Summary
Variation in the IL4 gene has been associated with parastic infections, but has not been studied in Bolivians infected with Trypanosoma cruzi. Our results suggest that variation at IL4 influences susceptibility to T. cruzi infection in Bolivians.
Keywords: Chagas disease, IL4, Trypanosoma cruzi infection, IgE, Bolivians
To the Editor
Chagas disease, caused by the parasite Trypanosoma cruzi (T. cruzi), affects 10 to 12 million people each year in Latin America, with Bolivia having the highest prevalence of infection (1 and references therein). In the chronic phase, Chagas infection may present as an indeterminate form in which 60% of infected individuals remain asymptomatic despite having positive serological reactions for T. cruzi. In the remaining 40% of Chagas disease patients, tissue inflammation leads to organ damage, affecting the cardiac, digestive, or nervous systems up to 25 years after initial infection. Several studies identified genetic markers for disease establishment and progression in Venezuelans, Brazilians, Peruvians, Colombians, and Mexicans 2, but no genetic studies have been conducted previously in Bolivians.
Cytokines produced in response to T. cruzi infection appear to modulate disease progression by enhancing or inhibiting parasite replication in a variety of cell types. In particular, the T helper 2 (Th2) cytokine interleukin 4 (IL-4) maintains inflammation and parasite persistence in Chagas disease 3, while Th1 cytokines maintain control of parasitism 4 but can also contribute toward the development of chronic myocarditis 5.
To determine whether genetic variation at the IL4 gene is associated with T. cruzi infection in Bolivians, we performed a re-sequencing study of an approximately 12 Kb region around the IL4 locus, including 470 base pairs (bp) of coding (exon) sequence, 368 bp of 5′ UTR, 82 bp of 3′ UTR and 11453 bp of intronic sequence. The study included 110 individuals from the Department of Cochabamba, Bolivia, with infection status serologically confirmed by two different diagnostic tests (HAI Chagas PolyChaco-Argentina and IFI Biocientifica, S.A., Argentina). Each subject was classified according to the serologic results as a case (positive serology) or a control (negative serology). Our sample included 74 cases (mean age 49, range 18-83 years, 47 female and 27 male) and 36 controls (mean age 45, range 18-73 years, 18 male and 18 female). DNA from one chimpanzee was used to infer the ancestral state at each polymorphic position.
We identified a total of 45 polymorphic sites (Table 1); all polymorphisms were in Hardy-Weinberg equilibrium. Only three variants were located in the coding region: two previously reported silent changes (Leu15Leu in exon 1 and Asp111 Asp in exon 3) and one novel amino acid replacement (Leu120Ser in exon 3). All three coding variants were observed only in cases. Sixteen variants (including the three exonic SNPs) were singletons, and all 16 occurred only in the cases. Two singletons were present in one individual; therefore, 15 out of 148 “case” chromosomes and none of the 72 “control” chromosomes carried singletons (Fisher exact test, P = 0.0066). The fact that all 16 singletons were confined to the case group is striking and suggests that rare alleles may contribute to altered IL4 expression and/or function and ultimately to disease susceptibility. Overall, 19 of the 45 polymorphisms (including 14 of the 16 singletons) are newly reported. Of the remaining variants with available frequency information (23 of 26), 20 have been previously reported in both African Americans and Europeans, one in African Americans only, one in Europeans only, and one in Asians only (Database of Single Nucleotide Polymorphisms (dbSNP), accessed July 2010). Chromatogram sequence traces of the twelve novel singletons not confirmed by multiple sequence reads are shown in the online repository (Figure E1).
Table 1.
Polymorphisms in the IL4 gene that are present in 110 Bolivians. The- 590 C/T SNP is shown on a shaded background. SNP position is in reference to AF395008 in GenBank. DAF, derived allele frequency.
| Location | SNP position | rs number | Alleles Ancestral/Derived | DAF | Alternate Name |
|---|---|---|---|---|---|
| Promoter/5′ region | 10889 | rs71645903 | C/G | <0.01 | |
| 10905 | rs71645904 | G/A | 0.19 | ||
| 10926 | rs71645905 | A/G | <0.01 | ||
| 10957 | rs10065221 | A/G | 0.02 | ||
| 10960 | rs10058157 | T/C | 0.02 | ||
| 11781 | rs71645906 | C/T | <0.01 | ||
|
| |||||
| 11903 | rs2243250 | C/T | 0.54 | −590 C/T | |
|
| |||||
| 12348 | rs17772853 | C/T | 0.02 | ||
|
| |||||
| Exon 1 | 12459 | rs2070874 | C/T | 0.53 | +33 C/T |
| 12536 | rs2243251 | A/G | <0.01 | Leu15Leu | |
|
| |||||
| Intron 2 | 13172 | rs71645907 | C/G | <0.01 | |
| 13239 | rs71645908 | C/T | <0.01 | ||
| 13322 | rs71645909 | A/G | <0.01 | ||
| 13571 | rs734244 | C/T | 0.59 | ||
| 13571 | rs71645910 | C/T | <0.01 | ||
| 13575 | rs71645911 | G/A | 0.01 | ||
| 15508 | rs2227284 | T/G | 0.50 | ||
| 15962 | rs2227282 | C/G | 0.53 | ||
| 16082 | rs2243263 | G/C | 0.48 | ||
| 16510 | rs71645912 | G/C | 0.03 | ||
| 16669 | rs2243267 | G/C | 0.52 | ||
| 16717 | rs71645913 | C/T | <0.01 | ||
| 16746 | rs2243268 | A/C | 0.51 | ||
| 16783 | rs9282745 | T/A | <0.01 | ||
| 16892 | rs2243270 | G/A | 0.66 | ||
| 17036 | rs2243308 | C/T | 0.01 | ||
| 17477 | rs71645914 | C/A | <lt0.01 | ||
| 17615 | rs2243274 | A/G | 0.49 | ||
|
| |||||
| Exon 3 | 18338 | rs35648164 | C/T | 0.01 | Asp111Asp |
| 18364 | rs71645915 | T/C | <0.01 | Leu120Ser | |
|
| |||||
| Intron 3 | 18383 | rs71645916 | C/A | <0.01 | |
| 190581 | 1/2or3 | 0.49 | 70 bp repeat | ||
| 19108 | rs71645917 | T/C | 0.06 | ||
| 19118 | rs71645918 | G/A | <0.01 | ||
| 19178 | rs2243281 | T/C | 0.14 | ||
| 19376 | rs2243283 | C/G | 0.06 | ||
| 19464 | rs71645919 | C/G | <0.01 | ||
| 19775 | rs2243284 | G/A | 0.50 | ||
| 19776 | rs2243285 | G/T | 0.17 | ||
| 20388 | rs71645920 | A/T | 0.01 | ||
| 20727 | rs2243288 | A/G | 0.59 | ||
| 20915 | rs2243289 | A/G | 0.53 | ||
| 20952 | rs2243290 | C/A | 0.56 | ||
|
| |||||
| 3′ region | 21659 | rs71645921 | A/G | <0.01 | |
| 21766 | rs2243291 | C/G | 0.48 | ||
| 22574 | rs2243293 | A/G | 0.69 | ||
| 22590 | rs2243294 | G/A | 0.31 | ||
The ancestral state of the 70 bp repeat (1 copy) is rare in humans (0.01 in Bolivians); the frequency of the minor (3 copy) allele is shown instead.
Polymorphism levels, as summarized by nucleotide diversity (π) and Watterson's estimator of the population mutation rate parameter θ (θW) are unremarkable and similar to genome-wide averages in both the cases (π = 0.00079 and θW = 0.00069) and controls (π = 0.00068 and θW = 0.00018). The Tajima's D (TD) statistic, summarizing information on the allele frequency spectrum, is expected to be near zero under the neutral equilibrium model. A positive value indicates an excess of intermediate frequency variants, whereas a negative value indicates a skew toward rare variants. TD values in cases and controls were compared to an empirical distribution of TD values in 318 genes re-sequenced in 24 African Americans and 23 Europeans (University of Washington–Fred Hutchinson Cancer Research Center Variation Discovery Resource, or Seattle SNPs). The TD value in cases (0.46) falls in the top 7% and 42% of TD values, but TD in controls (1.78) falls in the top 2% and 5% of TD values among African Americans and Europeans, respectively, suggesting an excess of intermediate frequency variants in controls.
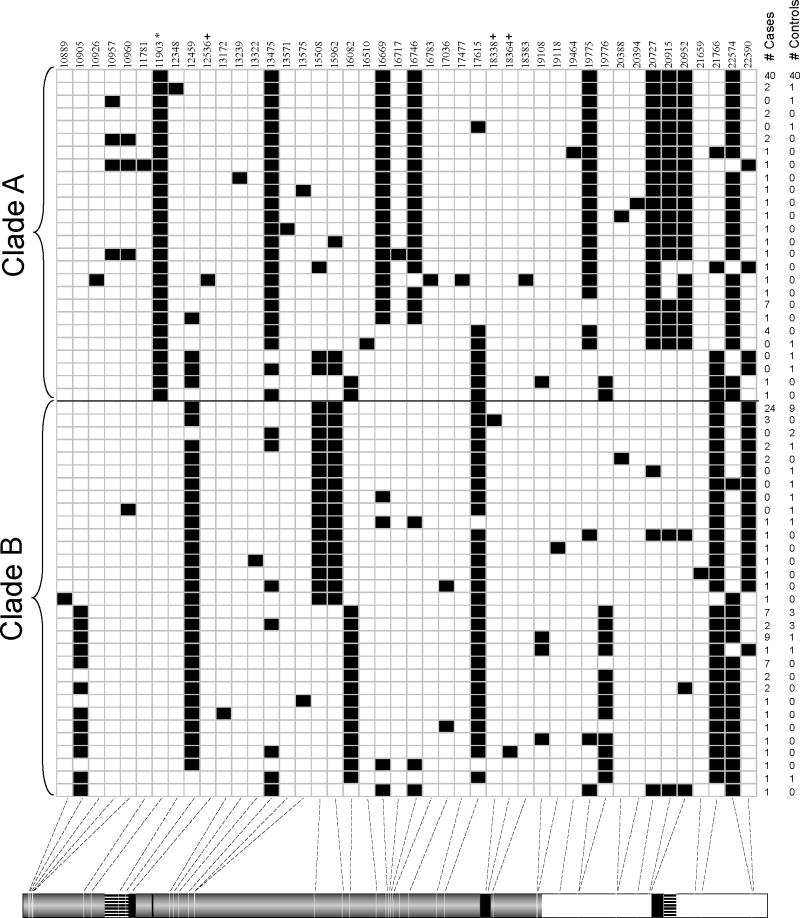
The IL4 gene is characterized by strong linkage disequilibrium (LD) in populations of European, Asian, and African descent, and a correspondingly strong haplotype structure. In Asian and European HapMap samples, each population has two major haplotype classes that are tagged by a SNP located in the promoter region (-590C/T; rs2243250), which has been associated with increased gene transcription and elevated total serum IgE levels in subjects with asthma and atopy 6, 7, as well as malaria 8. Curiously, the frequencies of the haplotype classes are essentially reversed in European and non-European HapMap (Phase I) samples, in which the derived -590T allele at SNP rs2243250 occurs at 16% frequency in Europeans (CEU), 75% in Asians (Chinese [CHB] and Japanese [JPT] combined), and 83% in Africans (Yorubans [YRI]). The frequency of the -590T allele is 54% in the Bolivian sample (Table 1), intermediate to the frequency in the HapMap samples. Because of the unusual haplotype structure in worldwide samples and the functional role associated with the -590C/T SNP 7, we stratified the haplotypes present in the Bolivians based on this SNP and refer to the two classes of haplotypes as Clade A (defined by the derived T allele) and Clade B (defined by the ancestral C allele) (Figure 1; rs2243250, position 11903). A visual representation of haplotypes at the IL4 locus is shown in Figure 1. In Bolivians, 49% of case chromosomes and 64% of control chromosomes belong to Clade A (Fisher exact test, P = 0.033; OR = 0.54, 95% CI 0.3001, 0.9554). The ancient estimated age of divergence for Clade A from Clade B (40,000 years ago, range=25,204-48,469; see online repository for details) places the split prior to the spread of modern humans into the new world. Clade A haplotypes form a relatively homogeneous group, in which nearly all observed haplotypes differ by only one or two SNPs from the common form despite the ancient estimate of divergence. Clade B haplotypes, on the other hand, are more heterogeneous and more divergent from each other (Figure 1 and online repository Figure E2). The combined observations of a relatively homogeneous group of haplotypes in the T. cruzi non-infected controls and a significant skew in the frequency spectrum toward intermediate frequency alleles in these individuals suggest that one or more variants on Clade A haplotypes provides protection against T. cruzi infection and, as a result, natural selection has favored the maintenance of these haplotypes in the Bolivian population. The selected SNP could be the functional -590C/T SNP or any other SNP that is in strong LD with -590C/T.
Figure 1.
Visual representation of haplotypes at the IL4 locus. Haplotypes are sorted by the derived (Clade A) or ancestral (Clade B) allele at -590C/T (designated by *). Three exonic SNPs are indicated by a “+”. White squares correspond to ancestral alleles and black squares to derived alleles. A schematic of the IL4 gene is shown at the bottom of the figure; filled rectangles show exons (drawn to scale).
Alleles or haplotypes associated with increased expression of the IL4 gene, and a correspondingly stronger Th2 immune response, could provide a substrate for natural selection. For example, approximately 20% of T. cruzi affected individuals ultimately develop the cardiac form of Chagas disease, which can result in heart failure. A study of IL-10 and IFN-γ responses in 111 individuals with either the indeterminate or the cardiac form of Chagas disease revealed that high levels of IFN-γ and low levels of IL-10 are associated with heart damage, with the highest levels of IFN-γ corresponding to the most severe heart pathologies 5. These results suggest that a strong Th1 response, while important in limiting parasite replication during the acute phase 4, can contribute to severe heart disease in the chronic phase. Consequently, during the indeterminate phase of infection, IL-4-producing T cells may be advantageous by balancing parasitism and tissue integrity, although this idea is at odds with studies showing a protective role for Th1 cytokine responses (and a counterproductive role for Th2 responses) with regard to control of parasite infection. Unfortunately, we do not have information other than T. cruzi infection status for the individuals in our study so we cannot make predictions about outcomes based on IL4 genotype or directly address these seemingly paradoxical findings. However, it would not be surprising if the balance between Th1/Th2/T regulatory cytokines has different (opposing) influences on the likelihood of infection with T. cruzi and on the subsequent morbidity associated with Chagas disease.
Our study is the first to characterize variation in the IL4 gene in Bolivians and to examine genetic associations between IL4 and Trypanosoma cruzi infection in any population. We selected this gene for our study because of the key role Th2 cytokines play in the immune response to intracellular pathogens, such as T. cruzi, and because a promoter polymorphism (C-590T; rs2243250) in this gene has been associated with differences in expression level 7 and Th2-mediated allergic diseases (reviewed in 9), and because other polymorphisms in IL4 have been associated with infection with other intracellular pathogens, including P. falciparum 8 and L. chagasi. Although our sample size is small in terms of association studies, our results suggest that the asthma/atopy associated -590T allele is a marker for IL4 haplotypes that confer protection against T. cruzi infection (positive serology). Further studies are warranted to clarify the relationship between genotype at this locus and the development of overt Chagas disease and subsequent clinical outcomes.
Supplementary Material
Acknowledgments
Supported by R01 HL072414. E.E.T. is supported by T32 HL007605.
Abbreviations Used
- IgE
immunoglobulin E
- IL4
interleukin 4
- LD
linkage disequilibrium
- MAF
minor allele frequency
- SNP
single nucleotide polymorphism
- Th1 or 2
T helper 1 or 2
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Outlook. Chagas Disease. Nature. 2010;465:S3–S22. doi: 10.1038/465S3a. [DOI] [PubMed] [Google Scholar]
- 2.Cunha-Neto E, Nogueira LG, Teixeira PC, Ramasawmy R, Drigo SA, Goldberg AC, et al. Immunological and non-immunological effects of cytokines and chemokines in the pathogenesis of chronic Chagas disease cardiomyopathy. Mem Inst Oswaldo Cruz. 2009;104(1):252–8. doi: 10.1590/s0074-02762009000900032. [DOI] [PubMed] [Google Scholar]
- 3.Ribeiro BM, Crema E, Rodrigues V., Jr Analysis of the cellular immune response in patients with the digestive and indeterminate forms of Chagas' disease. Hum Immunol. 2008;69:484–9. doi: 10.1016/j.humimm.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Reed SG. In vivo administration of recombinant IFN-gamma induces macrophage activation, and prevents acute disease, immune suppression, and death in experimental Trypanosoma cruzi infections. J Immunol. 1988;140:4342–7. [PubMed] [Google Scholar]
- 5.Gomes JA, Bahia-Oliveira LM, Rocha MO, Martins-Filho OA, Gazzinelli G, Correa-Oliveira R. Evidence that development of severe cardiomyopathy in human Chagas' disease is due to a Th1-specific immune response. Infect Immun. 2003;71:1185–93. doi: 10.1128/IAI.71.3.1185-1193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabesch M, Tzotcheva I, Carr D, Hofler C, Weiland SK, Fritzsch C, et al. A complete screening of the IL4 gene: novel polymorphisms and their association with asthma and IgE in childhood. J Allergy Clin Immunol. 2003;112:893–8. doi: 10.1016/j.jaci.2003.08.033. [DOI] [PubMed] [Google Scholar]
- 7.Rosenwasser LJ, Klemm DJ, Dresback JK, Inamura H, Mascali JJ, Klinnert M, et al. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy. 1995;25(2):74–8. doi: 10.1111/j.1365-2222.1995.tb00428.x. discussion 95-6. [DOI] [PubMed] [Google Scholar]
- 8.Verra F, Luoni G, Calissano C, Troye-Blomberg M, Perlmann P, Perlmann H, et al. IL4-589C/T polymorphism and IgE levels in severe malaria. Acta Trop. 2004;90:205–9. doi: 10.1016/j.actatropica.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006;7:95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.