Abstract
In the last decade, basic research in chemoreceptor genetics and neurobiology have revolutionized our understanding of individual differences in chemosensation. From an evolutionary perspective, chemosensory variations appear to have arisen in response to different living environments, generally in the avoidance of toxins and to better detect vital food sources. Today, it is often assumed that these differences may drive variable food preferences and choices, with downstream effects on health and wellness. A growing body of evidence indicates chemosensory variation is far more complex than previously believed. However, just because a genetic polymorphism results in altered receptor function in cultured cells or even behavioral phenotypes in the laboratory, this variation may not be sufficient to influence food choice in free living humans. Still, there is ample evidence to indicate allelic variation in TAS2R38 predicts variation in bitterness of synthetic pharmaceuticals (e.g., propylthiouracil) and natural plant compounds (e.g., goitrin), and this variation associates with differential intake of alcohol and vegetables. Further, this is only one of 25 unique bitter taste genes (TAS2Rs) in humans, and emerging evidence suggests other TAS2Rs may also contain polymorphisms that a functional with respect to ingestive behavior. For example, TAS2R16 polymorphisms are linked to the bitterness of naturally occurring plant compounds and alcoholic beverage intake, a TAS2R19 polymorphism predicts differences in quinine bitterness and grapefruit bitterness and liking, and TAS2R31 polymorphisms associate with differential bitterness of plant compounds like aristolochic acid and the sulfonyl amide sweeteners saccharin and acesulfame-K. More critically with respect to food choices, these polymorphisms may vary independently from each other within and across individuals, meaning a monolithic one-size-fits-all approach to bitterness needs to be abandoned. Nor are genetic differences restricted to bitterness. Perceptual variation has also been associated with polymorphisms in genes involved in odors associated with meat defects (boar taint), green/grassy notes, and cilantro, as well as umami and sweet tastes (TAS1R1/2/3). Here, a short primer on receptor genetics is provided, followed by a summary of current knowledge, and implications for human ingestive behavior are discussed.
1. Introduction
The term `taste' is often used by the public to describe flavor, the integrated sensation arising from the combination of input from the taste, olfactory and somatosensory systems (Delwiche, 2004; Mattes, 2009). `Taste' in this colloquial sense is reported by consumers as the greatest factor in food purchase decisions (IFIC, 2012). While `price' and `healthfulness' have grown in their influence in recent year, as of 2012, `taste' remains the top consideration in purchase decisions (IFIC, 2012). A substantial body of evidence indicates individuals differ greatly in flavor perception, due in part to genetic variations in chemoreceptor genes. These differences in biology have been implicated as drivers of food choice, highlighting the need for behavioral scientists, sensory practitioners and product developers to be aware of these differences. Collectively, these data also suggest variability across individuals is not a source of noise to be minimized in sensory testing; rather, this variation is what is interesting, both in its own right from a basic science standpoint, and in translation as a potential driver of differential food choices (i.e. market segmentation).
Recently it has become clear that variation in chemosensation is far more complex than previously believed. Of the 25 unique bitter taste genes in humans (TAS2Rs), the most studied is TAS2R38, which explains differential bitterness of synthetic thiourea compounds (e.g. propylthiouracil) and natural plant toxins (e.g. goitrin). Notably, TAS2R38 variation also associates with differential intake of both alcohol and vegetables. However, this is just one of many examples to emerge within the last few years. Nor are such differences limited to bitterness, as differences in chemosensory perception have also been documented for odor (OR7D4), as well as umami and sweet tastes (TAS1R1/TAS1R3 and TAS1R2/TAS1R3 respectively). Other evidence suggests variation in the gustin (CA6) and gustducin (GNAT3) genes may also associate with variation in the intensity of bitter and sweet sensations, respectively.
Functional variation in taste, smell and oral somatosensory systems may affect myriad sensory attributes elicited by food. Much of this variation has a genetic basis, and as personalized genomics becomes more feasible, it is likely that food manufacturers will target products to particular groups based on preferences predicted by genetics. The largest challenge will be to assemble a broad range of genetic variability into a small number of groups (segments) that manufacturers can target on the basis of differential preferences.
Additionally, growing evidence suggests `taste' receptors are expressed in many other tissues in the body (we place taste in quotes to emphasize activation of these receptors may not elicit a conscious perception). The degree to which extra-oral chemoreceptors influence metabolic processes or ingestive behavior via postprandial learning are just beginning to be explored; nor do we have a good understanding how polymorphisms in the genes encoding these receptors may influence learning or metabolism. This is an extremely active area of research (e.g. (Behrens & Meyerhof, 2010; Breer, Eberle, Frick, Haid, & Widmayer, 2012; Clark, Liggett, & Munger, 2012; Dotson, Vigues, Steinle, & Munger, 2010; Dotson, et al., 2008; Sclafani, Glass, Margolskee, & Glendinning, 2010)), so it is likely our understanding will increase greatly over the coming decade.
In this review, a brief overview on receptor genetics is provided, followed by a summary of current knowledge on functional variation to date with examples for taste, smell and oral somatosensation. Throughout, we highlight the implications for human ingestive behavior outside of the laboratory, as well as gaps in our current knowledge. The next two sections provide an overview of some foundational biological concepts relevant to chemoreceptor genetics; readers who are comfortable with these concepts may want to skip ahead to section 4.
2. Approaches from Quantitative and Molecular Genetics
Although learning and prior exposure may play a role, a substantial proportion of the observed variation in chemosensory perception may be attributed to genetic variation. Researchers have attempted to elucidate these mechanisms by using several distinct but complementary methods from quantitative and molecular genetics. These are detailed below in sections 2.2 and 2.3 after a brief discussion of nomenclature.
2.1 Imperfect relationships between phenotypes and genotypes
Understandably, many studies in the chemical senses confuse phenotypes and genotypes, so it is worth explicitly defining these terms here. According to the National Human Genome Research Institute, a phenotype is an observable trait within an individual. Typical examples would include height or blood type; the classic chemosensory example is the ability to taste (or not taste) phenylthiocarbamide (PTC) at low concentrations. From a behavioral perspective, anything that can be measured reliably and stably may be considered a potential phenotype (e.g. salt detection thresholds, suprathreshold bitterness intensity, sweet liking, food intake patterns, blood pressure, etc). In contrast, genotype refers to the collection of genes an individual carries, and in the case of a single nucleotide, it can refer to the two alleles an individual inherits from their mother and father at that specific position. Critically, the phenotype is the product of both genetics and the environment, and some observable traits are primarily genetic, while others are mostly due to environment. Also, an observable phenotype may arise from multiple diverse genotypes. Thus, phenotype–genotype associations may be imperfect, and underlying genotypes cannot always be inferred from the observed phenotype (e.g see Table 3 in (Hayes, Bartoshuk, Kidd, & Duffy, 2008)).
2.2 Quantitative Genetics – Twin Designs
Traditionally, twin studies were used to determine the genetic component of a trait (e.g., (Wysocki & Beauchamp, 1984)). While there are now many different twin study designs (see (van Dongen, Slagboom, Draisma, Martin, & Boomsma, 2012) for a review), in the classical twin study design, resemblance in monozygotic twins (`identical' twins who share all their segregating genetic material) is compared to that of dizygotic twins (`fraternal' twins who share half their genetic material on average). These types of designs allow heritability estimates to be calculated, and have been extremely important in determining the amount of variation of a trait that is attributable to genetic influences; in these models, the remaining variance is assumed to arise from environmental factors. For example, twin studies in the UK suggest that in females, the heritability is high (over 40%) for intake of certain foods such as garlic, coffee, and fruit and vegetables (Teucher, et al., 2007). Other groups have found a genetic influence on bread choice and frequency of consumption (Hasselbalch, et al., 2010) and even liking and consumption of chocolate and other sweet foods (Keskitalo, 2007 #461}. Food neophobia (a fear of new foods) also appears to have a strong heritable component (Cooke, Haworth, & Wardle, 2007; Knaapila, et al., 2011). However, while extremely useful, these designs do not provide information about the specific gene or genes responsible; rather, they provide a quantitative estimate of the overall heritability of a trait without identifying the molecular basis of the trait.
2.3 Molecular Genetics – Candidate Gene, Candidate SNP, and Genome Wide Association Studies
With the advent of PCR1 and subsequent advances in molecular biology, new genetic techniques became possible which allow identification of the individual genetic components that underlie these heritability estimates from quantitative genetics. These molecular approaches attempt to pinpoint the causal variation to a particular location within the genome. Candidate gene approaches require background knowledge of the mechanism of the trait of interest, and attempt to examine variation in a particular gene in relation to the trait or function of interest. In the case of chemosensory receptors, an example of this approach would be the examination of variations in putative odorant receptors in relation to olfactory ability (e.g. (Keller, Zhuang, Chi, Vosshall, & Matsunami, 2007)). Genome-wide association studies (GWAS) are slightly different from the candidate gene approach in that they do not require a priori knowledge. Rather, the phenotype or clinical outcome (for example, detection threshold) is examined in relation to thousands of variants on a microarray. Variants that associate with the measure of interest are identified, independent of theory or biological mechanism. Not all variants are examined, as some polymorphisms are in linkage disequilibrium (LD) with others. LD refers to the phenomena where loci that are in close proximity on the same chromosome are often inherited together. This means that one individual polymorphism may be highly predictive of others (i.e. the polymorphisms not statistically independent). These patterns can be exploited to make the process more efficient, as it means a smaller number of variants can be tested, if they are known to predict other unmeasured variants (Wilkening, Chen, Bermejo, & Canzian, 2009). However, due to the large number of individual mutations being tested, the chance of false-positives remains quite high. Accordingly, the correction of multiple comparisons means that an extremely small p-value is required to declare a relationship exists. However, this adjustment also means that a true association can sometimes be missed, which is where a candidate gene or candidate polymorphism approach has an advantage, as they are biologically informed rather than atheoretical like GWAS. Each approach has advantages and disadvantages, and both contribute to our understanding how genetic variation contributes to a specific trait of interest.
3. Brief Refresher on Receptor Biology
The lock-and-key model was proposed in 1894 by Emile Fisher to describe enzyme specificity, and it is still a useful way to conceptualize how molecules (ligands) bind to and activate chemoreceptors. Chemoreceptors are found in the mouth, nose and throat, serving as binding sites for myriad volatile and non-volatile chemicals in our environment. Transduction events mediated by these receptors cause signaling cascades that culminate in sensations of taste, smell and irritation. These receptors are made of proteins encoded by genes scattered across multiple chromosomes. Inherited variations in these genes may lead to differences in the translated protein, altering the functionality of the receptor, which may lead to either impaired or enhanced perception of individual chemicals across different people.
3.1 DNA to proteins
The sequence of nucleotides (A,G,C and T) in DNA (Deoxyribonucleic acid) is extremely similar across all humans (i.e., generally conserved), and any two unrelated individuals share about 99.9% of their respective genomes (Feuk, Carson, & Scherer, 2006). The `central dogma of molecular biology' originally advanced by Francis Crick is straightforward: DNA is read in groups of three nucleotides called codons, and these codons are transcribed to an intermediary molecule, Ribonucleic Acid (RNA). RNA is then translated to one amino acid, and the amino acids are joined together to form proteins.
Transcription of DNA into RNA is catalyzed by the enzyme RNA polymerase, which binds to the gene promoter. The promoter is a regulatory region of the gene that binds molecules that activate or repress transcription downstream of the promoter. Genes typically contain regions of introns and exons, and only the exons are expressed as proteins. The DNA double helix is opened into two strands: the sense strand and the antisense strand, and it is the antisense strand that is used as the template from which the complementary messenger RNA (mRNA) molecule is copied, resulting in an RNA molecule (strand) which contains the same information as the other `sense' strand of DNA. This RNA strand is then modified to remove the non-protein-coding introns, and join the exons together, in a post-transcriptional process known as splicing. It is possible to splice different areas of exons together, giving different `splice variants' which generate different forms of a protein (known as isoforms). This allows a single gene to encode more than one protein.
After splicing, mRNA forms a complex with tRNA (transfer RNA) and a ribosome. tRNA molecules recognize specific codons on the mRNA and bind specific amino acids. A peptide bond to the next amino acid is formed and the tRNA molecule moves on to the next codon to join another amino acid. Thus, a chain is formed, ending when a stop codon is reached.
3.2. Genetic differences across individuals: SNPs, Alleles, Haplotypes and CNVs
Spontaneous mutations may arise anywhere in the DNA, causing a change in the amino acid sequence. When such changes are inherited, and are consistently found in the population, they may be referred to as a single nucleotide polymorphism (SNP; pronounced snip). That is, a polymorphism (from Greek, meaning `many forms') is a specific location in DNA where more than one stable form exists across the population. The less common form is called the minor allele, and the proportion with which it occurs is known as the minor allele frequency (MAF). When there is linkage disequilibrium (LD) across multiple SNPs (see previous section), small groups of alleles may be inherited together such that the probability of having one allele is not statistically independent of the probably of having another nearby allele. When groups of alleles on a single strand are inherited this way, it is known as a haplotype. As DNA is double stranded, a pair of haplotypes forms a diplotype, much in the same way a pair of alleles form a genotype.
Depending on where in a codon the change is found, SNPs may result in the translation of a different amino acid. (A SNP does not always lead to an amino acid change, due to redundancy in amino acid coding. When this occurs, it is called a synonymous SNP; conversely, when a nucleotide change results in an amino acid change, it is termed a non-synonymous SNP.) The incorporation of a different amino acid in a polypeptide sequence can alter the function of the resultant protein in several different ways. If the change occurs in the binding pocket, this can affect the ability of the receptor to bind to the chemical (ligand). In the case of taste receptor proteins, the function of the receptor may be reduced or enhanced in different people (e.g., (Bufe, et al., 2005; Duffy, et al., 2004)).
SNPs in noncoding regions, such as promoter regions, can also affect receptor function. One example of this is the differential variation in sweet taste perception that is attributed to SNPs located in the promoter region of the TAS1R1 gene (Fushan, Simons, Slack, Manichaikul, & Drayna, 2009). There is also evidence suggesting SNPs within the promoter region of the gustducin gene (GNAT3) are responsible for some of the variation in sweet perception, although this is yet to be confirmed (Fushan, Simons, Slack, & Drayna, 2010).
Copy number variation (CNV) is another way in which the expression levels of genes and their resultant proteins can vary. Defined as sections of DNA longer than 1kb that are present in variable copies in a genome versus a reference genome, CNVs are stable, inheritable variations that occur when sections of DNA are repeated or deleted, (CNVs can arise through a variety of processes (reviewed by (Hastings, Lupski, Rosenberg, & Ira, 2009)). These can occur in a segment of a gene, one gene or multiple genes. In the case of chemoreceptors, variations in copy numbers can lead to differential expression of receptors, and variation in an individual's capability of detecting the sensation. A high degree of the variation in copy number (as much as 68%) occurs in areas encoding chemoreceptors (Wong, et al., 2007). This estimate suggests that copy number variations may be an important source of individual differences in chemosensation. Indeed, much of the variation in odor receptor (OR) genes are CNVs, and these can differ greatly from person to person. One recent investigation into CNV and smell demonstrated that the person with the greatest number of odorant receptors had 77 more receptors than the person with the fewest (Waszak, et al., 2010) (cited in Keller, 2011). Evolutionary biologists have theorized that the large variation in copy number in OR genes may have arisen from an initial expansion of the repertoire in response to the movement from sea to land, in order to detect air-borne odorants. Then, following the development of trichromatic vision, the sense of smell became less important, lowering the functional constraint on the OR genes, and thus humans have fewer intact copies than a number of other mammals. It appears humans may have undergone two separate `relaxation of constraint' events: old world monkeys and primates (trichromatic vision) have more pseudogenes than new world monkeys (Gilad et al 2004, Liman, 2012), and humans have more pseudogenes than old world monkeys and apes (Gilad et al, 2003, Liman, 2012). However, individual differences in copy number variation are an area that has not generally been well-studied, and very few CNVs have been functionally characterized. Thus, the remainder of the text below will primarily focus on other polymorphisms, mostly SNPs, that have been shown to influence chemosensation.
The following sections summarize some of the current knowledge on genetic variation in chemosensation with a particular emphasis on taste, as it is the best studied and understood to date.
4. Bitter Taste Differences
Variable bitter taste perception is the best-known example of genetic variation in oral sensation. In this section, we review the prototypical example as it relates to ingestive behavior, and then discuss several other more recent findings.
4.1 TAS2R38
The genetic basis for variability in the perception of bitterness was first demonstrated over 8 decades ago (Blakeslee, 1932). The idea that this variation might be driven by natural selection to avoid ingestion of bitter plant toxins is almost as old (see (Wooding, 2006; Wooding, et al., 2010) for reviews). A putative association between human food choice and genetic variation came later (e.g. (Fischer, Griffin, & Kaplan, 1963; Glanville & Kaplan, 1965)). In 2003, the molecular genetics underlying this long studied trait were finally uncovered (Kim, et al., 2003), and influences on ingestive behavior were documented almost immediately (Duffy, et al., 2004). Comprehensive reviews have recently been published on TAS2R38 variation and food choice (Feeney, 2011; Tepper, 2008) so we will only present a few examples here.
4.1.1 Molecular Genetics of TAS2R38
In 2003, Kim and colleagues mapped the ability to detect the thiourea (N-C=S) drug phenylthiocarbamide (PTC) to the TAS2R38 gene (HGNC: 9584; formerly PTC) on chromosome 7 (Kim, et al., 2003). They reported 3 SNPs that form two common haplotypes (Proline–Alanine–Valine, and Alanine–Valine–Isoleucine), and associate with individual differences in detection thresholds. These haplotypes are named for the amino acid substitutions Ala49Pro, Val262Ala, and Ile296Val. The PAV haplotype is ancestral, conferring the ability to detect thiourea (N-C=S) compounds while the AVI variant is less functional. Shortly thereafter, this finding was extended to beyond threshold phenotyping to include suprathreshold bitterness of propylthiouracil (PROP) (Duffy, et al., 2004), the other thiourea compound commonly used in taste research. Subsequently, the ability of PROP to activate hT2R38 was demonstrated in vitro, and differences in suprathreshold bitterness were confirmed in vivo (Bufe, et al., 2005). For a discussion on the difference between threshold and suprathreshold measurement and their respective phenotypes, see (Duffy, Hayes, Bartoshuk, & Snyder, 2009; Hayes & Keast, 2011).
The concept of supertasting is thoroughly intertwined with PROP bitterness, both in the public consciousness, and in the scientific literature (e.g (Bartoshuk, Duffy, & Miller, 1994)). However, it is important to note that elevated chemosensory response across diverse stimuli – the modern understanding of supertasting – can be separated from the definition based on PROP bitterness (Hayes & Keast, 2011). Critically however, TAS2R38 variation does not explain supertasting (Hayes, et al., 2008). Thus, it is inappropriate to talk about `genetic supertasters', either in reference to PAV homozygotes or in general. Detailed discussion of generalized supertasting (hypergeusia) (Hayes & Keast, 2011) and the influence of supertasting on ingestive behavior are beyond the scope of this review, and can be found elsewhere (see (Duffy, 2007; Feeney, 2011)). In the following section, we focus on TAS2R38 gene data as it relates to ingestive behavior.
4.1.2 TAS2R38 variation and alcohol use
The first evidence that TAS2R38 variation associates with alcohol intake came in 2004, when Duffy and colleagues reported annual alcohol intake (based on a summed quantity-frequency measure) associated with TAS2R38 diplotype in a cohort of primarily European ancestry (Duffy, et al., 2004). Specifically, PAV homozygotes consumed less alcohol than heterozygotes, who consumed less that AVI homozygotes, Notably, intake was heteroscedastic across the groups as TAS2R38 diplotype appeared to provide a ceiling, but not a floor; some individuals consumed little or no alcohol regardless of diplotype, but the heaviest drinking PAV homozygote still consumed far less than the heaviest drinking AVI. This finding was generally confirmed in a separate cohort of European-Americans when Hayes et al. reported that both intake frequency of alcoholic beverages and total alcohol intake differed by TAS2R38 diplotype (Hayes, et al., 2011). However, unlike the earlier cohort described by Duffy and colleagues, the intake of heterozygotes was not intermediate between the two homozygous groups. Thus, it is unresolved as to whether the effect of TAS2R38 variation on alcohol intake is dominant or additive. In a cohort of head and neck cancer patients who were predominantly white (presumably European American ancestry), Dotson and colleagues (Dotson, Wallace, Bartoshuk, & Logan, 2012) found the Ala49Pro SNP rs713598 was associated with frequency of consuming drinks containing alcohol and heavy drinking (defined as 6 or more drinks in a day), but not number of typical drinks consumed on a drinking day.
4.1.3 TAS2R38 variation and alcohol misuse and abuse
Wang and colleagues (Wang, et al., 2007) explored TAS2R38 diplotype as a predictor of Maxdrinks in the Collaborative Study of the Genetics of Alcoholism (COGA) cohort. COGA is a large family based study consisting of alcoholics, their family members, and controls from the community (Edenberg & Foroud, 2006). In contrast to the retrospective frequency (Hayes, et al., 2011) and quantity-frequency (Duffy, et al., 2004) measures described above, the Maxdrinks phenotype is assessed by asking “What is the largest number of drinks you have ever had in a 24-hour period?'). Thus, Maxdrinks is a quantitative trait measuring alcohol misuse rather than habitual use or a clinical diagnosis of dependence. In the COGA cohort, TAS2R38 diplotype associated with Maxdrinks in African-Americans but not European-Americans (Wang, et al., 2007). Because Maxdrinks measures the heaviest lifetime consumption rather than typical alcohol intake, a failure to observe an effect among European-Americans does not directly contradict the results of Duffy et al. (2004) and Hayes et al. (2011). It is also worth noting however that environment can severely attenuate genetic influences on alcohol intake (Dick, Rose, Viken, Kaprio, & Koskenvuo, 2001), which suggests unmeasured environmental factors may partially explain differences across studies.
4.1.4 TAS2R38 variation and vegetable bitterness, liking and intake
Reports linking TAS2R38 variation to vegetable intake are mixed. In 2006, Sandell and Breslin reported glucosinolate-containing vegetables evoked more bitterness for PAV homozygotes than for heterozygotes, who reported more bitterness than AVI homozygotes (Sandell & Breslin, 2006), although they did not measure liking or intake. Since increased vegetable bitterness associates with lower intake (e.g.(Dinehart, Hayes, Bartoshuk, Lanier, & Duffy, 2006), it is not surprising three subsequent studies found significant relationships between TAS2R38 variation and vegetable intake. In the Italian arm of the European Prospective Investigation into Cancer and Nutrition (EPIC) study, PA_ carriers (heterozygotes and PA_ homozygotes) ate less cruciferous vegetables than AV_ homozygotes (Sacerdote, et al., 2007). In young adults in the Northeastern United States, Duffy and colleagues found TAS2R38 haplotype associated with differences in intake across multiple assessment methods (Duffy, et al., 2010). Regardless of whether vegetable intake was estimated via 5 nonconsecutive food records, or a retrospective food frequency questionnaire, the PAV carriers consumed fewer vegetable servings and ate vegetables less often than AVI homozygotes. Likewise, in an outpatient study of older postmenopausal Brazilian women, Pro49 allele homozygotes were more likely to be non-consumers of bitter vegetables like chard and arugula (Colares-Bento, et al., 2012). However, not all studies support an association between TAS2R38 variation and vegetable intake (Gorovic, et al., 2011; Timpson, et al., 2005); the reasons for these null findings are unknown, but may relate to cultural foodways and habits that override genetic influences of liking and intake. Notably, there is a gap in the literature regarding hedonic response: no one has tested whether TAS2R38 variation explains differences in liking of sampled vegetables. However, this would seem to be a reasonable assumption, given the large body of evidence linking vegetable bitterness, liking and intake with thiourea phenotypes (see (Feeney, 2011) for a review).
4.2 TAS2R16
4.2.1 Evolutionary basis and molecular genetics of TAS2R16 variation
Receptors encoded by the TAS2R16 gene (HGNC: 14921) are relatively narrowly tuned, responding primarily to compounds containing a beta-glucopyranoside moiety such as salicin, amygdalin and other similar plant derived compounds (Bufe, Hofmann, Krautwurst, Raguse, & Meyerhof, 2002; Meyerhof, et al., 2010). Plants that produce cyanogenic glucosides do so to protect themselves against herbivores, as enzymatic cleavage of these compounds liberates cyanide. Presumably, there was strong evolutionary pressure to detect beta-glucopyranoside containing compounds. The human TAS2R16 gene contains several polymorphic loci, two of which give rise to nonsynonymous amino acid substitutions; critically, one of these altered residues enhances receptor activation in vitro (Soranzo, et al., 2005). The Lys172 variant is more responsive to natural plant-derived toxins than the ancestral Asn172 variant, suggesting positive selection may have occurred to help early humans avoid cyanogenic toxins (Soranzo, et al., 2005). Notably, the less responsive Asn172 is more common in individuals of African ancestry compared to those of European or Asian ancestry, the vast majority of whom carry the Lys172 allele. Similar small changes in TAS2R16 also explain differences in beta-glucopyranoside response across species (Imai, et al., 2012).
4.2.2 TAS2R16 variation and alcohol intake in humans
In addition to differential response to beta-glycopyranosides in vitro and in vivo, clinical and laboratory data have implicated polymorphisms in or near TAS2R16 in alcohol use and abuse. TAS2R16 is proximal to a region on chromosome 7 repeatedly implicated in alcohol dependence in the Collaborative Study of the Genetics of Alcoholism (COGA) cohort via genome wide association. Given this proximity, Hinrichs and colleagues explored whether TAS2R16 contains additional loci associated with alcohol dependence (Hinrichs, et al., 2006). They found the coding SNP rs846664, which results in the Asn172Lys substitution described above, associates with alcohol dependence across multiple diagnostic criteria. As the minor allele frequency of Asn172Lys varies dramatically across ancestry (MAF of 0.6% for European-Americans versus 26% for African-Americans), the clinical relevance of this SNP is mostly restricted to those of African ancestry, although non-African individuals who carry the Asn172 allele are also at higher risk for alcohol dependence. Subsequent analysis extended this association to include the Maxdrinks phenotype (see section 4.1.3), which is a separate phenotype based on alcohol use history rather than a diagnosis of dependence (Wang, et al., 2007).
Associations between alcohol abuse and variation in bitter taste are presumably due to protective effects on habitual intake, and these effects may transient, operating only during critical developmental windows. For example, phenotypic markers of TAS2R variation predict beer intake, but only during the first year of drinking (Intranuovo & Powers, 1998). Likewise, modeling of gene-gene interactions suggest bitter taste genes may act as a stage gate for other genetic risk factors; that is, if you never learn to drink alcohol because of an aversive taste, other risk alleles may be largely irrelevant (Mustavich, Miller, Kidd, & Zhao, 2010). Thus, our group and others have explored the potential role of other SNPs near this region as predictors of alcohol use, rather than abuse. Unlike the rs846664 SNP described above, the rs846672 SNP has a much higher Minor Allele Frequency in European-Americans (28%), and is located 4.5 kilobases downstream from TAS2R16. In a laboratory study where self reported alcohol intake was collected using a validated semiquantitative food frequency questionnaire, this SNP was a significant predictor of both intake frequency and overall intake in a cohort of primarily European ancestry (Hayes, et al., 2011). The high intake group drank alcohol twice as frequently as the low intake group. Although rapid progress has been made on the structure function relationships for hT2R16 and its' ligands in the past 5 years (eg (Meyerhof, et al., 2010; Sakurai, Misaka, Ishiguro, et al., 2010; Sakurai, Misaka, Ueno, et al., 2010)), the basis for these behavioral data are unknown, as ethanol has not been tested as an agonist in cultured cell expression systems.
4.3 TAS2R31
4.3.1 Bitterness of sulfonyl amide sweetners
In spite of strong consumer demand for sweetness in the absence of calories, the utility of non-nutritive sweeteners is often impaired by undesirable side tastes. The sulfonyl amide sweeteners saccharin and acesulfame potassium (AceK) have a concentration-dependent bitter side taste that varies across individuals (Horne, Lawless, Speirs, & Sposato, 2002; Schiffman, Booth, Losee, Pecore, & Warwick, 1995; Schiffman, Reilly, & Clark, 1979). As might be expected, this variable bitterness predicts acceptability (Kamerud & Delwiche, 2007).
4.3.2 Variation in TAS2R31 and sweetener bitterness
The genes TAS2R31 (HGNC: 19113; formerly TAS2R44) and TAS2R43 (HGNC: 18875) are part of a highly related subfamily of 5 TAS2Rs located on chromosome 12. Their role in perception of saccharin and AceK bitterness was first identified in 2004 (Kuhn, et al., 2004). Expressing these genes in embryonic kidney cells resulted in calcium influx when the cells were treated with saccharin, AceK, and aristolochic acid, a purely bitter toxin found in plants in the Birthwort family (Kuhn, et al., 2004). Using saccharin and aloin (from aloe), Pronin and colleagues demonstrated the presence of Tryptophan at amino acid residue 35 in hT2R31 (Arg35Trp) associates with increased receptor activation in vitro and lower recognition thresholds in vivo (Pronin, et al., 2007). Subsequently, Roudnitsky and colleagues explored these genes and other related TAS2Rs on chromosome 12, generating long-range haplotypes across these genes (Roudnitzky, et al., 2011). One common haplotype (out of seven common haplotypes) associated with the bitterness of AceK and saccharin. By mutating individual amino acid residues in artificial chimeric receptors in vitro, they demonstrated the Arg35Trp (R35W) polymorphism in TAS2R31 was causal. Notably, however, they also found that other mutations in TAS2R31 abolished the ability of hT2R31 to respond to AceK and saccharin, even when tryptophan was present at position 35. Notably, the studies described above generally (but not exclusively) use threshold methods to define human taste phenotypes. Data from our laboratory confirms that polymorphisms in TAS2R31 also explain differences in the perceived intensity (suprathreshold bitterness) elicited by AceK (Allen, McGeary, Knopik, & Hayes, 2013), but not RebaudiosideA (Allen, McGeary, & Hayes, 2013).
Other compounds known to activate hT2R31 in functional expression systems include amarogentin, arborescin, arglabin, caffeine, chloramphenicol, falcarindiol, grossheimin, helicin, quinine, cromolyn, denatonium benzoate, and diphenidiol (Meyerhof, et al., 2010). However, just because a compound is known to activate hT2R31 receptors encoded by TAS2R31 does not imply perception of these compounds will vary across individuals, as the 25 TAS2Rs found in humans appear to include substantial functional redundancy. For example, diphenidiol activates 15 different hT2Rs and quinine activates 9. Thus, individuals who express low-functioning hT2R31 variants may still have a normal phenotype if functional recovery is provided by another redundant receptor. The question of whether TAS2R31 variation predicts the acceptability of sulfonyl amide-sweetened beverages remains untested. Such studies will require large sample sizes and whole gene sequencing of multiple TAS2Rs to deal with the potential for functional recovery discussed above, and to account for a loss of function due to rare variants in TAS2R31.
4.4 A multigene haplotype on chromosome 7
4.4.1 Coffee Bitterness varies with a haplotype across TAS2R3, TAs2R4, and TAS2R5
Coffee bitterness appears to vary across a haplotype on TAS2R3, TAS2R4 and TAS2R5 (HGNC: 14910, 14911, and 19912, respectively), which are located close to each other on chromosome 7. In this region (7q31.3–q32), four SNPs (rs765007, rs2234001, rs2227264, rs2234012) form three common haplotypes: CCGT homozygotes (26% of individuals), heterozygotes (41%) and homozygotes TGAG (22%) (Hayes, et al., 2011). The TGAG homozygotes report twice as much bitterness as the CCGT homozygotes, and the heterozygotes also report significantly greater bitterness than CCGT homozygotes (Hayes, et al., 2011).
4.4.2 Coffee Liking and Intake may not vary with genetics
Notably, this haplotype did not predict coffee liking or intake among a cohort that was predominantly middle aged, presumably due to postingestive conditioning/learning over many years. The causal SNP for this variation in bitterness is unknown: two of the SNPs result in amino acid substitutions (rs2234001 in TAS2R4, Val96Leu; and rs2227264 in TAS2R5, Ser26Ile) while the other two rs765007 (TAS2R3) and rs2234012 (TAS2R5) occur in the 5' untranslated region (5' UTR), potentially altering protein expression (Calvo, Pagliarini, & Mootha, 2009). Also, in the absence of in vitro verification that these SNPs are functional, it is possible none of them are causal, and are merely in linkage disequilibrium with an unknown causal polymorphism that is nearby. Given the absence of an effect on liking, one would not expect effects on intake; however, there is no data formally testing this relationship to date. Alternatively, this variation might predict liking and/or intake, but only in individuals that have not yet been conditioned to like coffee via repeated exposure to the positive post-ingestive effects associated with coffee intake (e.g. young adults who are still learning to drink coffee).
4.5 TAS2R19 polymorphisms appear to be functional
4.5.1 The rs10772420 SNP in TAS2R19 associates with quinine bitterness, grapefruit bitterness and grapefruit liking
Evidence that TAS2R19 (HGNC: 19108; formerly TAS2R48) contains a functional polymorphism first emerged in 2009 when Duffy and coworkers (Duffy, Hayes, Sullivan, & Faghri, 2009) reported liking ratings for grapefruit juice in surveyed liking ratings for grapefruit juice varied with a SNP in TAS2R48. Subsequently, genome wide association by Reed and colleagues showed that the same SNP (rs10772420) explained variation in the bitterness of quinine (Reed, et al., 2010). The resulting amino acid substitution, Arg299Cys (rs10772420) accounted for a small but significant amount of the trait variance in the genome wide analysis, and the A allele (Cys299) was associated with greater quinine bitterness (perceived suprathreshold intensity) for both discovery and replication cohorts.
Subsequent evidence extended these findings to include sampled unsweetened white grapefruit juice (Hayes, et al., 2011). The Cys299 homozygotes (AA individuals) reported much greater bitterness than either heterogyzotes (AG individuals) or Arg299 homozygotes (GG individuals), who did not differ from each other. As would be expected from mixture suppression (Lawless, 1979), sweetness showed the reverse pattern, although the differences were not significant. This pattern of increased bitterness and decreased sweetness also had the expected effect on liking: the Cys299 homozygotes had significantly lower liking ratings that the Arg299 homozygotes. This is wholly consistent with prior work showing endogenous sweetness and bitterness make statistically independent contributions to grapefruit juice liking (Lanier, Hayes, & Duffy, 2005). However, it remains unknown whether these phenotypic differences are sufficient to influence the consumption of quinine containing beverages (i.e. tonic water), grapefruit or grapefruit juice.
Moreover, these data are somewhat complicated by evidence that neither quinine nor the grapefruit bitterants limonin and naringin activate hT2R19 receptors in functional expression assays in vitro (Meyerhof, et al., 2010). As is true with any association design, significant associations may result from linkage disequalibrium with other unmeasured polymorphisms. For example, Roudnitzky et al. (2011) recently demonstrated that while 30 SNPs across 5 distinct TAS2R genes all associated with AcesulfameK thresholds, this variation can be attributed to a single allele in a single gene (Arg35Trp in TAS2R31). Thus, it is possible the rs10772420 SNP in TAS2R19 is not causal, but is merely acting as a tag SNP for another causal SNP on a nearby gene.
4.5.2 Recent molecular data provide new insights into older psychophysical data
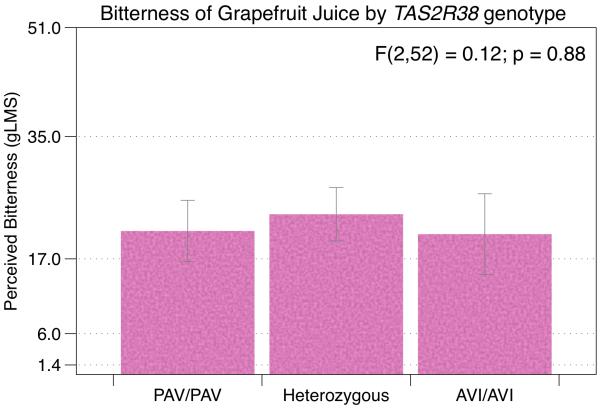
Prior to the discovery of the TAS2R genes and their receptors, several studies explored the relationship between grapefruit bitterness and the bitter probe propylthiouracil (PROP), with conflicting results (Drewnowski, Henderson, & Shore, 1997; Smagghe & Louis-Sylvestre, 1998). Drewnowski and colleagues found liking for grapefruit juice (surveyed) and sampled naringin (the predominant bitter flavonoid found in grapefruit) in water was significantly lower in those who found PROP to be the most bitter (Drewnowski, et al., 1997). As expected, the naringin bitterness-liking relationship and the PROP bitterness-liking relationship were both negative and significant, while the relationship between naringin liking and PROP liking was significant and positive. At the time, this led to the conclusion that the liking for grapefruit (and naringin) is therefore related to genetics underlying PROP variation. However, it is difficult to reconcile these data with modern molecular evidence. Specifically, PROP does not activate TASR19 (Meyerhof, et al., 2010) and grapefruit juice bitterness does not differ by TAS2R38 genotype (Figure 1; unpublished data from (Hayes, et al., 2011)). It may be that the relationship observed by Drewnowski and colleagues reflects some sort of generalized supertasting (i.e. hypergeusia; see (Hayes & Keast, 2011) rather than a specific effect of TAS2R38 variation. Indeed, PROP bitterness (i.e., the phenotype) captures variation in suprathreshold intensity across qualities even after controlling for TAS2R38 genotype (Hayes, et al., 2008). Notably, naringin has not been shown to activate TAS2R19in vitro (Meyerhof, et al., 2010) but this may be due to technical difficulties with naringin as a ligand in heterologous expression systems, or the TAS2R19 SNP acting as a tag SNP for another polymorphism.
Figure 1.
Unpublished data from Hayes et al. 2011 showing that the bitterness of unsweetened grapefruit juice does not vary by TAS2R38 genotype.
In direct contrast to the Drewnowski data, Smagghe and Louis Sylvestre (Drewnowski, et al., 1997; Smagghe & Louis-Sylvestre, 1998) reported those who were less sensitive to PROP (via detection thresholds, not suprathreshold intensity) reported greater suprathreshold bitterness from sampled naringin. At the time, Bartoshuk speculated this paradoxical finding was due to a reversal artifact based on the choice of psychophysical scale (Bartoshuk, 2000). However, insight from modern molecular data provides another alternative explanation for these conflicting accounts; in the Smagghe study, the numbers of PROP tasters and nontasters phenotyped for naringin bitterness were quite small (n=20 in each group). If Cys299 carriers were overrepresented among the PROP nontaster group by random chance, this could have driven a spurious, `paradoxical' relationship with PROP. Ironically, had the situation been reversed (with Cys299 carriers being overrepresented in the PROP taster group, again by chance), this would have been seen as support for the prevailing paradigm at the time (that PROP bitterness should predict grapefruit bitterness). Collectively, this suggests earlier work on correlations between various bitter compounds in studies with small sample sizes need to be carefully reconsidered in light of strong evidence that bitter response/sensitivity to real foods is not a monolithic up or down trait (Hayes, et al., 2011), as illustrated by the data described throughout this section
5. Sweet taste Differences
Sweetness, like bitterness, is transduced via G-protein coupled receptors (GPCRs). The sweet receptor is a heterodimer formed from the protein products of two genes, TAS1R2 and TAS1R3. Since the first sweetener-binding model was proposed in 1914, many different models of sweet receptor and ligand interactions have been proposed, which have evolved over time as the knowledge in this area has grown (See (DuBois, 2011; Hayes, 2008) for comprehensive reviews). The advancement of x-ray crystallography allowed molecular docking models and site-directed mutagenesis to identify three potential ligand-binding sites within the sweet receptor (Cui, et al., 2006). The most current model builds on these data, suggesting that 5 receptor sites within the T1R2/T1R3 dimer exist, for different structural classes of sweetener (Morini, Bassoli, & Temussi Piero, 2008). This model would also explain synergistic effects noted in some sweetener interactions (DuBois, 2011; Hayes, 2008).
SNPs in the genes encoding these binding-site regions could potentially affect receptor function, although no empirical evidence of this has been reported to date. However, as noted above, perceptual differences are not limited to SNPs within protein-coding regions of the genes, as recent evidence suggests that SNPs in the promoter regions of some genes may also affect the perception of sucrose.
5.1 TAS1R3 promoter SNPs may predict discriminability between low concentrations of sucrose
Fushan and colleagues (Fushan, et al., 2009) examined sucrose perception, asking 144 participants to rank the intensity a series of 9 sucrose solutions in an R-index task. Genetic analyses suggested that SNPs within the promoter regions of the TAS1R3 gene (rather than the sequence associated with one of the ligand binding regions of the receptor) were associated with the measured phenotype (Fushan, et al., 2009). Specifically, two C/T SNPs (rs307355 and rs35744813) in the promoter region (situated at -1572 and -1266 upstream of the coding sequence respectively) were associated with altered sweetness perception. Individuals with T alleles at both loci had reduced sweetness perception compared to those who were homozygous for the C allele at both loci. The authors noted the two lower-sensitivity T alleles are more prevalent in African populations, and postulated a link with geographical location and availability of carbohydrate-rich plants in the survival of early humans. Plants such as sugar cane are more likely to grow in tropical climates, so higher sensitivity to sucrose may not have been as important for survival to these populations as to Europeans living in colder climates, where the sensitive form of the allele is more common. However, this work has not been replicated (Hayes, McGeary, Grenga, & Swift, 2010), and it remains to be determined whether this variation may influence liking or intake of sweet foods.
5.2 GNAT3 and sucrose intensity
Gustducin is a G-protein involved in the signaling of sweet, bitter and umami tastes; its release is thought to be triggered by tastant binding, to either hT1R or hT2Rs (Chandrashekar, Hoon, Ryba, & Zuker, 2006). GNAT3, located on chromosome 7, encodes a subunit of gustducin, and a number of SNPs have been identified within it that have been linked to differences in sucrose perception (Fushan, et al., 2010). This report, based on the same cohort and phenotyping strategy as their earlier 2009 report (144 participants and ROC analysis of scores of ranked sucrose solutions), suggests that in addition to the variation in sweet perception explained by SNPs in TAS1R3, additional variation can be explained by eleven SNPs in the gustducin gene. The most individually significant of these SNPs is rs7792845; because it is located 10 kb upstream of the coding region, it presumably modifies the promoter region of the gene. Again, there remains a knowledge gap as to whether this variation is sufficient to influence food liking or consumption of sweet foods.
5.3 Heritability of Variation in Sweet liking
Liking for sweetness is thought to be innate, not learned, as evidence shows that newborns increase their suck rate in response to increased sucrose concentrations, and young children relax in the presence of strange adults when sucrose is given (Maller & Desor, 1973). Although this preference for sweetness is instinctive, perception of sweetness can vary across people (e.g.(Hayes & Duffy, 2007), as can the level of sweetness that is preferred (e.g., (Hayes & Duffy, 2008; Lundgren, et al., 1978). Multiple studies suggest preference for sweet stimuli is heritable in both children and adults. Mennella and colleagues (2005) examined heritability of sweet preference in children and their parents and found that preference for sweet foods (cereals and beverages) was heritable, although they failed to observe a heritable component for sucrose solutions tested in the laboratory. In 4–7 year old children, a strong genetic component was found in the preference for different sweetened grape solutions (Bretz, et al., 2006), while Keskitalo and coworkers (Keskitalo, Tuorila, et al., 2007) found roughly half of the variation in hedonic scores for a 20% w/v sucrose solution in adults aged 17–80 was attributable to genetic factors. However, the molecular basis of this heritability remains unknown, precluding the sorts of candidate gene and candidate SNP studies described above for bitterness. Notable, a region on chromosome 16 has been implicated in sweet preferences (Keskitalo, Knaapila, et al., 2007), but a specific gene was not identified and this region does not contain any genes known to be involved in taste.
5.4 Intake of Sweet Foods and Sugar
A strong genetic component has also been implicated in the consumption of sweet foods (Keskitalo, Knaapila, et al., 2007), and variations in non-oral chemoreceptors have been implicated in dietary intakes of sugar. For example, variation in GLUT2, a glucose transporter gene involved in glucose sensing in the brain, has been liked to sugar intakes (Eny, Wolever, Fontaine-Bisson, & El-Sohemy, 2008). However, only one report to date has linked variation in a sweet taste receptor to sugar intake. Eny and colleagues (Eny, Wolever, Corey, & El-Sohemy, 2010) examined dietary intakes in relation to two common SNPs in the sweet receptor subunit gene TAS1R1: Ile191Val (a variation in a potential ligand binding pocket) and Ser9Cys (a variation in a region believed to be important for signaling). They observed Ile191Val variation associated with dietary sugar intake in overweight individuals, with Val191 individuals consuming less daily sugar than their Ile191 counterparts. The mechanism through which this SNP is associated with intake is not yet known; one possibility is that this is a tag SNP, and simply linked to the functional SNP of interest. Alternatively, it is possible this SNP leads to a reduction in receptor function, requiring increased intake in order to get the same sugar `hit' as individuals with the fully functional receptor. Additional work is needed to both replicate these findings and to work out the putative mechanism by which they influence intake.
6. Influence of Polymorphisms on Umami Savory Sensations
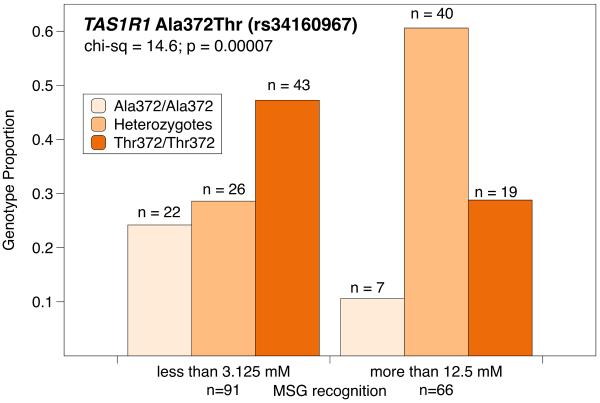
Umami, from the Japanese word for deliciousness, describes the savory, meaty taste elicited by L-glutamate, first isolated from seaweed in 1908. The taste receptor for umami is a heterodimer encoded by the TAS1R1 and TAS1R3 genes (the hT1R3 protein is shared by the sweet and umami heterodimer complexes). The hT1R1/hT1R3 dimer is also stimulated by 5' ribonucleotides, and is therefore believed to have evolved as an indicator of dietary sources of amino acids. Evidence suggests that inter-individual perceptual differences exist for umami, with taste thresholds differing up to 5 fold (Lugaz, Pillias, & Faurion, 2002), and, similar to bitter taste perception, a fraction of the population can be described as `taste blind' to umami. Lugaz and colleagues (2002) estimate the prevalence to be approximately 4%, although this estimate should be used cautiously, as this is still a poorly studied trait. Shigemura and colleagues (Shigemura, Shirosaki, Sanematsu, Yoshida, & Ninomiya, 2009) examined 17 TAS1R1 / TAS1R3 SNPs in 254 Japanese subjects in relation to the detection thresholds for IMP (a purine nucleomonophosphate) and MSG, compounds known to elicit umami tastes. Eight of these SNPs were previously reported (Kim, Wooding, Ricci, Jorde, & Drayna, 2005), while 9 were new polymorphisms identified by Shigemura and coworkers. Of these, three SNPs, at amino acid residue 12 and 372 in TAS1R1 and 757 in TAS1R3 were deemed to be `common' and were investigated in relation to recognition thresholds. The TAS1R1 SNP at position 12 was not significantly associated with umami perception, while the TAS1R1 Ala372Thr SNP (rs34160967) appeared to be functional, with the 372Thr allele conferring increased sensitivity to umami. (This relationship was only significant for MSG and a mixture of MSG and IMP, but not for IMP alone). The TAS1R3 Arg757Cys SNP (rs307377) also appeared to be functional for all umami tastants, with the Cys757 allele resulting in reduced function (higher thresholds). Considered together, these results led the authors to conclude that the TAS1R1/R3 heterodimer may have separate binding areas for IMP and MSG, and that the TAS1R3 757 SNP may lie in the area for IMP while the TAS1R1 372 SNP may be important for MSG binding.
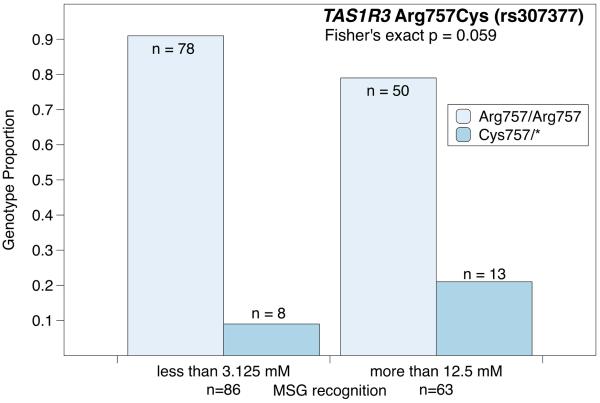
In Figures 2 and 3 data from Shigemura et al. are reanalyzed in `Low' and `High' threshold groups for MSG. In Figure 2, individuals with one or two copies of the Cys757 variant are over represented among the insensitive (high threshold) individuals. In Figure 3, Thr372 homozygotes are overrepresented in the MSG sensitive (low threshold) individuals. Moreover, it appears that these two SNPs are not in linkage disequilibrium, and they appear to have offsetting effects. That is, having the TAS1R1 Thr372 (low) SNP and the TAS1R3 Arg757 (normal) SNP pulls an individual's threshold down, while having the TAS1R1 Ala372 (normal) SNP and the TAS1R3 Cys757 (high) SNP pulls an individual's threshold up. Raliou et al., (2011) confirmed this role of the Arg757Cys SNP in umami perception, via a heterologous system, and observed that the Cys757 variant reduced activation of the heterodimer in response to MSG. As with many of the examples detailed above, it is still unknown whether this variation is sufficient to influence liking or intake of glutamate rich foods.
Figure 2.
Reanalysis of data from Shigemura et al 2009 indicates Cys757 carriers for the TAS1R3 SNP (rs307377) may be overrepresented among those with elevated MSG recognition thresholds.
Figure 3.
Data from Shigemura also show Thr372 homozygotes (G/G) for the TAS1R3 SNP (rs34160967) are overrepresented in the group with low MSG recognition thresholds.
7. Salty and Sour Taste – Minimal Evidence to Date
Variation in the perception of sour and of salt taste have not received the same degree of attention as the three GPCR mediated prototypical tastes. Twin studies suggest salt perception appears to be more dependent on environmental influences than genetic factors (Wise, Hansen, Reed, & Breslin, 2007). Additionally, salt preferences are strongly influenced by physiological state (Wald and Lesham 2003), and are more variable in women than men (Hayes, Sullivan, & Duffy, 2010), presumably due to hormonal cycling. Other studies have demonstrated that sensory habituation to high sodium levels may increase the liking of high-sodium foods (Bertino, Beauchamp, & Engelman, 1982; Blais, et al., 1986). Collectively, difficulty in defining a stable phenotype for salt taste or preference impairs the ability to explore genetic variation, and it suggests any genetic influences on ingestive behavior may be swamped by environmental factors. Indeed, previous twin studies have failed to observe any heritablity for either salt taste or preference (Greene, Desor, & Maller, 1975). Nonetheless, very recent data suggest that variations in TRPV1 and SCNN1B may influence the perception sodium chloride (Dias, et al., 2013). This is a very new area of research and if replicated, it is unknown whether such variation is sufficient to alter liking or intake of salty foods.
Likewise, relatively little is known about genetic influences on sour taste perception, or differences in liking or intake of sour foods. Twin studies suggest a substantial amount of the variance in citric acid thresholds can be attributed to genetic factors (Wise, et al., 2007); whether this is due to receptor differences, or other factors like salivary buffering capacity is unknown. Also, sour taste liking is quite variable, but there does not appear to be a strong relationship between sourness intensity and affective response to sour foods (Liem, Westerbeek, Wolterink, Kok, & de Graaf, 2004; Tornwall, et al., 2012). A candidate sour receptor PKD2L1 has been identified (Ishimaru, et al., 2006), but knocking out the PKD2L1 gene in mice reduces but does not eliminate sour taste response (Horio, et al., 2011), suggesting other unknown receptors also play a role in mammaliam sour taste.
8. SNPs in the Gustin (CA6) gene may have non-quality specific effects on taste
Gustin, a zinc metalloprotein, is thought to be a trophic factor involved in the development of taste buds (Henkin, Martin, & Agarwal, 1999). Variations in the gustin gene CA6 were recently reported to associate with some of the bitterness of PROP (Calo, et al., 2011). As mentioned above in section 4, most of the observed variation in PROP bitterness can be explained by variation in the TAS2R38 gene (Bufe, et al., 2005; Kim, et al., 2003; Prodi, et al., 2004); the remainder of the unexplained variation may be due to other unknown factors that may or may not have a genetic basis. Some authors have postulated that either the density or total number of fungiform papillae may account for the remainder of the unexplained variation, but reports have been inconsistent. Recent work by Calo and colleagues (Calo, et al., 2011) revisits the idea that differences in tongue anatomy are involved in differences in taste perception. They observed a SNP within the gustin (CA6) gene (HGNC: 1380) were also associated with PROP bitterness intensity. An A>G substitution at base pair 31479 (rs227433) explained additional variance in PROP bitterness after accounting for effects of TAS2R38; the major allele `A' was associated with PROP sensitivity, and the `G' was associated with nontasting. When both TAS2R38 haplotypes and the gustin SNP were combined, they accounted for just under 60% of the variation in PROP response in their sample, suggesting additional unknown factors still play a role. It is thought that the variation in this gene affects the zinc-binding capacity in the resultant protein (Padiglia, et al., 2010) and thus the AA form allows zinc to fully bind. An individual's nutritional status in relation to zinc is a possible regulatory factor that could also potentially explain some of the variation in PROP response.
Because the putative nature of this link with taste perception occurs via a mechanism is not limited to bitterness alone, variations in the gustin gene could be important for taste function in general. More work is needed to determine whether gustin SNPs associate with variation in fungiform papillae density, or with the perceived intensity of other tastants. Further research is also needed to determine whether variation in the gustin gene influences food liking or intake.
9. Tactile Sensations
9.1 Creaminess perception
Creaminess is an important quality in many types of foods, and these sensations can arise from varying levels of fat or starch in a food. Chocolate and ice cream are two highly liked foods, and part of their appeal has been linked to their dynamic texture as they melt in the mouth (Hyde & Witherly, 1993). As they melt, they decrease in oral viscosity, and the degree of this `thinning' in viscosity is linked to their perceived creaminess and to consumer sensory acceptance (Prindiville, Marshall, & Heymann, 1999; Prindiville, Marshall, & Heymann, 2000).
9.1.1 Copy number variation in the salivary amylase gene
Differences in perceived creaminess have been associated with levels of salivary alpha amylase. The glucose monomers in starch are connected by 1,4- glycosidic bonds, which are cleaved by amylase while food is chewed. Salivary alpha amylase (a protein) is encoded by the AMY1 gene, and there is substantial variation in the number of copies of this gene an individual has, ranging from 2 – 15 diploid copies. The concentration of oral salivary amylase is proportional to the number of copies, and it is hypothesized that the greater the enzyme concentration in saliva, the faster these bonds are broken. This copy number variation appear to have arisen in evolutionary response to differing levels of starch in the diet of different populations (Perry, et al., 2007). Subsequently, Mandel and co-workers ((Mandel, Peyrot des Gachons, Plank, Alarcon, & Breslin, 2010) demonstrated that individuals with copy number variation (CNV) of the AMY1 gene had different levels of salivary amylase present, and that perception of viscosity thinning of an oral starch sample over time was related to the level of oral amylase, although they were unable find evidence directly associating the number of copies of the AMY1 gene. As with any meditational variability, the lack of a direct effect may simply result from a lack of power. The degree to which CNV in AMY1 associates with liking of creamy starch based foods, or selection of a diet high in starchy foods remains unknown, and requires further study.
9.1.2 TAS2R38 variation and creaminess
Other genetic factors may also be involved in creaminess perception. For example, is evidence that fungiform papillae density, which can vary from person to person, plays a role in perceived creaminess from different foods, such as salad dressing (Tepper & Nurse, 1997). Because papillae density associates with PROP bitterness, and PROP bitterness associates with TAS2R38 genotype, this has led some investigators to suggest potential associations between creaminess perception and variation in TAS2R38. However, more recent data indicate that fungiform papillae density is independent of TAS2R38 (Hayes, et al., 2008), suggesting that reports linking creaminess and PROP bitterness (Hayes & Duffy, 2007; Tepper & Nurse, 1997) may have been due to PROP being a marker for overall taste intensity (hypergeusia) (Hayes & Keast, 2011), and not due to variation in TAS2R38. This is an important illustration of the distinction between genotype and phenotype, raised in section 2.1. Many studies indicate elevated PROP bitterness associates with perceived creaminess, and many other studies indicate PROP bitterness is largely (but not entirely) determined by TAS2R38 genotype. However, hT2R38 is a narrowly tuned bitter taste receptor, so there is no simple, biologically plausible mechanism by which TAS2R38 genotype should influence creaminess perception. Thus, the ability of PROP bitterness (the phenotype) to predict creaminess is likely due to other factors independent of TAS2R genetics.
9.2 Astringency
Astringency is a sensation described as a drying and/or puckering of the mouth, often caused by consumption of the naturally-occurring plant flavonoid phenols such as tannins (Lee & Lawless, 1991), which are present in many common beverages like tea, coffee, wine and beer. When such beverages are consumed, the tannins bind to salivary proteins within the mouth, causing aggregation, and leading to the perception of a dry mouth (Kallithraka, Bakker, & Clifford, 1998). This sensation can lead to the rejection of beverages in some consumers (Lesschaeve & Noble, 2005).
9.2.1 Differences in salivary protein content influences astringency perception and liking of high tannin foods
Individuals may differ in their levels of salivary protein, which affects the amount of astringency they perceive from tannic acid (Horne, Hayes, & Lawless, 2002). The degree of perceived astringency may be a strong driver of preference and liking in many cases. Dinella and colleagues (Dinnella, Recchia, Tuorila, & Monteleone, 2011) grouped individuals into low, medium and high responders, based on their salivary protein content, and the high responder group reported significantly more astringency from apple, grape and carrot juices containing added tannic acid, compared to the low and medium responders. Across all three juices, liking scores also differed by responder group. Nor were differences limited to sampled beverages with manipulated tannin content; in the same study, preference for raw chicory was also lower in the high responder groups, and food such as ripe pear and banana, which elicit less astringency, were more liked.
Critically, the mechanisms underlying these differences in astringency perception may be genetic; a recent Finnish study in monozygotic and dizygotic twins measured salivary protein levels, and sensory responses to tannic acid spiked and control (unspiked) apple juice (Tornwall, et al., 2011). Their data suggest the level of salivary protein is genetically determined, and liking and frequency of use of astringent foods are heritable. Notably, they used naïve assessors and ask them to rate the `intensity' of a tannic spiked apple juice versus unspiked juice; the term `astringency' was not mentioned or defined for participants. Presumably, similar results would be obtained from assessors trained to evaluate astringency. Unfortunately, as with any twin design, the biological basis of this variation is unknown, and no candidate genes have been identified thus far.
Astringency perception may also be dependent on salivary flow rates, which also vary across individuals (Horne, Hayes, et al., 2002; Lesschaeve & Noble, 2005). For example, individuals with low salivary flow rates rated wine as more intensely astringent, and found that the astringency lasted for a longer time than high-salivary flow individuals for the same wine samples. Although other studies have shown similar results in red (Ishikawa & Noble, 1995) and white wine (Fischer, 1994), not all studies agree (e.g (Guinard, Zoumas-Morse, & Walchak, 1997)). It is possible that these different results could stem from a lack of a clear, universal definition of astringency. Methodological issues in the measurement of astringency pose a further problem, as this sensation builds up over time during sampling. Furthermore, astringency from beverages depends on the food with which they are eaten (Less & Noble, 2005). This emphasizes highlights the point that high quality psychophysics are needed to define phenotypes, and that subtle variation across labs can potentially obscure effects. It remains unknown whether salivary flow rates are heritable, and if it is, what the genetic basis of this variation is. Given observed differences in perception as a function of flow rates, this would seem to be a fruitful area for future research.
10. Odor
Neurons that are present in the olfactory epithelium transmit electrical impulses when the receptors they express are stimulated (Hasin-Brumshtein, Lancet, & Olender, 2009). Each neuron expresses one odorant receptor (OR) gene, monoallelically (Chess, Simon, Cedar, & Axel, 1994). These are transmitted to synaptic complexes, and each stimulus (odorant) is a unique combination of activated units (Hasin-Brumshtein, et al., 2009). Early work hypothesized that smells were interpreted through a `primary' or fundamental odor system (Amoore, 1977). Today, it is apparent that the relationship between odorants and ORs is complex (multiple ORs bind can recognize one odorant molecule, and multiple molecules can be recognized by one OR). Due to the complicated nature of this relationship, current data supports a cross fiber patterning model, which suggests primary odors do not exist (Niimura, 2012).
Although the pathway of odor perception has not yet been fully elucidated, it is clear that sensory `blindness' to certain compounds is not limited to taste; some compounds which are strong-smelling to some individuals appear odorless to others. This phenomenon has been termed `specific anosmia' (Amoore, 1977), and is known to exist for a variety of different stimuli. Early evidence for this came from Blakeslee, who showed that when presented pink and red verbena flowers, approximately 1/3 of those tested could smell only the pink flowers, while the remainder could only smell the red flowers (Blakeslee, 1918). Another, somewhat infamous, example is that of the smell from butyl mercaptan, a compound similar to that of the foul-smelling odor emitted by a skunk, which is odorless to a fraction of the population (Gates, 1946, cited in (Patterson & Lauder, 1948). Initial genetic studies indicated that specific anosmias were hereditary, and they were inherited as a recessive characteristic (they also suggested a second type of anosmia, `delayed onset', which appeared to be inherited as a dominant trait) (Patterson & Lauder, 1948). The genetic basis for specific anosmia may arise from mutations in receptor genes involved in the different odors, although to date, only few examples have been characterized.
Odorant receptors are encoded by a superfamily of OR genes, one of the largest gene repertoires in the human genome. To date, 851 OR genes have been identified in humans, of which just approximately 384 are known to be functional (Zhang & Firestein, 2009). The remainder are pseudogenes: pseudogenes are genetic sequences which have most of the characteristics of a gene but which have a disabling feature such as a frameshift or stop codon in the coding sequence, or are missing a promotor region. They are generally believed to be genes which have lost their function over time due to relaxation of selective pressure. In ORs, these loss-of-function events are thought to have occurred relatively recently, and thus, there is substantial variation in the level of inter-individual OR gene functionality (Menashe, Man, Lancet, & Gilad, 2003; Zhang & Firestein, 2009). That is, the number of psuedogenes may vary from person to person – currently at least 60 loci known to exist, the true number is likely to be higher and will be determined as further individual genomes are sequenced. Although it is currently unknown if differences in pseudoegenization lead to differential olfactory sensitivity, phenotypic variation in the sense of smell is commonplace. This may be driven by Copy Number Variation, which is particularly high in the OR gene family compared to the rest of the genome and is responsible for most of the inter-individual genomic variation. Because these patterns are observed in mammals with trichromatic vision, this has lead to speculation that this is an evolutionary relic from the corresponding reduction in the functional constraint on the sense of smell. That is, as color vision evolved, smell became less important for survival.
Few CNVs have been functionally characterized in relation to olfactory ability and as such, SNP variations account for some of the classic examples of olfactory anosmia (e.g., OR7D4 and androstenone) (Keller, et al., 2007). Another classic example is the perception of methanethiol, an odorous metabolite found in the urine of some individuals after asparagus consumption. Detection of this compound is associated with a SNP near the odor receptor gene OR2M7 (Eriksson, et al., 2010). Other specific anosmias are thought to have a genetic basis, but here we focus on several examples especially relevant to food. Data on isovaleric acid (cheesy) and urinary metabolites of asparagus are reviewed elsewhere (Newcomb, Xia, & Reed, 2012).
10.1 Androstenone and OR7D4
Androstenone is a steroid derived from testosterone, which elicits a strong odor in some individuals. The ability to smell androstenone varies across people; some describe it as `foul smelling', `urinous' and `sweaty', while others report it is `sweet-smelling' or `floral', and finally, to some, it is undetectable. Early work showing differences in androstenone detection thresholds between monzygotic and dizygotic twins suggested this was a genetically determined trait (Wysocki & Beauchamp, 1984). A recent study in a sample of Finnish twins examined responses to a range of odors, which included androstenone and isovaleric acid, along with chocolate, cinnamon, turpentine, and isovaleric acid (Knaapila, 2008). Their data suggest the environmental factors influencing odor perception (of the tested odorants) was stronger than the genetic component. However, all the odors were lumped together in this model, and this approach does not speak to individual genes or polymorphisms, which are the primary focus of this review.
Conversely, a GWAS approach conducted in 2007 indicated that SNPs in the OR receptor gene OR7D4 are partly responsible for differential ability to smell androstenone (Keller, et al., 2007). Two amino acid changes within OR7D4, R88W and T133M, alters the ability to detect androstenone, and a closely related compound, androstatiedone. The `R' form at position 88 and the `T' form at position 133 are associated with normal perception, while the `W' and `M' forms respectively are associated with impaired androstenone detection. Together, these substitutions account for approximately 40% of variation in the ability to smell androstenone.
Of specific relevance to food, androstenone is produced by male pigs (boars), and is often present in their skin, leading to a pork defect known as ` boar taint', where the androstenone odor has been described as `sweaty, `ammonia', `dirty', `silage' and `acrid', among others. In one recent study, anstrostenone-containing meat was found to be less acceptable to subjects with two copies of the RT variant of the OR7D4 receptor, indicating that this genetic variation affects food preferences (Lunde, et al., 2012). The existence of this taint in pork is already a particular problem for pork producers, and one that may become may become more worrisome in Europe, where a proposed ban on pig castration due to animal cruelty issues could lead to a rise in androstenone levels in pork, and subsequently reduce consumer acceptance. In Norway for example, approximately 40% of the population is sensitive to this odor (Lunde, et al., 2012). It is unclear whether the proportion of androstenone-sensitive individuals varies in different countries, but if so, it may impact the areas from which pork products are imported to different countries, with consequential economic considerations (Lunde, et al., 2012).
10.2 Cis-3-hexen-1-ol, green/grassy odors, and food acceptability
Cis-3-hexen-1-ol (C3HEX) is the prototypical odorant associated with `green' or `grassy' odors, and C3HEX is present in a range of fruit and vegetables. Differences in C3HEX detection thresholds span two orders of magnitude, and this variation has been associated with polymorphisms in OR2J3. The amino acid substitutions T113A and R226Q, are associated with in an impaired ability to smell C3HEX and may account for a quarter of the variation in vivo. (However, the haplotypes do not respond in vitro to C3HEX, so they may merely be in linkage disequilibrium with the causal SNPs).
More critically, variation in the ability to detect C3HEX is linked to food preferences. In a comprehensive investigation, Jaeger and colleagues demonstrated that this variation was important for preference using a range of foods including tomato juice, hummus, and Japanese green tea (Jaeger, et al., 2012). Samples spiked with C3HEX received lower acceptability ratings than the `normal' samples, and the degree of the negative response was higher in those who were C3HEX-responsive. As C3HEX is found in a vast array of plant-derived foods, this has wide implications for the food industry in terms of inter-individual food preferences. The challenge for the next phase of this area of research will be not only to elucidate the other functional genetic variations in chemosensory receptors but also to examine how these variations interact, to determine the effect on eating behavior.
10.3 Cilantro Aversions
Cilantro (also known as coriander leaf) has been described as `polarizing', as some individuals have an extreme dislike for this herb. Flavor descriptions range from `fragrant' and citrussy' to `soapy' and `insect or bug-like' (Mauer & El-Sohemy, 2012). The idea that the variation in the `soapy' character from cilantro is genetically-determined is not new (e.g. (Hertz, 2004). Recent GWAS have attempted to identify the SNPs involved. Data from the Toronto Nutrigenomics and Health study in 316 females indicates a role for both an odorant receptor variant (rs7277172 in OR4N5) and a SNP in a bitter taste receptor (rs427871 near TAS2R1), as 75% of participants who were homozygous for the minor alleles of both SNPs reported disliking cilantro, while none of those who homozygous for the major alleles reported disliking this herb (Mauer 2011, Master's thesis, available online). However, more recent evidence in a much larger group of subjects (11,851) suggests that a genetic variation (rs7921001) in the odorant receptor gene OR6A2 on chromosome 11 may be responsible for the detection of soapiness and disliking of cilantro (Eriksson et al, 2012). As food preferences are influenced by a wide range of factors, including exposure and cultural influences, this may account for some of the differences in these two reports. Future research using all of these putative SNPs may further ascertain their role in cilantro flavor and preference.
11. Summary and Conclusions
Biological differences in chemosensory ability from SNPs and CNVs are both commonplace and highly variable from person to person. This appears to be partly a result of evolutionary processes that have driven us to detect a vast array of potential toxins as well as potential foodstuffs, through both gustation, olfaction, and oral somatosensation. This is exemplified by the fact that although many SNPs are associated with differences in perception across multiple taste qualities, by far the most diverse is the variation in bitter taste receptor genes, likely due to the increased selective advantage in the conferred ability to discriminate a wide range of potential toxins at low levels. This discriminatory facility is strengthened by the fact that single compound can contain multiple distinct moieties that bind to separate TAS2Rs. For example, allylglucosinolate (also called sinigrin) contributes to the bitterness of brassica vegetables such as Brussels sprouts and cauliflower (Engel, Baty, Le Corre, Souchon, & Martin, 2002; van Doorn, et al., 1998). Critically, allylglucosinolate contains both a beta-glucopyranoside ring and a thiourea moiety (N-C=S), which activate hT2R16 and hT2R38 respectively (Meyerhof, et al., 2010). This demonstrates both the diversity of bitter tastants within food and how individuals may perceive a range of bitter tastes differently depending on different genetic variations. As such, the historical treatment of `bitter' as a uniform trait exemplified by a single compound is no longer tenable, and should not be used as strategy to screen sensory panelists. Instead, we suggest that sensory practitioners use the actual food of interest be used for panelist screening; hops for studies on beer, cocoa polyphenols for chocolate, etc.
Further, the influence of genetic variation on food sensations, liking and intake are not limited to bitterness. Nor are these effects limited to taste; recent data suggest differences in olfactory ability may also impact food preference, and should be considered, particularly where volatile compounds are important in the flavor aspect of a food product. This is an emerging area of research, and more work is needed; it will be interesting to see how the next challenges will be met.
In conclusion, inter-individual genetic differences have arisen in response to the changing environment as part of the evolutionary process. In the modern world, such variations affect how the surrounding environment is perceived, and have important implications for individual food preference and intake. The challenge that now lies ahead for sensory researchers is to establish how various combinations of chemosensory differences may interact to both influence and predict food choice.
Supplementary Material
Highlights
Genetic variation in chemosensation is more common than previously thought
This variation is known to alter sensations from and preference for food
Recent data suggests these differences extend beyond taste to include odor and touch
We summarize current knowledge, emphasizing human behavioral data
Acknowledgements
This review was based on an oral presentation at the 9th Pangborn Sensory Science Symposium in Toronto, Canada by the lead author. The scope has been expanded to the present form and refined with the assistance of the coauthors. This work was funded by a National Institutes of Health grant from the National Institute National of Deafness and Communication Disorders [DC010904] to JEH, United States Department of Agriculture Hatch Project PEN04332 funds, and funds from the Pennsylvania State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
PCR is the Polymerase Chain Reaction, which makes it possible to amplify DNA. Kary Mullis and Michael Smith shared the 1993 Nobel Prize in Chemistry for their work on PCR.
References
- Allen AL, McGeary JE, Hayes JE. Rebaudioside A and Rebaudioside D bitterness do not covary with Acesulfame K bitterness or polymorphisms in TAS2R9 and TAS2R31. Chemosens Percept. 2013 doi: 10.1007/s12078-013-9149-9. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AL, McGeary JE, Knopik VS, Hayes JE. Bitterness of the Non-nutritive Sweetener Acesulfame Potassium Varies With Polymorphisms in TAS2R9 and TAS2R31. Chem Senses. 2013;38(5):379–389. doi: 10.1093/chemse/bjt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoore JE. Specific anosmia and the concept of primary odors. Chem Senses. 1977;2(3):267–281. [Google Scholar]
- Bartoshuk LM. Comparing sensory experiences across individuals: recent psychophysical advances illuminate genetic variation in taste perception. Chem Senses. 2000;25(4):447–460. doi: 10.1093/chemse/25.4.447. [DOI] [PubMed] [Google Scholar]
- Bartoshuk LM, Duffy VB, Miller IJ. PTC/PROP tasting: anatomy, psychophysics, and sex effects. Physiol Behav. 1994;56(6):1165–1171. doi: 10.1016/0031-9384(94)90361-1. [DOI] [PubMed] [Google Scholar]
- Behrens M, Meyerhof W. Oral and extraoral bitter taste receptors. Results Probl Cell Differ. 2010;52:87–99. doi: 10.1007/978-3-642-14426-4_8. [DOI] [PubMed] [Google Scholar]
- Bertino M, Beauchamp GK, Engelman K. Long-Term Reduction in Dietary-Sodium Alters the Taste of Salt. American Journal of Clinical Nutrition. 1982;36(6):1134–1144. doi: 10.1093/ajcn/36.6.1134. [DOI] [PubMed] [Google Scholar]
- Blais CA, Pangborn RM, Borhani NO, Ferrell MF, Prineas RJ, Laing B. Effect of Dietary-Sodium Restriction on Taste Responses to Sodium-Chloride - a Longitudinal-Study. American Journal of Clinical Nutrition. 1986;44(2):232–243. doi: 10.1093/ajcn/44.2.232. [DOI] [PubMed] [Google Scholar]
- Blakeslee AF. Genetics of Sensory Thresholds: Taste for Phenyl Thio Carbamide. Proc Natl Acad Sci U S A. 1918;18(1):120–130. doi: 10.1073/pnas.18.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee AF. Genetics of sensory thresholds: Taste for phenyl thio carbamide. Proceedings of the National Academy of Sciences. 1932;18:120–130. doi: 10.1073/pnas.18.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breer H, Eberle J, Frick C, Haid D, Widmayer P. Gastrointestinal chemosensation: chemosensory cells in the alimentary tract. Histochem Cell Biol. 2012;138(1):13–24. doi: 10.1007/s00418-012-0954-z. [DOI] [PubMed] [Google Scholar]
- Bretz WA, Corby PMA, Melo MR, Coelho MQ, Costa SM, Robinson M, Schork NJ, Drewnowski A, Hart TC. Heritability estimates for dental caries and sucrose sweetness preference. Arch Oral Biol. 2006;51(12):1156–1160. doi: 10.1016/j.archoralbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 2005;15(4):322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 2002;32(3):397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- Calo C, Padiglia A, Zonza A, Corrias L, Contu P, Tepper BJ, Barbarossa IT. Polymorphisms in TAS2R38 and the taste bud trophic factor, gustin gene co-operate in modulating PROP taste phenotype. Physiol Behav. 2011 doi: 10.1016/j.physbeh.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A. 2009;106(18):7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Hoon MA, Ryba NJP, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444(7117):288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic Inactivation Regulates Olfactory Receptor Gene-Expression. Cell. 1994;78(5):823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Clark AA, Liggett SB, Munger SD. Extraoral bitter taste receptors as mediators of off-target drug effects. FASEB J. 2012;26(12):4827–4831. doi: 10.1096/fj.12-215087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colares-Bento FC, Souza VC, Toledo JO, Moraes CF, Alho CS, Lima RM, Cordova C, Nobrega OT. Implication of the G145C polymorphism (rs713598) of the TAS2r38 gene on food consumption by Brazilian older women. Arch Gerontol Geriatr. 2012;54(2):e13–18. doi: 10.1016/j.archger.2011.05.019. [DOI] [PubMed] [Google Scholar]
- Cooke LJ, Haworth CMA, Wardle J. Genetic and environmental influences on children's food neophobia. American Journal of Clinical Nutrition. 2007;86(2):428–433. doi: 10.1093/ajcn/86.2.428. [DOI] [PubMed] [Google Scholar]
- Cui M, Jiang P, Maillet E, Max M, Margolskee R, Osman O. The Heterodimeric Sweet Taste Receptor has Multiple Potential Ligand Binding Sites. Current Pharmaceutical Design. 2006;12(35):4591–4600. doi: 10.2174/138161206779010350. [DOI] [PubMed] [Google Scholar]
- Delwiche JF. The impact of perceptual interactions on perceived flavor. Food Qual Pref. 2004;15:137–146. [Google Scholar]
- Dias AG, Rousseau D, Duizer L, Cockburn M, Chiu W, Nielsen D, El-Sohemy A. Genetic Variation in Putative Salt Taste Receptors and Salt Taste Perception in Humans. Chemical Senses. 2013;38(2):137–145. doi: 10.1093/chemse/bjs090. [DOI] [PubMed] [Google Scholar]
- Dick DM, Rose RJ, Viken RJ, Kaprio J, Koskenvuo M. Exploring gene-environment interactions: socioregional moderation of alcohol use. J Abnorm Psychol. 2001;110(4):625–632. doi: 10.1037//0021-843x.110.4.625. [DOI] [PubMed] [Google Scholar]
- Dinehart ME, Hayes JE, Bartoshuk LM, Lanier SL, Duffy VB. Bitter taste markers explain variability in vegetable sweetness, bitterness, and intake. Physiol Behav. 2006;87(2):304–313. doi: 10.1016/j.physbeh.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Dinnella C, Recchia A, Tuorila H, Monteleone E. Individual astringency responsiveness affects the acceptance of phenol-rich foods. Appetite. 2011;56(3):633–642. doi: 10.1016/j.appet.2011.02.017. [DOI] [PubMed] [Google Scholar]
- Dotson CD, Vigues S, Steinle NI, Munger SD. T1R and T2R receptors: the modulation of incretin hormones and potential targets for the treatment of type 2 diabetes mellitus. Curr Opin Investig Drugs. 2010;11(4):447–454. [PMC free article] [PubMed] [Google Scholar]
- Dotson CD, Wallace MR, Bartoshuk LM, Logan HL. Variation in the Gene TAS2R13 is Associated with Differences in Alcohol Consumption in Patients with Head and Neck Cancer. Chemical Senses. 2012;37(8):737–744. doi: 10.1093/chemse/bjs063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson CD, Zhang L, Xu H, Shin YK, Vigues S, Ott SH, Elson AE, Choi HJ, Shaw H, Egan JM, Mitchell BD, Li X, Steinle NI, Munger SD. Bitter taste receptors influence glucose homeostasis. PLoS One. 2008;3(12):e3974. doi: 10.1371/journal.pone.0003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewnowski A, Henderson SA, Shore AB. Taste responses to naringin, a flavonoid, and the acceptance of grapefruit juice are related to genetic sensitivity to 6-n-propylthiouracil. Am J Clin Nutr. 1997;66(2):391–397. doi: 10.1093/ajcn/66.2.391. [DOI] [PubMed] [Google Scholar]
- DuBois GE. Validity of early indirect models of taste active sites and advances in new taste technologies enabled by improved models. Flavour and Fragrance Journal. 2011;26(4):239–253. [Google Scholar]
- Duffy VB. Variation in oral sensation: implications for diet and health. Curr Opin Gastroenterol. 2007;23(2):171–177. doi: 10.1097/MOG.0b013e3280147d50. [DOI] [PubMed] [Google Scholar]
- Duffy VB, Davidson AC, Kidd JR, Kidd KK, Speed WC, Pakstis AJ, Reed DR, Snyder DJ, Bartoshuk LM. Bitter receptor gene (TAS2R38), 6-n propylthiouracil (PROP) bitterness and alcohol intake. Alcohol Clin Exp Res. 2004;28(11):1629–1637. doi: 10.1097/01.ALC.0000145789.55183.D4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy VB, Hayes JE, Bartoshuk LM, Snyder DJ. Taste: Vertebrate Psychophysics. In: Squire LR, editor. Encyclopedia of Neuroscience. Academic Press; Oxford: 2009. pp. 881–886. [Google Scholar]
- Duffy VB, Hayes JE, Davidson AC, Kidd JK, Kidd KK, Bartoshuk LM. Vegetable Intake in College-Aged Adults Is Explained by Oral Sensory Phenotypes and TAS2R38 Genotype. Chemosensory Perception. 2010;3(3):137–148. doi: 10.1007/s12078-010-9079-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy VB, Hayes JE, Sullivan BS, Faghri P. Surveying Food/Beverage Liking: A Tool for Epidemiological Studies to Connect Chemosensation with Health Outcomes. Ann NY Acad Sci. 2009;1170:558–568. doi: 10.1111/j.1749-6632.2009.04593.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addict Biol. 2006;11(3–4):386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Engel E, Baty C, Le Corre D, Souchon I, Martin N. Flavor-active compounds potentially implicated in cooked cauliflower acceptance. J Agric Food Chem. 2002;50(22):6459–6467. doi: 10.1021/jf025579u. [DOI] [PubMed] [Google Scholar]
- Eny KM, Wolever TM, Corey PN, El-Sohemy A. Genetic variation in TAS1R2 (Ile191Val) is associated with consumption of sugars in overweight and obese individuals in 2 distinct populations. Am J Clin Nutr. 2010;92(6):1501–1510. doi: 10.3945/ajcn.2010.29836. [DOI] [PubMed] [Google Scholar]
- Eny KM, Wolever TMS, Fontaine-Bisson B, El-Sohemy A. Genetic variant in the glucose transporter type 2 is associated with higher intakes of sugars in two distinct populations. Physiological Genomics. 2008;33(3):355–360. doi: 10.1152/physiolgenomics.00148.2007. [DOI] [PubMed] [Google Scholar]
- Eriksson N, Macpherson JM, Tung JY, Hon LS, Naughton B, Saxonov S, Avey L, Wojcicki A, Pe'er I, Mountain J. Web-Based, Participant-Driven Studies Yield Novel Genetic Associations for Common Traits. PLoS ONE. 2010;6(6) doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney E. The impact of bitter perception and genotypic variation of TAS2R38 on food choice. Nutrition Bulletin. 2011;36(1):20–33. [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Fischer R, Griffin F, Kaplan AR. Taste Thresholds, Cigarette Smoking, and Food Dislikes. Med Exp Int J Exp Med. 1963;210:151–167. doi: 10.1159/000135346. [DOI] [PubMed] [Google Scholar]
- Fushan AA, Simons CT, Slack JP, Drayna D. Association between common variation in genes encoding sweet taste signaling components and human sucrose perception. Chem Senses. 2010;35(7):579–592. doi: 10.1093/chemse/bjq063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushan AA, Simons CT, Slack JP, Manichaikul A, Drayna D. Allelic polymorphism within the TAS1R3 promoter is associated with human taste sensitivity to sucrose. Curr Biol. 2009;19(15):1288–1293. doi: 10.1016/j.cub.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville EV, Kaplan AR. Food Preference and Sensitivity of Taste for Bitter Compounds. Nature. 1965;205:851–853. [Google Scholar]
- Gorovic N, Afzal S, Tjonneland A, Overvad K, Vogel U, Albrechtsen C, Poulsen HE. Genetic variation in the hTAS2R38 taste receptor and brassica vegetable intake. Scand J Clin Lab Invest. 2011;71(4):274–279. doi: 10.3109/00365513.2011.559553. [DOI] [PubMed] [Google Scholar]
- Greene LS, Desor JA, Maller O. Heredity and experience: their relative importance in the development of taste preference in man. J Comp Physiol Psychol. 1975;89(3):279–284. doi: 10.1037/h0076802. [DOI] [PubMed] [Google Scholar]
- Guinard JX, Zoumas-Morse C, Walchak C. Relation between parotid saliva flow and composition and the perception of gustatory and trigeminal stimuli in foods. Physiol Behav. 1997;63(1):109–118. doi: 10.1016/s0031-9384(97)00399-5. [DOI] [PubMed] [Google Scholar]
- Hasin-Brumshtein Y, Lancet D, Olender T. Human olfaction: from genomic variation to phenotypic diversity. Trends Genet. 2009;25(4):178–184. doi: 10.1016/j.tig.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Hasselbalch AL, Silventoinen K, Keskitalo K, Pietilainen KH, Rissanen A, Heitmann BL, Kyvik KO, Sorensen TIA, Kaprio J. Twin Study of Heritability of Eating Bread in Danish and Finnish Men and Women. Twin Research and Human Genetics. 2010;13(2):163–167. doi: 10.1375/twin.13.2.163. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10(8):551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE. Transdisciplinary Perspectives on Sweetness. Chemosensory Perception. 2008;1(1):48–57. [Google Scholar]
- Hayes JE, Bartoshuk LM, Kidd JK, Duffy VB. Supertasting and PROP Bitterness Depends on More Than the TAS2R38 Gene. Chem Senses. 2008;33(3):255–265. doi: 10.1093/chemse/bjm084. Epub 221 January 2008, doi:2010.1093/chemse/bjm2084. [DOI] [PubMed] [Google Scholar]
- Hayes JE, Duffy VB. Revisiting sugar fat mixtures: sweetness and creaminess vary with phenotypic markers of oral sensation. Chem Senses. 2007;32(3):225–236. doi: 10.1093/chemse/bjl050. First published January 224, 2007, doi:2010.1093/chemse/bjl2050. [DOI] [PubMed] [Google Scholar]
- Hayes JE, Duffy VB. Oral sensory phenotype identifies level of sugar and fat required for maximal liking. Physiol Behav. 2008;95(1–2):77–87. doi: 10.1016/j.physbeh.2008.04.023. Epub 2008 May 2002. doi:2010.1016/j.physbeh.2008.2004.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Keast RSJ. Two decades of supertasting: where do we stand? Physiology & Behavior. 2011;104(5):1072–1074. doi: 10.1016/j.physbeh.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, McGeary JE, Grenga A, Swift RM. Do TAS1R3 promoter region SNP rs35744813 A allele carriers show a reduced response to concentrated sucrose? St. Pete's Beach; Florida: 2010. [ABSTRACT], AChemS XXXII. [Google Scholar]
- Hayes JE, Sullivan BS, Duffy VB. Explaining variability in sodium intake through oral sensory phenotype, salt sensation and liking. Physiol Behav. 2010;100(4):369–380. doi: 10.1016/j.physbeh.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Wallace MR, Knopik VS, Herbstman DM, Bartoshuk LM, Duffy VB. Allelic Variation in TAS2R Bitter Receptor Genes Associates with Variation in Sensations from and Ingestive Behaviors toward Common Bitter Beverages in Adults. Chem Senses. 2011;36(3):311–319. doi: 10.1093/chemse/bjq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin RI, Martin BM, Agarwal RP. Efficacy of exogenous oral zinc in treatment of patients with carbonic anhydrase VI deficiency. Am J Med Sci. 1999;318(6):392–405. doi: 10.1097/00000441-199912000-00006. [DOI] [PubMed] [Google Scholar]
- Hinrichs AL, Wang JC, Bufe B, Kwon JM, Budde J, Allen R, Bertelsen S, Evans W, Dick D, Rice J, Foroud T, Nurnberger J, Tischfield JA, Kuperman S, Crowe R, Hesselbrock V, Schuckit M, Almasy L, Porjesz B, Edenberg HJ, Begleiter H, Meyerhof W, Bierut LJ, Goate AM. Functional variant in a bitter-taste receptor (hTAS2R16) influences risk of alcohol dependence. Am J Hum Genet. 2006;78(1):103–111. doi: 10.1086/499253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio N, Yoshida R, Yasumatsu K, Yanagawa Y, Ishimaru Y, Matsunami H, Ninomiya Y. Sour taste responses in mice lacking PKD channels. PLoS One. 2011;6(5):e20007. doi: 10.1371/journal.pone.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne J, Hayes J, Lawless HT. Turbidity as a measure of salivary protein reactions with astringent substances. Chem Senses. 2002;27(7):653–659. doi: 10.1093/chemse/27.7.653. [DOI] [PubMed] [Google Scholar]
- Horne J, Lawless HT, Speirs W, Sposato D. Bitter taste of saccharin and acesulfame-k. Chem Senses. 2002;27(1):31–38. doi: 10.1093/chemse/27.1.31. [DOI] [PubMed] [Google Scholar]
- Hyde RJ, Witherly SA. Dynamic contrast: a sensory contribution to palatability. Appetite. 1993;21(1):1–16. doi: 10.1006/appe.1993.1032. [DOI] [PubMed] [Google Scholar]
- Imai H, Suzuki N, Ishimaru Y, Sakurai T, Yin L, Pan W, Abe K, Misaka T, Hirai H. Functional diversity of bitter taste receptor TAS2R16 in primates. Biol Lett. 2012;8(4):652–656. doi: 10.1098/rsbl.2011.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intranuovo LR, Powers AS. The perceived bitterness of beer and 6-n-propylthiouracil (PROP) taste sensitivity. Ann N Y Acad Sci. 1998;855:813–815. doi: 10.1111/j.1749-6632.1998.tb10665.x. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Noble AC. Temporal Perception of Astringency and Sweetness in Red Wine. Food Quality and Preference. 1995;6(1):27–33. [Google Scholar]
- Ishimaru Y, Inada H, Kubota M, Zhuang H, Tominaga M, Matsunami H. Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc Natl Acad Sci U S A. 2006;103(33):12569–12574. doi: 10.1073/pnas.0602702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger SR, Pineau B, Bava CM, Atkinson KR, AMcRae JF, Axten LG, Chheang SL, Beresford MK, Peng M, Paisley AG, Reinbach HC, Rouse SA, Wohlers MW, Jia Y, Newcomb RD. Investigation of the impact of sensitivity to cis-3-hexen-1-ol (green/grassy) on food acceptability and selection\J. Food Quality and Preference. 2012;24(2):230–242. [Google Scholar]
- Kallithraka S, Bakker J, Clifford MN. Evidence that salivary proteins are involved in astringency. Journal of Sensory Studies. 1998;13(1):29–43. [Google Scholar]
- Kamerud JK, Delwiche JF. Individual Differences in Perceived Bitterness Predict Liking of Sweeteners. Chem Senses. 2007 doi: 10.1093/chemse/bjm050. [DOI] [PubMed] [Google Scholar]
- Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. Genetic variation in a human odorant receptor alters odour perception. Nature. 2007;449(7161):468–472. doi: 10.1038/nature06162. [DOI] [PubMed] [Google Scholar]
- Keskitalo K, Knaapila A, Kallela M, Palotie A, Wessman M, Sammalisto S, Peltonen L, Tuorila H, Perola M. Sweet taste preferences are partly genetically determined: identification of a trait locus on chromosome 16. Am J Clin Nutr. 2007;86(1):55–63. doi: 10.1093/ajcn/86.1.55. [DOI] [PubMed] [Google Scholar]
- Keskitalo K, Tuorila H, Spector TD, Cherkas LF, Knaapila A, Silventoinen K, Perola M. Same genetic components underlie different measures of sweet taste preference. Am J Clin Nutr. 2007;86(6):1663–1669. doi: 10.1093/ajcn/86.5.1663. [DOI] [PubMed] [Google Scholar]
- Kim U, Wooding S, Ricci D, Jorde LB, Drayna D. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Hum Mutat. 2005;26(3):199–204. doi: 10.1002/humu.20203. [DOI] [PubMed] [Google Scholar]
- Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299(5610):1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- Knaapila A, Silventoinen K, Broms U, Rose RJ, Perola M, Kaprio J, Tuorila HM. Food Neophobia in Young Adults: Genetic Architecture and Relation to Personality, Pleasantness and Use Frequency of Foods, and Body Mass Index-A Twin Study. Behavior Genetics. 2011;41(4):512–521. doi: 10.1007/s10519-010-9403-8. [DOI] [PubMed] [Google Scholar]
- Kuhn C, Bufe B, Winnig M, Hofmann T, Frank O, Behrens M, Lewtschenko T, Slack JP, Ward CD, Meyerhof W. Bitter taste receptors for saccharin and acesulfame K. J Neurosci. 2004;24(45):10260–10265. doi: 10.1523/JNEUROSCI.1225-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier SA, Hayes JE, Duffy VB. Sweet and bitter tastes of alcoholic beverages mediate alcohol intake in of-age undergraduates. Physiol Behav. 2005;83(5):821–831. doi: 10.1016/j.physbeh.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Lawless HT. Evidence for neural inhibition in bittersweet taste mixtures. Journal of Comparative & Physiological Psychology. 1979;93(3):538–547. doi: 10.1037/h0077582. [DOI] [PubMed] [Google Scholar]
- Lee CB, Lawless HT. Time-course of astringent sensations. Chemical Senses. 1991;16(3):225–238. [Google Scholar]
- Lesschaeve I, Noble AC. Polyphenols: factors influencing their sensory properties and their effects on food and beverage preferences. Am J Clin Nutr. 2005;81(1 Suppl):330S–335S. doi: 10.1093/ajcn/81.1.330S. [DOI] [PubMed] [Google Scholar]
- Liem DG, Westerbeek A, Wolterink S, Kok FJ, de Graaf C. Sour taste preferences of children relate to preference for novel and intense stimuli. Chem Senses. 2004;29(8):713–720. doi: 10.1093/chemse/bjh077. [DOI] [PubMed] [Google Scholar]
- Lugaz O, Pillias AM, Faurion A. A new specific ageusia: some humans cannot taste L-glutamate. Chem Senses. 2002;27(2):105–115. doi: 10.1093/chemse/27.2.105. [DOI] [PubMed] [Google Scholar]
- Lunde K, Egelandsdal B, Skuterud E, Mainland JD, Lea T, Hersleth M, Matsunami H. Genetic variation of an odorant receptor OR7D4 and sensory perception of cooked meat containing androstenone. PLoS One. 2012;7(5):e35259. doi: 10.1371/journal.pone.0035259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren B, Jonsson B, Pangborn RM, Sontag AM, Barylko-Pikielna N, Pietrzak E, Garruti RS, Chaib MA, Yoshida M. Taste discrimination vs hedonic response to sucrose in coffee beverage. An interlaboratory study. Chem Senses Flav. 1978;3(3):249–265. [Google Scholar]
- Maller O, Desor JA. Effect of taste on ingestion by human newborns. Symp Oral Sens Percept. 1973:279–291. [PubMed] [Google Scholar]
- Mandel AL, Peyrot des Gachons C, Plank KL, Alarcon S, Breslin PA. Individual differences in AMY1 gene copy number, salivary alpha-amylase levels, and the perception of oral starch. PLoS One. 2010;5(10):e13352. doi: 10.1371/journal.pone.0013352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes RD. Is there a Fatty Acid Taste? Annu Rev Nutr. 2009;29:305–327. doi: 10.1146/annurev-nutr-080508-141108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer L, El-Sohemy A. Prevalence of cilantro (Coriandrum sativum) disliking among different ethnocultural groups. Flavour. 2012;1(1):1–5. [Google Scholar]
- Menashe I, Man O, Lancet D, Gilad Y. Different noses for different people. Nat Genet. 2003;34(2):143–144. doi: 10.1038/ng1160. [DOI] [PubMed] [Google Scholar]
- Mennella JA, Pepino MY, Reed DR. Genetic and environmental determinants of bitter perception and sweet preferences. Pediatrics. 2005;115(2):e216–222. doi: 10.1542/peds.2004-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses. 2010;35(2):157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- Morini G, Bassoli A, Temussi Piero A. Multiple Receptors or Multiple Sites? Modeling the Human T1R2-T1R3 Sweet Taste Receptor Sweetness and Sweeteners. 2008:147–161. [Google Scholar]
- Mustavich LF, Miller P, Kidd KK, Zhao H. Using a pharmacokinetic model to relate an individual's susceptibility to alcohol dependence to genotypes. Hum Hered. 2010;70(3):177–193. doi: 10.1159/000317056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb R, Xia M, Reed D. Heritable differences in chemosensory ability among humans. Flavour. 2012;1(1):9. [Google Scholar]
- Niimura Y. Evolution of Chemosensory Receptor Genes in Primates and Other Mammals. In: Hirohisa H. Imai, Go Y., editors. Post-genome Biology of Primates. Springer; Tokyo: 2012. pp. 43–63. [Google Scholar]
- Padiglia A, Zonza A, Atzori E, Chillotti C, Calo C, Tepper BJ, Barbarossa IT. Sensitivity to 6-n-propylthiouracil is associated with gustin (carbonic anhydrase VI) gene polymorphism, salivary zinc, and body mass index in humans. Am J Clin Nutr. 2010;92(3):539–545. doi: 10.3945/ajcn.2010.29418. [DOI] [PubMed] [Google Scholar]
- Patterson PM, Lauder BA. THE INCIDENCE AND PROBABLE INHERITANCE OF SMELL BLINDNESS: To Normal Butyl Mercaptan. J Hered. 1948;39(10):295–297. doi: 10.1093/oxfordjournals.jhered.a105774. [DOI] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, Carter NP, Lee C, Stone AC. Diet and the evolution of human amylase gene copy number variation. Nature Genetics. 2007;39(10):1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prindiville EA, Marshall RT, Heymann H. Effect of Milk Fat on the Sensory Properties of Chocolate Ice Cream. Journal of Dairy Science. 1999;82(7):1425–1432. doi: 10.3168/jds.S0022-0302(00)75105-8. [DOI] [PubMed] [Google Scholar]
- Prindiville EA, Marshall RT, Heymann H. Effect of milk fat, cocoa butter, and whey protein fat replacers on the sensory properties of lowfat and nonfat chocolate ice cream. Journal of Dairy Science. 2000;83:2216–2223. doi: 10.3168/jds.S0022-0302(00)75105-8. [DOI] [PubMed] [Google Scholar]
- Prodi DA, Drayna D, Forabosco P, Palmas MA, Maestrale GB, Piras D, Pirastu M, Angius A. Bitter taste study in a sardinian genetic isolate supports the association of phenylthiocarbamide sensitivity to the TAS2R38 bitter receptor gene. Chem Senses. 2004;29(8):697–702. doi: 10.1093/chemse/bjh074. [DOI] [PubMed] [Google Scholar]
- Pronin AN, Xu H, Tang H, Zhang L, Li Q, Li X. Specific alleles of bitter receptor genes influence human sensitivity to the bitterness of aloin and saccharin. Curr Biol. 2007;17(16):1403–1408. doi: 10.1016/j.cub.2007.07.046. [DOI] [PubMed] [Google Scholar]
- Reed DR, Zhu G, Breslin PA, Duke FF, Henders AK, Campbell MJ, Montgomery GW, Medland SE, Martin NG, Wright MJ. The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudnitzky N, Bufe B, Thalmann S, Kuhn C, Gunn HC, Xing C, Crider BP, Behrens M, Meyerhof W, Wooding SP. Genomic, genetic and functional dissection of bitter taste responses to artificial sweeteners. Hum Mol Genet. 2011;20(17):3437–3449. doi: 10.1093/hmg/ddr252. [DOI] [PubMed] [Google Scholar]
- Sacerdote C, Guarrera S, Smith GD, Grioni S, Krogh V, Masala G, Mattiello A, Palli D, Panico S, Tumino R, Veglia F, Matullo G, Vineis P. Lactase persistence and bitter taste response: instrumental variables and mendelian randomization in epidemiologic studies of dietary factors and cancer risk. Am J Epidemiol. 2007;166(5):576–581. doi: 10.1093/aje/kwm113. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Misaka T, Ishiguro M, Masuda K, Sugawara T, Ito K, Kobayashi T, Matsuo S, Ishimaru Y, Asakura T, Abe K. Characterization of the beta-D-Glucopyranoside Binding Site of the Human Bitter Taste Receptor hTAS2R16. Journal of Biological Chemistry. 2010;285(36):28373–28378. doi: 10.1074/jbc.M110.144444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Misaka T, Ueno Y, Ishiguro M, Matsuo S, Ishimaru Y, Asakura T, Abe K. The human bitter taste receptor, hTAS2R16, discriminates slight differences in the configuration of disaccharides. Biochem Biophys Res Commun. 2010;402(4):595–601. doi: 10.1016/j.bbrc.2010.10.059. [DOI] [PubMed] [Google Scholar]
- Sandell MA, Breslin PA. Variability in a taste-receptor gene determines whether we taste toxins in food. Curr Biol. 2006;16(18):R792–794. doi: 10.1016/j.cub.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Schiffman SS, Booth BJ, Losee ML, Pecore SD, Warwick ZS. Bitterness of sweeteners as a function of concentration. Brain Res Bull. 1995;36(5):505–513. doi: 10.1016/0361-9230(94)00225-p. [DOI] [PubMed] [Google Scholar]
- Schiffman SS, Reilly DA, Clark TB., 3rd Qualitative differences among sweeteners. Physiol Behav. 1979;23(1):1–9. doi: 10.1016/0031-9384(79)90113-6. [DOI] [PubMed] [Google Scholar]
- Sclafani A, Glass DS, Margolskee RF, Glendinning JI. Gut T1R3 sweet taste receptors do not mediate sucrose-conditioned flavor preferences in mice. Am J Physiol Regul Integr Comp Physiol. 2010;299(6):R1643–1650. doi: 10.1152/ajpregu.00495.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemura N, Shirosaki S, Sanematsu K, Yoshida R, Ninomiya Y. Genetic and molecular basis of individual differences in human umami taste perception. PLoS One. 2009;4(8):e6717. doi: 10.1371/journal.pone.0006717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagghe K, Louis-Sylvestre J. Influence of PROP-sensitivity on taste perceptions and hedonics in French women. A study performed without retronasal olfaction. Appetite. 1998;30(3):325–339. doi: 10.1006/appe.1997.0145. [DOI] [PubMed] [Google Scholar]
- Soranzo N, Bufe B, Sabeti PC, Wilson JF, Weale ME, Marguerie R, Meyerhof W, Goldstein DB. Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr Biol. 2005;15(14):1257–1265. doi: 10.1016/j.cub.2005.06.042. [DOI] [PubMed] [Google Scholar]
- Tepper BJ. Nutritional Implications of Genetic Taste Variation: The Role of PROP Sensitivity and Other Taste Phenotypes. Annu Rev Nutr. 2008 doi: 10.1146/annurev.nutr.28.061807.155458. [DOI] [PubMed] [Google Scholar]
- Tepper BJ, Nurse RJ. Fat perception is related to PROP taster status. Physiol Behav. 1997;61(6):949–954. doi: 10.1016/s0031-9384(96)00608-7. [DOI] [PubMed] [Google Scholar]
- Teucher B, Skinner J, Skidmore PML, Cassidy A, Fairweather-Tait SJ, Hooper L, Roe MA, Foxall R, Oyston SL, Cherkas LF, Perks UC, Spector TD, MacGregor AJ. Dietary patterns and heritability of food choice in a UK female twin cohort. Twin Research and Human Genetics. 2007;10(5):734–748. doi: 10.1375/twin.10.5.734. [DOI] [PubMed] [Google Scholar]
- Timpson NJ, Christensen M, Lawlor DA, Gaunt TR, Day IN, Smith GD. TAS2R38 (phenylthiocarbamide) haplotypes, coronary heart disease traits, and eating behavior in the British Women's Heart and Health Study. Am J Clin Nutr. 2005;81(5):1005–1011. doi: 10.1093/ajcn/81.5.1005. [DOI] [PubMed] [Google Scholar]
- Tornwall O, Dinnella C, Keskitalo-Vuokko K, Silventoinen K, Perola M, Monteleone E, Kaprio J, Tuorila H. Astringency Perception and Heritability Among Young Finnish Twins. Chemosensory Perception. 2011;4(4):134–144. [Google Scholar]
- Tornwall O, Silventoinen K, Keskitalo-Vuokko K, Perola M, Kaprio J, Tuorila H. Genetic contribution to sour taste preference. Appetite. 2012;58(2):687–694. doi: 10.1016/j.appet.2011.12.020. [DOI] [PubMed] [Google Scholar]
- van Dongen J, Slagboom PE, Draisma HHM, Martin NG, Boomsma DI. The continuing value of twin studies in the omics era. Nat Rev Genet. 2012 doi: 10.1038/nrg3243. [DOI] [PubMed] [Google Scholar]
- van Doorn HE, van der Kruk GC, van Holst GJ, Raaijmakers-Ruijs NCME, Postma E, Groeneweg B, Jongen WHF. The glucosinolates sinigrin and progoitrin are important determinants for taste preference and bitterness of brussels sprouts. Journal of the Science of Food and Agriculture. 1998;78(1):30–38. [Google Scholar]
- Wang JC, Hinrichs AL, Bertelsen S, Stock H, Budde JP, Dick DM, Bucholz KK, Rice J, Saccone N, Edenberg HJ, Hesselbrock V, Kuperman S, Schuckit MA, Bierut LJ, Goate AM. Functional Variants in TAS2R38 and TAS2R16 Influence Alcohol Consumption in High-Risk Families of African-American Origin. Alcohol Clin Exp Res. 2007;31(2):209–215. doi: 10.1111/j.1530-0277.2006.00297.x. [DOI] [PubMed] [Google Scholar]
- Waszak SM, Hasin Y, Zichner T, Olender T, Keydar I, Khen M, Stütz AM, Schlattl A, Lancet D, Korbel JO. Systematic Inference of Copy-Number Genotypes from Personal Genome Sequencing Data Reveals Extensive Olfactory Receptor Gene Content Diversity. PLoS Comput Biol. 2010;6(11):e1000988. doi: 10.1371/journal.pcbi.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkening S, Chen BW, Bermejo JL, Canzian F. Is there still a need for candidate gene approaches in the era of genome-wide association studies? Genomics. 2009;93(5):415–419. doi: 10.1016/j.ygeno.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Wise PM, Hansen JL, Reed DR, Breslin PAS. Twin study of the heritability of recognition thresholds for sour and salty taste. Chemical Senses. 2007;32(8):749–754. doi: 10.1093/chemse/bjm042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, deLeeuw RJ, Dosanjh NS, Kimm LR, Cheng Z, Horsman DE, MacAulay C, Ng RT, Brown CJ, Eichler EE, Lam WL. A Comprehensive Analysis of Common Copy-Number Variations in the Human Genome. American journal of human genetics. 2007;80(1):91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S. Phenylthiocarbamide: a 75-year adventure in genetics and natural selection. Genetics. 2006;172(4):2015–2023. doi: 10.1093/genetics/172.4.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S, Gunn H, Ramos P, Thalmann S, Xing C, Meyerhof W. Genetics and Bitter Taste Responses to Goitrin, a Plant Toxin Found in Vegetables. Chem Senses. 2010 doi: 10.1093/chemse/bjq061. [DOI] [PubMed] [Google Scholar]
- Wysocki CJ, Beauchamp GK. Ability to smell androstenone is genetically determined. Proc Natl Acad Sci U S A. 1984;81(15):4899–4902. doi: 10.1073/pnas.81.15.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Firestein S. Genomics of Olfactory Receptors Chemosensory systems in mammals, fishes, and insects. Vol. 47. Springer-Verlag; Berlin Heidelberg: 2009. pp. 239–255. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.