol. Cell. 2005;20:709. doi: 10.1016/j.molcel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Bordeleau ME, Matthews J, Wojnar JM, Lindqvist L, Novae O, Jankowsky E, Sonenberg N, Northcote P, Teesdale-Spittle P, Pelletier J. Proc Natl Acad Sci U S A. 2005;102:10460. doi: 10.1073/pnas.0504249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawatani M, Okumura H, Honda K, Kanoh N, Muroi M, Dohmae N, Takami M, Kitagawa M, Futamura Y, Imoto M, Osada H. Proc Natl Acad Sci U S A. 2008;105:11691. doi: 10.1073/pnas.0712239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, Tani T, Horinouchi S, Yoshida M. Nat Chem Biol. 2007;3:576. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 11.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. Nature. 2000;403:623. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 12.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Nature. 2002;415:141. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 13.Yates JR, Ruse CI, Nakorchevsky A. Annu Rev Biomed Eng. 2009;11:49. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 14.Schulze WX, Usadel B. Annu Rev Plant Biol. 2010;61:491. doi: 10.1146/annurev-arplant-042809-112132. [DOI] [PubMed] [Google Scholar]
- 15.Cox J, Mann M. Annu Rev Biochem. 2011;80:273. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- 16.Miyagi M, Rao KC. Mass Spectrom Rev. 2007;26:121. doi: 10.1002/mas.20116. [DOI] [PubMed] [Google Scholar]
- 17.Ong SE, Kratchmarova I, Mann M. J Proteome Res. 2003;2:173. doi: 10.1021/pr0255708. [DOI] [PubMed] [Google Scholar]
- 18.Ong SE, Schenone M, Margolin AA, Li X, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, Tolliday NJ, Koehler AN, Marcaurelle LA, Golub TR, Gould RJ, Schreiber SL, Carr SA. Proc Natl Acad Sci U S A. 2009;106:461. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. Nat Biotechnol. 2005;23:329. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 20.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Nat Biotechnol. 2007;25:10354. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 21.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F. Nature. 2009;461:614. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 22.Qume M. Methods Mol Biol. 1999;106:3. doi: 10.1385/0-89603-530-1:3. [DOI] [PubMed] [Google Scholar]
- 23.Hulme EC, Trevethick MA. Br J Pharmacol. 2010;161:1219. doi: 10.1111/j.1476-5381.2009.00604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hulme EC. Methods Mol Biol. 1999;106:139. doi: 10.1385/0-89603-530-1:139. [DOI] [PubMed] [Google Scholar]
- 25.Benovic JL, Shorr RG, Caron MG, Lefkowitz RJ. Biochemistry. 1984;23:4510. doi: 10.1021/bi00315a002. [DOI] [PubMed] [Google Scholar]
- 26.Shorr RG, Strohsacker MW, Lavin TN, Lefkowitz RJ, Caron MG. J Biol Chem. 1982;257:12341. [PubMed] [Google Scholar]
- 27.Shorr RG, Lefkowitz RJ, Caron MG. J Biol Chem. 1981;256:5820. [PubMed] [Google Scholar]
- 28.Cho TM, Hasegawa J, Ge BL, Loh HH. Proc Natl Acad Sci U S A. 1986;83:4138. doi: 10.1073/pnas.83.12.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama H, Shikano H, Aoyama T, Amano T, Kanaoka Y. FEBS Lett. 1986;208:278. doi: 10.1016/0014-5793(86)81032-8. [DOI] [PubMed] [Google Scholar]
- 30.Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. Science. 1984;226:544. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 31.Siekierka JJ, Hung SH, Poe M, Lin CS, Sigal NH. Nature. 1989;341:755. doi: 10.1038/341755a0. [DOI] [PubMed] [Google Scholar]
- 32.Heitman J, Mowa NR, Hall MN. Science. 1991;253:905. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 33.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. Cell. 1994;78:35. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 34.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. Nature. 1994;369:756. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 35.Koltin Y, Faucette L, Bergsma DJ, Levy MA, Cafferkey R, Koser PL, Johnson RK, Livi GP. Mol Cell Biol. 1991;11:1718. doi: 10.1128/mcb.11.3.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bierer BE, Mattila PS, Standaert RF, Herzenberg LA, Burakoff SJ, Crabtree G, Schreiber SL. Proc Natl Acad Sci U S A. 1990;87:9231. doi: 10.1073/pnas.87.23.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Mowa NR, Hall MN. Cell. 1993;73:585. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 38.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Science. 1999;285:901. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 39.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Nature. 2002;418:387. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 40.Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW. Nat Genet. 1999;21:278. doi: 10.1038/6791. [DOI] [PubMed] [Google Scholar]
- 41.Baetz K, McHardy L, Gable K, Tarling T, Reberioux D, Bryan J, Andersen RJ, Dunn T, Hieter P, Roberge M. Proc Natl Acad Sci U S A. 2004;101:4525. doi: 10.1073/pnas.0307122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW. Proc Natl Acad Sci U S A. 2004;101:793. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Gamier P, Prestwich GD, Leonardson A, Garrett-Engele P, Rush CM, Bard M, Schimmack G, Phillips JW, Roberts CJ, Shoemaker DD. Cell. 2004;116:121. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- 44.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, Lorenz TC, Payne GS, Ishihara S, Ohya Y, Andrews B, Hughes TR, Frey BJ, Graham TR, Andersen RJ, Boone C. Cell. 2006;126:611. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 45.Luesch H, Wu TY, Ren P, Gray NS, Schultz PG, Supek F. Chem Biol. 2005;12 doi: 10.1016/j.chembiol.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 46.Hoon S, Smith AM, Wallace IM, Suresh S, Miranda M, Fung E, Proctor M, Shokat KM, Zhang C, Davis RW, Giaever G, St Onge RP, Nislow C. Nat Chem Biol. 2008;4:498. doi: 10.1038/nchembio.100. [DOI] [PubMed] [Google Scholar]
- 47.Bartolomei MS, Corden JL. Mol Cell Biol. 1987;7:586. doi: 10.1128/mcb.7.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Funanage VL. Mol Cell Biol. 1982;2:467. doi: 10.1128/mcb.2.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan JP, Colon ME, Beebe LA, Melancon P. J Cell Biol. 1994;126:65. doi: 10.1083/jcb.126.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. Exp Hematol. 2003;31:1007. [PubMed] [Google Scholar]
- 51.Whitehead I, Kirk H, Kay R. Mol Cell Biol. 1995;15:704. doi: 10.1128/mcb.15.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen DW, Liang XJ, Suzuki T, Gottesman MM. Mol Pharmacol. 2006;69:1383. doi: 10.1124/mol.105.017525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fields S, Song O. Nature. 1989;340:245. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 54.Licitra EJ, Liu JO. Proc. Natl. Acad. Sci. USA. 1996;93:12817. doi: 10.1073/pnas.93.23.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker K, Bleczinski C, Lin H, Salazar-Jimenez G, Sengupta D, Krane S, Cornish VW. Proc Natl Acad Sci U S A. 2002;99:16537. doi: 10.1073/pnas.262420099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Becker F, Murthi K, Smith C, Come J, Costa-Roldan N, Kaufmann C, Hanke U, Degenhart C, Baumann S, Wallner W, Huber A, Dedier S, Dill S, Kinsman D, Hediger M, Bockovich N, Meier-Ewert S, Kluge AF, Kley N. Chem Biol. 2004;11:211. doi: 10.1016/j.chembiol.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 57.Chidley C, Haruki H, Pedersen MG, Muller E, Johnsson K. Nat Chem Biol. 2011;7:375. doi: 10.1038/nchembio.557. [DOI] [PubMed] [Google Scholar]
- 58.Schiff PB, Fant J, Horwitz SB. Nature. 1979;277:665. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- 59.Schiff PB, Horwitz SB. Proc Natl Acad Sci U S A. 1980;77:1561. doi: 10.1073/pnas.77.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garrison JL, Kunkel EJ, Hegde RS, Taunton J. Nature. 2005;436:285. doi: 10.1038/nature03821. [DOI] [PubMed] [Google Scholar]
- 61.Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Science. 1999;286:971. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 62.Tamm I, Hand R, Caliguiri LA. J Cell Biol. 1976;69:229. doi: 10.1083/jcb.69.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sehgal PB, Derman E, Molloy GR, Tamm I, Darnell JE. Science. 1976;194:431. doi: 10.1126/science.982026. [DOI] [PubMed] [Google Scholar]
- 64.Sehgal PB, Darnell JE, Jr, Tamm I. Cell. 1976;9:473. doi: 10.1016/0092-8674(76)90092-1. [DOI] [PubMed] [Google Scholar]
- 65.Zandomeni R, Mittleman B, Bunick D, Ackerman S, Weinmann R. Proc Natl Acad Sci U S A. 1982;79:3167. doi: 10.1073/pnas.79.10.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zandomeni R, Bunick D, Ackerman S, Mittleman B, Weinmann R. J Mol Biol. 1983;167:561. doi: 10.1016/s0022-2836(83)80098-9. [DOI] [PubMed] [Google Scholar]
- 67.Chodosh LA, Fire A, Samuels M, Sharp PA. J Biol Chem. 1989;264:2250. [PubMed] [Google Scholar]
- 68.Marshall NF, Price DH. Mol Cell Biol. 1992;12:2078. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kephart DD, Marshall NF, Price DH. Mol Cell Biol. 1992;12:2067. doi: 10.1128/mcb.12.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marshall NF, Price DW. J Biol Chem. 1995;270:12335. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 71.Marshall NF, Peng J, Xie Z, Price DW. J Biol Chem. 1996;271:27176. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 72.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Genes Dev. 1997;11:2622. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. EMBO J. 1998;17:7395. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Misumi Y, Miki K, Takatsuki A, Tamura G, Ikehara Y. J Biol Chem. 1986;261:11398. [PubMed] [Google Scholar]
- 75.Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y. J. Biol Chem. 1988;263:18545. [PubMed] [Google Scholar]
- 76.Lippincott-Schwartz J, Donaldson JG, Schweizer A, Berger EG, Hauri HP, Yuan LC, Klausner RD. Cell. 1990;60:821. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- 77.Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Cell. 1989;56:801. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wood SA, Park JE, Brown WJ. Cell. 1991;67:591. doi: 10.1016/0092-8674(91)90533-5. [DOI] [PubMed] [Google Scholar]
- 79.Lippincott-Schwartz J, Yuan L, Tipper C, Amherdt M, Orci L, Klausner RD. Cell. 1991;67:601. doi: 10.1016/0092-8674(91)90534-6. [DOI] [PubMed] [Google Scholar]
- 80.Donaldson JG, Lippincott-Schwartz J, Bloom GS, Kreis TE, Klausner RD. J Cell Biol. 1990;111:2293. doi: 10.1083/jcb.111.6.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Donaldson JG, Lippincott-Schwartz J, Klausner RT. J Cell Biol. 1991;112:579. doi: 10.1083/jcb.112.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Serafini T, Stenbeck G, Brecht A, Lottspeich F, Orci L, Rothman JE, Wieland FT. Nature. 1991;349:215. doi: 10.1038/349215a0. [DOI] [PubMed] [Google Scholar]
- 83.Orci L, Tagaya M, Amherdt M, Perrelet A, Donaldson JG, Lippincott-Schwartz J, Klausner RD, Rothman JE. Cell. 1991;64:1183. doi: 10.1016/0092-8674(91)90273-2. [DOI] [PubMed] [Google Scholar]
- 84.Stearns T, Willingham MC, Botstein D, Kahn RA. Proc Natl Acad Sci U S A. 1990;87:1238. doi: 10.1073/pnas.87.3.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Serafini T, Orci L, Amherdt M, Brunner M, Kahn RA, Rothman JE. Cell. 1991;67:239. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- 86.Donaldson JG, Cassel D, Kahn RA, Klausner RD. Proc Natl Acad Sci U S A. 1992;89:6408. doi: 10.1073/pnas.89.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Donaldson JG, Kahn RA, Lippincott-Schwartz J, Klausner RD. Science. 1991;254:1197. doi: 10.1126/science.1957170. [DOI] [PubMed] [Google Scholar]
- 88.Donaldson JG, Finazzi D, Klausner RD. Nature. 1992;360:350. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- 89.Morinaga N, Tsai SC, Moss J, Vaughan M. Proc Natl Acad Sci U S A. 1996;93:12856. doi: 10.1073/pnas.93.23.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morinaga N, Moss J, Vaughan M. Proc Natl Acad Sci U S A. 1997;94:12926. doi: 10.1073/pnas.94.24.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Togawa A, Morinaga N, Ogasawara M, Moss J, Vaughan M. J Biol Chem. 1999;274:12308. doi: 10.1074/jbc.274.18.12308. [DOI] [PubMed] [Google Scholar]
- 92.Peyroche A, Paris S, Jackson CL. Nature. 1996;384:479. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- 93.Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL. Mol Cell. 1999;3:275. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- 94.Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, Goodrich JA, Liu JO, et al. Nat Chem Biol. 2011;7:182. doi: 10.1038/nchembio.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kupchan SM, Court WA, Dailey RG, Jr, Gilmore CJ, Bryan RF. J. Am. Chem. Soc. 1972;94:7194. doi: 10.1021/ja00775a078. [DOI] [PubMed] [Google Scholar]
- 96.Leuenroth SJ, Okuhara D, Shotwell JD, Markowitz GS, Yu Z, Somlo S, Crews CM. Proc. Natl. Acad. Sci. USA. 2007;104:4389. doi: 10.1073/pnas.0700499104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McCallum C, Kwon S, Leavitt P, Shen DM, Liu W, Gurnett A. Immunobiology. 2007;212:549. doi: 10.1016/j.imbio.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 98.Corson TW, Cavga H, Aberle N, Crews CM. Chembiochem. 2011;12:1767. doi: 10.1002/cbic.201100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McCallum C, Kwon S, Leavitt P, Shoop W, Michael B, Felcetto T, Zaller D, O'Neill E, Frantz-Wattley B, Thompson C, Forrest G, Carballo-Jane E, Gurnett A. Therapy. 2005;2:261. [Google Scholar]
- 100.Vispe S, DeVries L, Creancier L, Besse J, Breand S, Hobson DJ, Svejstrup JQ, Annereau JP, Cussac D, Dumontet C, Guilbaud N, Barret JM, Bailly C. Mol. Cancer Ther. 2009;8:2780. doi: 10.1158/1535-7163.MCT-09-0549. [DOI] [PubMed] [Google Scholar]
- 101.Creese I, Snyder SH. J Pharmacol Exp Ther. 1973;194:205. [PubMed] [Google Scholar]
- 102.Speirs AL. Lancet. 1962;1:303. doi: 10.1016/s0140-6736(62)91248-5. [DOI] [PubMed] [Google Scholar]
- 103.Taussig HB. Sci Am. 1962;207:29. doi: 10.1038/scientificamerican0862-29. [DOI] [PubMed] [Google Scholar]