Abstract
Background
Genome-wide association studies have implicated single nucleotide polymorphisms (SNPs) in 7 genes as glioma risk factors, including 2 (TERT, RTEL1) involved in telomerase structure/function. We examined associations of these 7 established glioma risk loci with age at diagnosis among patients with glioma.
Methods
SNP genotype data were available for 2286 Caucasian glioma patients from the University of California, San Francisco (n = 1434) and the Mayo Clinic (n = 852). Regression analyses were performed to test for associations between “number of risk alleles” and “age at diagnosis,” adjusted for sex and study site and stratified by tumor grade/histology where appropriate.
Results
Four SNPs were significantly associated with age at diagnosis. Carrying a greater number of risk alleles at rs55705857 (CCDC26) and at rs498872 (PHLDB1) was associated with younger age at diagnosis (P = 1.4 × 10−22 and P = 9.5 × 10−7, respectively). These SNPs are stronger risk factors for oligodendroglial tumors, which tend to occur in younger patients, and their association with age at diagnosis varied across tumor subtypes. In contrast, carrying more risk alleles at rs2736100 (TERT) and at rs6010620 (RTEL1) was associated with older age at diagnosis (P = 6.2 × 10−4 and P = 2.5 × 10−4, respectively). These SNPs are risk factors for all glioma grades/histologies, and their association with age at diagnosis was consistent across tumor subgroups.
Conclusions
Carrying a greater number of risk alleles might be expected to decrease age at diagnosis. However, glioma susceptibility conferred by variation in telomerase-related genes did not follow this pattern. This supports the hypothesis that telomerase-related mechanisms of telomere maintenance are more associated with gliomas that develop later in life than those utilizing telomerase-independent mechanisms (ie, alternative lengthening of telomeres).
Keywords: age at diagnosis, glioma, single nucleotide polymorphism, telomerase, telomere
Glioma, the most common central nervous system cancer in adults, generally has poor prognosis. Glioblastoma, the most common and most aggressive form of glioma, has a median patient survival time of just 15 months from diagnosis under current standard of care.1 While prognosis is better for low-grade astrocytic tumors and tumors with an oligodendroglial component, over time these tumors all progress to high-grade glioma.2
Gliomagenesis is a complex and multifaceted process influenced by both inherited and acquired genetic variation. Glioma risk loci in 7 genes have been confirmed in genome-wide association studies.3–6 Several of these risk loci are found in genes previously implicated in gliomagenesis due to their mutation in glioma-associated hereditary cancer syndromes (TP53, CDKN2B/ANRIL) or their alteration in glioma tumors (TP53, CDKN2B/ANRIL, EGFR).7–9 Risk loci in 2 genes involved in telomerase structure and function (TERT [telomerase reverse transcriptase] and RTEL1 [regulator of telomere elongation helicase 1]) had not been implicated in gliomagenesis prior to genome-wide association studies, but telomerase activation has been observed in ∼90% of all human cancers.10
Telomeres act as a protective cap at the end of chromosomes but are progressively shortened during mitotic divisions.11 Telomere depletion ultimately leads to replicative senescence, limiting the proliferative capacity of cells. With activation of telomerase, an enzyme that adds DNA sequence repeats to telomeres, dividing cells can replace lost telomeric DNA and continue proliferating.10 TERT is a key component of human telomerase, and RTEL1 is needed to allow telomerase-dependent telomere extension to proceed effectively.12,13 Of the tumors that do not maintain telomere length through activation of telomerase, a significant subset activates a secondary pathway: alternative lengthening of telomeres (ALT).14
Inheriting an increased number of glioma risk alleles might be expected to decrease age at diagnosis among glioma patients by reducing the number of somatic mutations an individual must acquire to initiate tumor formation or by facilitating the accumulation of such mutations. To investigate this, we examined the associations of known glioma risk loci with age at diagnosis in glioma patients from the University of California, San Francisco (UCSF) and the Mayo Clinic. Caucasian glioma patients were genotyped at single nucleotide polymorphisms (SNPs) in 7 genes associated with glioma risk in previous genome-wide association studies, including variants in TERT, EGFR, CCDC26, CDKN2B/ANRIL, PHLDB1, TP53, and RTEL1. Associations were also calculated within histologic subgroups, stratified by tumor IDH-mutation status, and pooled across study sites. Interactions between age and risk SNPs were also examined in case-control comparisons.
Materials and Methods
Study Population
This study included European-ancestry glioma patients and controls from UCSF (1434 cases, 1114 controls) and the Mayo Clinic (852 cases, 789 controls). Both participating institutions received institutional review board approval, and informed consent was obtained from subjects. Patient recruitment methods have been described in detail elsewhere.3,15 Pathology review was performed as previously described.3,16
SNP Selection
SNPs in 7 different genes have been significantly associated with glioma risk in previous genome-wide case-control studies.3–6 We chose the SNP most strongly associated with glioma risk in previous case-control analyses for all association testing described in this paper.15,17 Thus, we analyzed only 1 glioma risk SNP from each region.
Genotyping
Genotyped on GoldenGate custom genotyping arrays (Illumina) were 810 UCSF cases, all Mayo cases, 512 UCSF controls, and all Mayo controls. GoldenGate genotyping was performed by the UCSF Genome Center and Mayo Genotyping core facilities as previously described.15 Samples were submitted in 96-well plates containing intra- and interplate replicates to ensure genotype reproducibility.
Genotypes for an additional 606 UCSF cases and 602 controls were extracted from an Illumina 370k genome-wide SNP chip genotyped as part of a previous study.3 The 370k platform does not contain a probe for rs55705857 (CCDC26) or rs78378222 (TP53). Therefore, genotype data were available for only 810 total UCSF cases and 512 UCSF controls at rs55705857. However, 461 additional UCSF cases were directly genotyped at rs78378222 (TP53) as previously described,6 for a total of 1271 UCSF cases and 512 UCSF controls with genotype data at this locus.
For both study sites, samples with genotyping array call rates <95% were excluded from analysis. SNPs with genotyping call rates <95% in any site were excluded from all analyses. To exclude poorly genotyped SNPs, any SNP with a Hardy-Weinberg equilibrium P-value <.001 in controls, stratified by site, was removed from further analyses.
Statistical Analysis of SNP Associations
For the case-only analyses, the correlation between number of risk alleles and age at diagnosis was assessed using Pearson's correlation coefficient calculated in SAS v9.1.3, stratified by study site. Regression analyses were conducted using linear regression in PLINK v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/), adjusted for sex and study site, assuming an allelic additive model in which the regression coefficient represents the effect of each extra copy of the risk allele.18 Because at 3 loci the glioma risk allele is not the minor allele (rs2736100, rs11979158, rs6010620), the reference allele was forced using the “—reference-allele” command so that regression coefficients corresponded to the change in age at diagnosis associated with each additional copy of the glioma risk allele, as defined by previous case-control studies. Linear regression coefficients were also calculated in analyses stratified by tumor histology, and heterogeneity across histologic strata was assessed using Cochran's Q and I2. For all linear regression models, residual plots, including normal probability plots, were examined for departures from normality, excess skew, and kurtosis. All reported P-values are 2-sided. For the primary study results listed in Table 2, a total of 11 statistical comparisons were considered (1 comparison per SNP for the 3 loci displaying no heterogeneity of effect across tumor types; 2 comparisons per SNP for the 4 loci displaying significant heterogeneity of effect across tumor types). A strict Bonferroni correction for these 11 comparisons corresponds to an adjusted significance threshold of 4.5 × 10−3 (0.05/11).
Table 2.
Association between age at diagnosis and number of risk alleles at known glioma risk loci in combined UCSF Adult Glioma Study and Mayo Clinic patients, stratified by tumor cell type
| SNP | Chromosome | Gene | Risk Allele | All Gliomas |
Purely Astrocytic Tumorsa |
Tumors With an Oligo Componentb |
Heterogeneity Testsc |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect (SE)d | Pe | Effect (SE)d | Pe | Effect (SE)d | Pe | P | I2 | ||||
| Rs2736100 | 5 | TERT | C | 1.47 (0.43) | 6.2 × 10−4 | 1.38 (0.48) | 4.3 × 10−3 | 0.85 (0.74) | 0.25 | 0.55 | 0 |
| Rs11979158 | 7 | EGFR | A | 0.48 (0.60) | 0.43 | 0.78 (0.68) | 0.25 | −1.37 (1.05) | 0.19 | 0.086 | 66.16 |
| Rs55705857 | 8 | CCDC26 | G | - | - | −7.25 (0.93) | 1.2 × 10−14 | −2.81 (0.94) | 3.1 × 10−3 | 8.0 × 10−4 | 91.15 |
| Rs1412829 | 9 | CDKN2B/ANRIL | G | - | - | 0.55 (0.47) | 0.24 | −1.91 (0.77) | 0.013 | 6.2 × 10−3 | 86.66 |
| Rs498872 | 11 | PHLDB1 | A | - | - | −2.30 (0.48) | 2.1 × 10−6 | 0.25 (0.73) | 0.73 | 3.5 × 10−3 | 88.31 |
| Rs78378222 | 17 | TP53 | C | - | - | −2.49 (1.35) | 0.065 | 4.25 (2.45) | 0.084 | 0.016 | 82.77 |
| Rs6010620 | 20 | RTEL1 | G | 2.02 (0.55) | 2.5 × 10−4 | 1.80 (0.63) | 4.3 × 10−3 | 0.81 (0.91) | 0.37 | 0.37 | 0 |
aIncludes glioblastomas (n = 1217), and grades II–III astrocytomas (n = 555).
bIncludes oligodendrogliomas (n = 277) and mixed oligoastrocytomas (n = 230).
cP-values from Cochran’s Q-statistic, testing for heterogeneity in beta across strata of tumor cell type (purely astrocytic vs oligodendroglial). I2 can range from 0 to 100, where larger numbers indicate a greater level of heterogeneity across histology strata.
dEffect size (in y) is generated from a linear regression model where age at diagnosis is the dependent variable and number of risk alleles is the independent variable, controlling for sex and study site. Positive values indicate older age at diagnosis with an increasing number of risk alleles. Negative values indicate younger age at diagnosis with an increasing number of risk alleles.
eP-values are 2-sided and are derived from the regression model (H0: beta = 0). A total of 11 statistical comparisons are considered (1 comparison per SNP for the 3 loci displaying no heterogeneity of effect across tumor types; 2 comparisons per SNP for the 4 loci displaying significant heterogeneity of effect across tumor types). A strict Bonferroni correction for these 11 comparisons corresponds to an adjusted significance threshold of 4.5 × 10−3 (0.05/11). P-values in bold were considered statistically significant.
Case-control association statistics for rs55705857 (CCDC26), rs498872 (PHLDB1), rs2736100 (TERT), and rs6010620 (RTEL1) were calculated using logistic regression in PLINK, adjusting for sex and study site. In order to assess variation in SNP effect size at different patient ages, glioma cases and controls were divided into 5 age strata, in years: <40, 40–49, 50–59, 60–69, and 70+. Reported case-control associations are for an allelic additive model, where odds ratios are for each additional copy of the known risk allele.
Assessment of IDH-Mutation Status
UCSF tumor specimens were sequenced to identify IDH1 and IDH2 mutations using previously described methods.19 The region spanning the R132 codon of IDH1 and the region spanning the R172 codon of IDH2 were amplified by PCR with M13-tagged primers to facilitate amplification and sequencing. Products were run on a 1.5% agarose gel and subsequently sequenced in both directions at the UCSF Genomics Core Facility according to the manufacturer's protocol. Sequences were analyzed with Applied Biosystems Sequence Scanner Software v1.0. Mayo tumor specimens were assayed for IDH1 mutations using pyrosequencing and for IDH2 mutations using both pyrosequencing and Sanger sequencing as previously described.20
Results
A total of 2286 glioma patients (1434 UCSF, 852 Mayo) and 1903 controls (1114 UCSF, 789 Mayo) had acceptable genotyping call rates and were included in our analyses. Subject characteristics, including histopathologic classification of glioma cases, are outlined in Table 1. The 7 SNPs reported in this study passed all call-rate and Hardy-Weinberg equilibrium thresholds.
Table 1.
Demographic and tumor histology characteristics of the UCSF Adult Glioma Study and the Mayo Clinic glioma patients and controls used in the genetic association analyses
| UCSF Samples |
Mayo Samples |
Combined |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| n | Mean Age, yr (SD) | % Male | n | Mean Age, yr (SD) | % Male | n | Mean Age, yr (SD) | % Male | |
| Glioblastoma | 887 | 56.6 (12.1) | 64.7 | 330 | 55.7 (12.4) | 62.7 | 1219 | 56.4 (12.2) | 64.2 |
| Anaplastic astrocytoma | 170 | 46.9 (15.0) | 55.9 | 188 | 48.4 (14.8) | 54.8 | 358 | 47.7 (14.9) | 55.3 |
| Grade II astrocytoma | 115 | 42.5 (13.5) | 66.1 | 70 | 42.1 (13.6) | 55.7 | 185 | 42.4 (13.5) | 62.2 |
| Mixed oligoastrocytoma | 64 | 39.0 (11.3) | 59.4 | 166 | 39.0 (11.5) | 54.8 | 230 | 39.0 (11.4) | 56.1 |
| Oligodendroglioma | 179 | 45.0 (12.1) | 54.7 | 98 | 41.6 (10.7) | 51.0 | 277 | 43.8 (11.7) | 53.4 |
| All histologies | 1434a | 52.0 (14.0) | 62.3 | 852 | 48.1 (14.4) | 57.5 | 2288 | 50.5 (14.3) | 60.5 |
| Controls | 1114 | 56.9 (15.2) | 54.4 | 789 | 49.8 (14.1) | 57.0 | 1903 | 53.9 (15.2) | 55.5 |
aNumbers by histologic type add to 1434 because 12 astrocytomas were of indeterminate grade and 7 gliomas had no histology information.
Cases were stratified into 2 groups: purely astrocytic tumors (glioblastoma, anaplastic astrocytoma, grade 2 astrocytoma) and tumors with an oligodendroglial component (oligodendroglioma and mixed oligoastrocytoma). Four SNPs were strongly and significantly associated with age at diagnosis in one or both subgroups (Table 2), and these results were consistent in UCSF and Mayo patients (Table S1). The CCDC26 risk allele was associated with a 7.25-year decreased age at diagnosis in the astrocytic subgroup (95% confidence interval [CI] = 5.44–9.07 yr; P = 1.2 × 10−14) and a 2.81-year decreased age at diagnosis in the oligodendroglial tumor subgroup (95% CI = 0.96–4.65 yr; P = 3.1 × 10−3) (Table 2). Rs55705857 in CCDC26 is known to be a risk factor primarily for oligodendroglial and IDH-mutated gliomas,15 but a significant association with decreased age at diagnosis was observed in all grade and histology subgroups (Table S2), and also when analysis was restricted to IDH-mutant tumors (Table S3). Because the magnitude of this effect appeared stronger in the astrocytic tumor subgroups than in the oligodendroglial subgroups (Table S2), analyses are presented stratified by tumor cell type.
Carrying a greater number of rs498872 risk alleles in PHLDB1 was associated with a 2.30-year younger age at diagnosis in the combined regression analysis of all astrocytic tumors (95% CI = 1.35–3.25 yr; P = 2.1 × 10−6). There was statistically significant heterogeneity in this effect between the purely astrocytic tumors and tumors with an oligodendroglial component (PHET = 3.5 × 10−3), where no significant association was observed (P = .73).
While risk alleles in CCDC26 and PHLDB1 were associated with a reduced age at diagnosis, risk alleles in the telomerase-related genes TERT and RTEL1 were associated with an increased age at diagnosis across all tumor grades and histologies (Table 2 and Table S2). Among all glioma patients, controlling for sex and study site, each additional copy of the rs2736100 risk allele in TERT was associated with a 1.47-year increased age at diagnosis (95% CI = 0.63–2.32 yr; P = 6.2 × 10−4). This association was consistent in the astrocytic and oligodendroglial tumor subgroups (I2 = 0.0).
Carrying a greater number of risk alleles at the other telomerase-related SNP, rs6010620 in RTEL1, was associated with a 2.02-year increased age at diagnosis in analysis of all gliomas (95% CI = 0.94–3.10 yr; P = 2.5 × 10−4). Like the TERT association, this association did not display heterogeneity across grades or histologies (I2 = 0.0) (Table 2 and Table S2).
Because TERT and RTEL1 function within a common pathway, the associations of rs2736100 and rs6010620 with age at diagnosis were also modeled jointly to assess whether the observed associations were independent or possibly synergistic in nature. Including both SNPs in a regression model did not attenuate associations, indicating that both rs2736100 and rs6010620 are independently associated with age at diagnosis. Inclusion of an interaction term in the model (rs2736100 genotype*rs6010620 genotype) did not reveal the presence of any significant effect modification (P = .78). Combining the risk allele dosage into a single ordinal variable representing the total number of risk SNPs in a telomerase-related gene (range, 0–4) supported this conclusion, as each additional risk SNP was associated with a 1.72-year older age at diagnosis (95% CI = 1.06–2.37 yr; P = 2.5 × 10−7) (Table 3).
Table 3.
Association between age at diagnosis and number of risk alleles in a telomerase-related gene (TERT or RTEL1) in combined UCSF Adult Glioma Study and Mayo Clinic glioma patients
| Number of Glioma Risk Alleles in a Telomerase-Related Gene |
Effect (SE) | P | |||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |||
| n | 19 | 155 | 611 | 1010 | 491 | 1.72 (0.33) | 2.5 × 10–7 |
| Average age at diagnosis, yr | 41.7 | 47.1 | 49.4 | 50.9 | 52.5 | ||
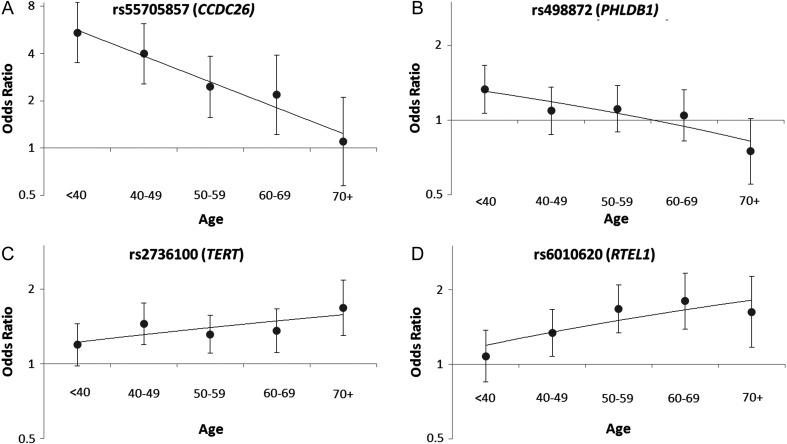
In order to determine whether the significant effects observed in the case-only analyses of CCDC26, PHLDB1, TERT, and RTEL1 were similar in case-control analyses, case-control SNP associations were calculated in 5 different age strata. Both rs55705857 (CCDC26) and rs498872 (PHLDB1) conferred the greatest risk for glioma in the youngest age group and the least risk for glioma in the oldest age group, consistent with the case-only associations previously discussed (Fig. 1A and B). Conversely, rs2736100 (TERT) conferred the greatest risk for glioma in the oldest age group (odds ratio [OR] = 1.68, 95% CI = 1.30–2.17) and the least risk for glioma in the youngest age group (OR = 1.19, 95% CI = 0.98–1.45) (Fig. 1C). Additionally, rs6010620 (RTEL1) conferred a greater risk for glioma in people aged ≥70 years compared with its effect in those <40 years (OR = 1.62, 95% CI = 1.07–2.26 vs OR = 1.08, 95% CI = 0.85–1.37, respectively) (Fig. 1D). The ORs observed for SNPs in TERT and RTEL1, which grow in magnitude with subject age, are consistent with the case-only associations previously discussed.
Fig. 1.
Changes in the magnitude of glioma risk associated with 4 SNPs across subject age strata in case-control analyses. Odds ratios for glioma were calculated in case-control analyses adjusted for sex and study site; 95% CIs appear around each effect estimate. The y axis is represented on a log-scale (base 2), ranging from 0.50 to 8.0 for Fig. 1A and 0.50 to 2.0 for Fig. 1B–1D.
Among patients with oligodendroglial tumors, an increased number of risk alleles at rs1412829 in CDKN2B/ANRIL was associated with a 1.91-year younger age at diagnosis (95% CI = 0.41–3.42 yr; P = .013), but no association was observed in the astrocytic tumor group (PHET = 6.2 × 10−3) (Table 2). Although we have previously shown that rs1412829 confers increased risk for IDH wild-type tumors but not IDH-mutant tumors,17 the rs1412829 glioma risk allele was also associated with a decreased age at diagnosis among patients with IDH-mutant tumors (Table S3).
The effect of the glioma risk allele in TP53 on age at diagnosis also appeared to differ across the astrocytic/oligodendroglial strata (PHET = 0.016), but power to detect stratified associations at this locus is limited because the risk allele frequency is just 3.2% in cases (Table 2).
Discussion
SNP rs55705857 (CCDC26) is a risk factor for oligodendroglial tumors and also for IDH-mutated astrocytomas, but not IDH wild-type astrocytomas.15 While this SNP is associated with a younger age at diagnosis in all glioma strata in our sample, the effect is significantly more pronounced in the purely astrocytic tumor group, consistent with the earlier age at diagnosis observed for IDH-mutated astrocytic tumors compared with IDH wild-type astrocytic tumors.21 Similarly, rs498872 (PHLDB1) is associated with risk for IDH-mutated tumors but not with risk for IDH wild-type tumors.22 In our analyses, this SNP was associated with a younger age at diagnosis only among astrocytic tumors. Adult oligodendroglial tumors typically develop earlier in life than adult astrocytic tumors. Therefore, the stronger effects of CCDC26 and PHLDB1 gene variants on age at diagnosis in patients with astrocytic tumors suggests that these tumors undergo gliomagenesis through a pathway similar to that of oligodendroglial tumors.
If there is a pathway of gliomagenesis shared by oligodendrogliomas and early-onset astrocytomas, IDH1/2 mutation is the obvious candidate marker of such a pathway. Mutations in IDH1/2, CIC, and FUBP1 all correlate with an oligodendroglial tumor histology, younger age at diagnosis, and better survival. Together, mutations in IDH1/2 and ATRX correlate with lower-grade astrocytomas and secondary glioblastomas, which have a younger age at onset than primary glioblastoma.23 Tumors with ATRX mutations nearly universally activate the ALT pathway, immortalizing cells through a telomerase-independent mechanism.23,24 ALT has been observed in 7%–15% of adult glioblastomas and oligodendrogliomas23–25 and in ∼75% of grades II and III astrocytomas and mixed oligoastrocytomas.23,26 Additionally, the ALT phenotype is associated with a younger age at diagnosis among patients with IDH-mutant glioma, irrespective of tumor grade.23 Our data support the hypothesis that tumors with an earlier age at diagnosis are more likely to maintain their telomeres through a telomerase-independent mechanism (eg, ALT) than are tumors with a later age at diagnosis.
Risk alleles in TERT and RTEL1, genes related to telomerase structure and function, were associated with a later age at diagnosis in glioma patients. These SNPs confer an increased risk for glioma, independent of tumor grade or histology.16,17 As a result, it may not be surprising that the effect of these SNPs on age at diagnosis is also consistent across grade, histology, and IDH-mutation strata. Specific variants in TERT have previously been associated with longer telomere length and increased lifespan in humans.27,28 The ability to maintain or lengthen telomeres confers resistance to replicative senescence and supports proliferative potential.10 A cell with increased telomere length, or increased telomerase activity, may have greater capacity to circumvent replicative senescence, a major mechanism of tumor suppression. Variants in RTEL1 and TERT may also place cells in a preactivated state, making them more liable to initiate telomerase activity following normal growth arrest and thereby achieving a critical step in cancer progression. Such a potential pathway would correspond with a tumor that develops later in life through acquired mutations and an inherent predisposition to eschew growth arrest.
It is worth noting that rs2736100 (TERT) and rs6010620 (RTEL1) have risk allele frequencies greater than 50% in our case sample. Although rs2736100 confers increased risk for lung cancer, testicular germ cell cancer, and glioma,29 the risk variant has an allele frequency of 52.7% in Caucasian HapMap samples. Rs6010620 in RTEL1 has an even higher risk allele frequency of 75.7% in Caucasian HapMap samples.30 Although the neoplastic diseases associated with these variants are for the most part postreproductive, and therefore the risk alleles are less susceptible to selection, it is uncommon to have such high cancer risk allele frequencies. Considering that variants in telomerase-related genes may increase individual lifespans but may also increase cellular lifespans in a manner that resists tumor suppression, both positive and negative selective pressures may influence allele frequency at these loci.
Because several cancers are associated with TERT SNPs and nearly 90% of neoplasms activate telomerase, understanding the role of these genes in carcinogenesis is of paramount importance. Our data indicate that patients with a greater number of TERT and RTEL1 risk SNPs develop glioma later in life. These results suggest that such tumors maintain telomeres through the canonical telomerase-based mechanism, distinct from the ALT mechanism associated with tumors appearing earlier in life. Because telomerase-based cancer therapeutics are currently undergoing clinical trials,10 our results may also be useful in identifying which patients can benefit most from enrollment in these studies.
Supplementary Material
Conflict of interest statement. None declared.
Funding
Work at UCSF was supported by the National Institutes of Health (grant nos R25CA112355, R01CA52689, and P50CA097257), as well as the National Brain Tumor Foundation, the UCSF Lewis Chair in Brain Tumor Research, and donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen. Work at the Mayo Clinic was supported by the National Institutes of Health (grant nos P50CA108961 and P30 CA15083), the National Institute of Neurological Disorders and Stroke (grant no. RC1NS068222Z), the Bernie and Edith Waterman Foundation, and the Ting Tsung and Wei Fong Chao Family Foundation.
The collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by the California Health and Safety Code, Section 103885; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000036C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement #1U58 DP000807-01 awarded to the Public Health Institute. The ideas and opinions expressed herein are those of the author(s), and endorsement by the State of California Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their contractors and subcontractors neither is intended nor should be inferred.
References
- 1.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Sanai N, Chang S, Berger MS. Low-grade gliomas in adults. J Neurosurg. 2011;115(5):948–965. doi: 10.3171/2011.7.JNS101238. [DOI] [PubMed] [Google Scholar]
- 3.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanson M, Hosking FJ, Shete S, et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Hum Mol Genet. 2011;20(14):2897–2904. doi: 10.1093/hmg/ddr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stacey SN, Sulem P, Jonasdottir A, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43(11):1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahuau M, Vidaud D, Jenkins RB, et al. Germ-line deletion involving the INK4 locus in familial proneness to melanoma and nervous system tumors. Cancer Res. 1998;58(11):2298–2303. [PubMed] [Google Scholar]
- 8.Malkin D, Li FP, Strong LC, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250(4985):1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 9.Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84(19):6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shay JW, Wright WE. Role of telomeres and telomerase in cancer. Semin Cancer Biol. 2011;21(6):349–353. doi: 10.1016/j.semcancer.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allsopp RC, Vaziri H, Patterson C, et al. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci U S A. 1992;89(21):10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uringa EJ, Lisaingo K, Pickett HA, et al. RTEL1 contributes to DNA replication and repair and telomere maintenance. Mol Biol Cell. 2012;23(14):2782–2792. doi: 10.1091/mbc.E12-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sfeir A, Kosiyatrakul ST, Hockemeyer D, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138(1):90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3(11):1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins RB, Xiao Y, Sicotte H, et al. A low-frequency 8q24.21 variant is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH mutation. Nat Genet. 2012;44(10):1122–1125. doi: 10.1038/ng.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins RB, Wrensch MR, Johnson D, et al. Distinct germ line polymorphisms underlie glioma morphologic heterogeneity. Cancer Genet. 2011;204(1):13–18. doi: 10.1016/j.cancergencyto.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh KM, Anderson E, Hansen HM, et al. Analysis of 60 reported glioma risk SNPs replicates published GWAS findings but fails to replicate associations from published candidate-gene studies. Genet Epidemiol. 2013;37(2):222–228. doi: 10.1002/gepi.21707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen BC, Smith AA, Zheng S, et al. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst. 2011;103(2):143–153. doi: 10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kipp BR, Voss JS, Kerr SE, et al. Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Hum Pathol. 2012 doi: 10.1016/j.humpath.2011.12.007. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 21.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice T, Zheng Z, Decker PA, Walsh KM, et al. Inherited variant on chromosome 11q23 increases susceptibility to IDH mutated but not IDH normal gliomas regardless of grade or histology. Neuro Oncol. 2013;15(5):535–541. doi: 10.1093/neuonc/nos324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiao Y, Killela PJ, Reitman ZJ, et al. Frequent ATRX, CIC, and FUBP1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heaphy CM, de Wilde RF, Jiao Y, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333(6041):425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald KL, McDonnell J, Muntoni A, et al. Presence of alternative lengthening of telomeres mechanism in patients with glioblastoma identifies a less aggressive tumor type with longer survival. J Neuropathol Exp Neurol. 2012;69(7):729–736. doi: 10.1097/NEN.0b013e3181e576cf. [DOI] [PubMed] [Google Scholar]
- 26.Henson JD, Hannay JA, McCarthy SW, et al. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin Cancer Res. 2005;11(1):217–225. [PubMed] [Google Scholar]
- 27.Mangino M, Hwang SJ, Spector TD, et al. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum Mol Genet. 2012;21(24):5385–5394. doi: 10.1093/hmg/dds382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atzmon G, Cho M, Cawthon RM, et al. Evolution in Health and Medicine Sackler Colloquium: genetic variation in human telomerase is associated with telomere length in Ashkenazi centenarians. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1710–1717. doi: 10.1073/pnas.0906191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mocellin S, Verdi D, Pooley KA, et al. Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J Natl Cancer Inst. 2012;104(11):840–854. doi: 10.1093/jnci/djs222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.