Dear Editor,
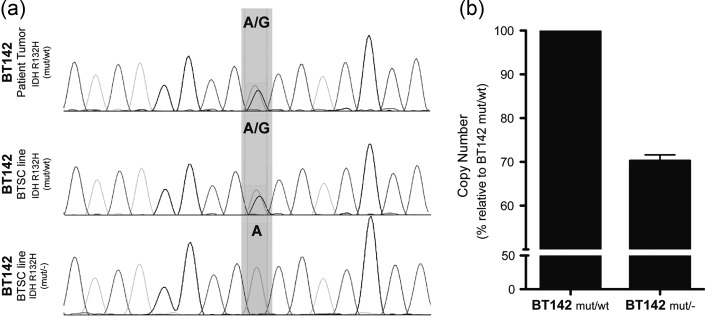
We report a novel follow-up observation pertaining to “An in vivo patient-derived model of endogenous IDH1-mutant glioma,” which was recently published in Neuro-Oncology.1 Since publication, we have observed the gradual and repeated loss of the wild-type IDH1 allele in vitro with retention of the mutant allele. Sequencing of IDH1 exon 4 from 3 independent late passage cultures showed homozygosity for the R132H allele (mut/-), whereas both mutant and wild-type alleles were present in the original line (mut/wt) and tumor (Fig. 1A). In addition, a decreased copy number was seen at the IDH locus, consistent with loss of the IDH1 wild-type allele (Fig. 1B). The American Type Culture Collection (ATCC) has independently observed this phenomenon in BT142. The ATCC is preparing to distribute the BT142 mut/-,2 while we test conditions that best preserve the heterozygous phenotype.
Fig. 1.
Loss of heterozygosity in favor of the mutant allele of IDH1 correlates with a decreased copy number of the IDH1 locus. (a) IDH sequencing on the IDH1-mutant anaplastic oligoastrocytoma and derived IDHmt brain tumor stem cell line (BT142). (b) Copy number assay of the IDH1 locus on heterozygote and homozygote BT142.
The loss of the wild-type allele has been reported in vivo in patients and has been shown to be similar to phenotypically wild-type IDH, resulting in decreased 2-hydroxyglutarate production,3,4 also observed in the BT142 mut/- line. This unforeseen change leading to a second cell line will be valuable for comparisons of the implications of mutant and wild-type IDH phenotypes on proliferation, tumorigenicity, and therapeutic resistance in a syngeneic setting.
Methods
Sequencing for IDH1 was performed as previously described.1 The TaqMan Copy Number Assay (Applied Biosystems) was used to assess copy-number variations. Briefly, genomic DNA was extracted using the DNeasy kit (Qiagen) and quantified using UV absorbance (A260/A280 ratio >1.7). The genomic DNA samples were diluted to 5 ng/μL in nuclease-free water; 20 ng of genomic DNA was mixed with the IDH1 TaqMan Copy Number Assay and the RNase P Reference Assay in a PCR plate, and quantitative real-time PCR was performed according to the manufacturer's instructions. The manufacturer's software, Copy Caller, was used for analysis.
References
- 1.Luchman HA, Stechishin OD, Dang NH, et al. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro Oncol. 2012;14:184–191. doi: 10.1093/neuonc/nor207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. American Type Culture Collection cat#ACS-1018.
- 3.Jin G, Reitman ZJ, Duncan CG, et al. Disruption of wild-type IDH1 suppresses D-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res. 2013;73:496–501. doi: 10.1158/0008-5472.CAN-12-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward PS, Lu C, Cross JR, et al. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem. 2013;288(6):3804–3815. doi: 10.1074/jbc.M112.435495. [DOI] [PMC free article] [PubMed] [Google Scholar]