Abstract
The pathogenesis of organ-specific autoimmune diseases such as Type 1 Diabetes (T1D) is regulated by genetic and environmental factors. There is increasing evidence that environmental factors acting at the intestinal level, with a special regard to the diverse bacterial species that constitute the microbiota, influence the course of autoimmune diseases in tissues outside the intestine both in humans and in preclinical models. In this review we recapitulate current knowledge on the intestinal immune system, its role in local and systemic immune responses and how multiple environmental factors can shape these responses with pathologic or beneficial outcomes for autoimmune diseases such T1D.
Keywords: Intestinal immunity, microbiota, diet, dendritic cells, regulatory T cells, Th17 cells, autoimmune diseases, type 1 diabetes
Introduction
The complexity of the immune system reflects the multiple, complementary and sometimes opposing functions that are necessary to maintain a healthy organism. These functions are strictly dependent on the heterogeneous and dynamic features of the surrounding environment, and range from defense against invasive pathogens to capacity of tolerating innocuous substances to which the body is constantly exposed and self-components of the body. Autoimmune diseases arise from defects of this delicate equilibrium that, throughout immune-mediated recognition and destruction of self-tissues, cause systemic and/or organ-specific inflammation and tissue destruction. The etiologic factors that alter immune tolerance towards self-issues and trigger autoimmunity are largely unknown. In most autoimmune conditions, a combination of both genetic and environmental influences has been reported, suggesting a complex multi-hit model of disease, in which genetically predisposed individuals develop autoimmunity following exposure to different environmental factors [1]. As environmental factors play a key role in the development of autoimmune dysfunction, mucosal surfaces that are constantly in contact with the environment such as the intestinal mucosa become a natural place to look for the protagonists of autoimmune diseases.
The intestinal immune system
Mucosal surfaces, such as the intestine, represent a barrier between the human body and the external environment. Most pathogens enter the body throughout the intestinal mucosa, and must be rapidly eliminated by a protective immune response. By contrast, the intestine is normally home to a variety of commensal bacteria and is continuously exposed to large quantities of food proteins. As a result, the intestinal immune system has to discriminate between invasive organisms and harmless antigens, inducing immunological tolerance towards the latter. The anatomical organization and peculiar cell subsets that are present in the gut mucosa reveal the complexity of its functions.
At the basis of a robust and regulated immune response in the gut lays the necessity for the precise recognition of what substances and organisms the mucosal surface is facing. The intestinal epithelium is lined by a single layer of columnar intestinal epithelial cells (IECs), whose function is not only that of physical barrier to luminal microbes. Indeed, IECs directly influence bacterial colonization via establishment of a protective mucus layer and secretion of antimicrobial peptides. Furthermore, they sample the intestinal microenvironment and modulate accordingly the immune response in the gut [5]. Along the extensive intestinal surface area, both IECs and innate immune cells, in particular dendritic cells (DCs), are equipped with a broad collection of pattern recognition receptors (PRRs), including Toll-like receptors (TLRs) [6] and nucleotide-binding oligomerization domain-containing protein (NOD)-like receptors (NLRs) [7]. DCs also express other classes of PRRs, such as C-type lectin receptors (CLRs), that can either induce independent signaling pathways or affect signaling through TLRs [8]. These receptors allow recognition of pathogen-associated molecular patterns (PAMPs), driving complementary functions in IECs and hematopoietic cells, aimed at the maintenance of intestinal homeostasis [9]. Basal PRR activation in the healthy intestine controls barrier function and influences commensal composition. On the other hand, aberrant PRR signaling, either the recognition of elevated pro-inflammatory stimuli [10,11] or the loss of immune modulatory ones [12,13], leads to significant changes in the activation status of the intestinal immune system and to the eventual pathophysiology of chronic inflammation and autoimmunity [14].
Innate and adaptive immunity in the GALT
The intestine is home to a variety of mononuclear phagocytes (DCs and macrophages) that elicit both innate and adaptive immune responses by acting as antigen-presenting cells (APCs). They reside in the gut-associated lymphoid tissue (GALT) that is composed of the Payer’s patches (PPs), mesenteric lymph nodes (MLNs) and the lamina propria tissue. APCs of the GALT have unique phenotypic and functional features compared to their counterparts resident in different peripheral tissues, thus suggesting a unique role of the gut mucosa APCs in shaping the immune response. In the small intestine APCs have been classified according to the expression of surface molecules such as CD11c (αX integrin), CD11b (αM integrin), CD103 (αE integrin) and the chemokine receptor CX3CR1, but their codification as bona fide DCs or macrophages remains elusive because of shared markers and similar morphology. The expression of CD103 characterizes small intestinal APCs capable of activating both effector and regulatory T lymphocytes and eliciting immune tolerance as well as protective immunity against pathogens [15,16]. In vitro studies demonstrated that small intestinal lamina propria CD103+ DCs, but not CD103- DCs, drive the differentiation of FoxP3+ inducible regulatory T (iTreg) cells through a mechanism that depends on TGF-β and retinoic acid [17,18]. Although the single epithelial layer represents a thin barrier between the intestinal lumen and the underlying lamina propria, these tolerogenic CD103+ DCs appear unable to directly sample microbial and diet-related products in the steady state and depend on small intestinal goblet cells (GCs) as sources of antigens from the intestinal lumen [5]. Pro-inflammatory immune responses occur when bacteria penetrate the epithelial layer, thus triggering activation of pattern recognition receptors on DCs. For example, TLR5-expressing CD11c+ CD11b+ CD103+ cells, upon stimulation by the bacterial component flagellin, elicit adaptive immunity by inducing the differentiation of naïve B cells into immunoglobulin A-producing plasma cells and promoting the differentiation of effector Th17 and Th1 cells [19].
As the number and variety of commensal microorganisms reach their climax in the colon of vertebrates, the mononuclear phagocytes that reside in the lamina propria of the large intestine (colon) play a key role for homeostatic coexistence and reciprocal interactions between the commensal microbiota and the immune system. The steady-state colonic lamina propria of mice contains two major subsets of macrophages, characterized by high expression of CX3CR1 chemokine receptor and low CD103 expression. These macrophages constitutively release high levels of the anti-inflammatory cytokine IL-10 in response to the microbiota and induce FoxP3+ Treg cells in the presence of TGF-β [20]. This suggests that, in the large intestine, the high phagocytic capacity of colonic macrophages coexists with the ability to prevent excessive inflammatory responses against the intestinal flora and maintain intestinal immune homeostasis.
FoxP3+ inducible regulatory T (iTreg) cells play a pivotal role in the maintenance of intestinal immune homeostasis. A critical factor that affects the choice between physiological unresponsiveness and inflammation in the gut is indeed represented by the balance between immune regulatory T cell subpopulations, which mainly include FoxP3+ iTregs and IL-10-secreting FoxP3- Tr1 cells, and pro-inflammatory T cell subsets, such as Th1 cells and Th17 cells. These T cell subpopulations are reciprocally regulated, with TGF-β as a common differentiation factor and other signals that selectively promote the differentiation of the effector Th1/Th17 or the regulatory FoxP3+/IL-10+ T cells subsets. As previously mentioned, intestinal APCs such as DCs and macrophages are critical for the T cell decision to become an effector or regulatory T cell in the gut mucosa. In fact, secretion of the pro-inflammatory cytokine IL-6 by intestinal DCs promotes Th17 cell lineage priming in mucosal tissues, as it overcomes the suppressive effects of TGF-β and retinoic acid produced by CD103+ DCs [21]. IL-10-producing regulatory T cells including both FoxP3+ Treg and FoxP3- Tr1 cells can directly modulate effector T cell differentiation by suppressing Th17 cell induction in the small intestine through an IL-10-mediated mechanism [22]. The gut microenvironment, which is known to be relatively hypoxic under physiologic conditions, may also influence the choice between Treg and Th17 cell differentiation through the hypoxia-inducible factor 1 (HIF-1), a key metabolic sensor that was recently shown to act at the transcriptional level to favor activation of RORγt and inhibit FoxP3, master gene regulators respectively of effector Th17 cells and FoxP3+ Treg cells [23].
Recent studies report that, beside reciprocal interactions between innate and adaptive immune cells, various bacterial species and their molecular components can differentially influence the course of the intestinal immune response, by selectively favoring the differentiation or function of specific T helper cell subsets. In the next sections, we will discuss how the intestinal microbiota affects local and systemic immune responses.
Shaping immunity in the gut: the impact of commensal microbiota on the development and function of the immune system
The human intestinal microbial compartment includes at least 1,000 distinct bacterial species, totaling about 1014 bacteria and containing 100-fold more genes than their human host [24,25]. The understanding of the populations and functions of commensal microorganisms was greatly amplified by the advent of new molecular techniques that allow sequencing and recognition of bacterial species that could not be detected by culture-dependent methods [26,27]. A large body of evidence for the critical role of the microbiota in shaping the immune response comes from the study of germ-free mice, which are completely deprived of intestinal commensals. It has long been acknowledged that, when compared with specific pathogen-free (SPF) animals, germ-free mice show numerous defects in the development of gut-associated lymphoid organs showing hypoplastic Peyer’s patches and reduced numbers of CD4+ T cells in the lamina propria [28]. Importantly, immunological anomalies of germ-free mice are not restricted to the intestinal mucosa and GALT, as their spleen and peripheral lymph nodes (other than gut draining mesenteric lymph nodes) are also affected and lack conventional T and B-cell zones [29]. A recent study established that not the mere presence of bacteria in the gut lumen but selected host-specific microbiota is essential for the maturation of the gut immune system. In fact, small intestines of germ-free mice colonized at birth with human or rat gut microbiota do not develop a normal gut immune system but remain similar to those of germ-free mice, with defective innate and adaptive immunity and increased susceptibility to gastrointestinal infections [30]. Hence, although similarities between the healthy human microbiota and that of other mammals exist at the phylum level [31], these findings suggest that the immune system of each mammalian host has coevolved with the microbiota and require specific bacterial species to develop structurally and functionally.
It is becoming increasingly clear that the commensal species inhabiting our mucosal surfaces are not only fundamental for the correct maturation of lymphoid organs but also play a role in determining the phenotype and functions of different immune cell subsets. Germ-free mice colonized with human gut microbiota had extensive changes in the transcriptional program of intestinal CD4+ T cells with reduced expression of IL-17 and IL-22 cytokines and overall reduction of the Th17 response [30]. This was related to the absence in the human microbiota of one specific bacterial strain, the segmented filamentous bacteria (SFB). SFB are indeed potent inducers of the Th17 response, as demonstrated by mono-colonization experiments, in which SFB was sufficient to trigger the differentiation of Th17 cells and expression of IL-17 and IL-22 in the lamina propria of mice lacking any other commensal species [32,33]. The serum amyloid A (SAA), expressed in the lamina propria of mice after SFB colonization, could favor Th17 differentiation by directly stimulating intestinal DCs to secrete Th17-inducing cytokines IL-6 and IL-23. However, the bacterial component/s and host receptors that regulate SAA expression in the gut mucosa upon SFB colonization remain elusive. The genome of SFB encodes four types of flagellins, three of which are recognized by TLR5 [34], thus suggesting that the flagellin-TLR5 axis may be involved in the SFB-driven Th17 differentiation induced by intestinal CD11c+ CD11b+ CD103+ DCs [15,19]. The capacity of commensal bacteria products to synergize with TLR agonists (PAMPs from pathogens) and modulate intestinal DC function and T cell immunity has been extensively studied. For example, some in vitro studies found that a combination of Lactobacillus casei and the TLR3 agonist poly (I : C) is a potent promoter of Th1 responses [35].
Intestinal microbiota not only induce effector Th17/Th1 differentiation. In fact, there is a large body of evidence that selected bacterial species modulate (both positively and negatively) differentiation of regulatory T cells in the intestines. Bacteroides fragilis was able to prevent intestinal inflammation in a model of experimental colitis, and its protective effect was dependent on polysaccharide A (PSA) contained in its capsule [36]. PSA signals directly through TLR2 expressed on CD4+ T cells to induce differentiation of FoxP3+ Treg cells and prevent Th17 expansion and colitis with an IL-10-mediated mechanism [13,37]. Furthermore, a defined cocktail of 46 Clostridium strains, spore-forming bacteria that are indigenous to the murine intestinal microbiota, promoted accumulation of FoxP3+ Treg cells in the colonic lamina propria. Colonization of germ-free mice with this consortium of bacteria triggered TGF-β expression by intestinal epithelial cells, thus establishing optimal conditions for FoxP3+ Treg cell differentiation and function, with a mechanism that did not rely on canonical pattern recognition through TLRs or NLRs [38]. TLR9 engagement by the gut microbiota DNA played an opposite effect and negatively modulated Treg cell differentiation in the small intestine. In fact, in the absence of TLR9 triggering (TLR9-/- mice), there was an increased intestinal FoxP3+ Treg cell differentiation and reduced Th1 and Th17 function. Moreover, the presence of microbiota DNA inhibited the capacity of lamina propria DCs to induce Treg cell conversion in vitro [39].
In summary, it is now clear that the gut microbiota affects intestinal immune homeostasis by altering the balance between effector and regulatory T cell subsets through different mechanisms and play a crucial role for the maintenance of a healthy immune system in the gut.
Beyond the intestine: from gut to systemic immunity
In healthy mammals, the impact of the microbiota on the immune system and, specifically, on the equilibrium between effector and regulatory T cell subsets extends beyond the intestinal environment. The physical interaction between live microbiota species and immune cells takes place in the gut mucosa where commensal bacteria as well as pathogens are confined in the intestinal lumen by anatomical barriers like the epithelium and mucus layer. However, bacterial components can be captured by mucosal DCs through dendrites reaching the intestinal lumen and transported to the mesenteric lymph nodes, where they are presented to T cells for the establishment of local protective immune responses [40]. The pancreatic lymph nodes (PLNs), mesenteric lymph nodes draining pancreatic tissues, are important sites for the induction of gut-driven immune responses, as demonstrated by the ability of DCs loaded with orally administered ovalbumin to prime both OT-I and OT-II TCR-transgenic cells in the PLNs [41]. Although the systemic immune system is believed to remain ignorant of gut-derived antigens and immunity, it is theoretically possible that effector and regulatory T cells differentiated in the GALT as well as intestinal DCs modulated within the intestine and possibly loaded with bacterial antigens can reach secondary lymphoid organs other than mesenteric lymph nodes and affect immunity at systemic level.
Alternatively, bacterial molecules and metabolic products can directly penetrate the host tissues and influence systemic immune responses. For instance, peptidoglycan derived from the commensal microbiota was found outside the gut and, specifically, in the sera and bone marrow cells, where it primed the innate immunity by enhancing neutrophil function via Nod1 signaling [42]. The short chain fatty acids (SCFAs) are the metabolic products of dietary fiber fermentation by commensal microbiota and, upon ingestion of large amount of fibers, are rapidly absorbed and found in systemic circulation where they modulate innate immunity by different mechanisms. For example, they negatively regulate neutrophil function through binding of the G-protein coupled receptor 43 (GPR43) [12]. Acetate and propionate, two SCFAs produced by colonic bacteria of the Bacteroidetes phylum, induced apoptosis in neutrophils and negatively regulate their chemotaxis by reducing surface expression of the pro-inflammatory receptors C5aR and CXCR2 [12]. Butyrate, a SCFA mostly produced by Firmicutes species, is a major energy source for colonic intestinal epithelial cells and induces mucin synthesis helping maintaining the integrity of the gut epithelium [43]. The systemic innate immune system is therefore intimately connected with the gut microbiota and its metabolic products.
The influence of gut immunity on autoimmune diseases at sites distant from the intestine
The link between the intestinal environment, gut immunity and autoimmune diseases occurring outside the intestine is increasingly recognized. Specifically, the role of the microbiota in modulating autoimmunity at sites distant from the intestinal mucosa is currently the object of intensive studies. Recent reports demonstrated that microbiota modulate pathogenesis of autoimmune diseases in the central nervous system (CNS) in the experimental model of multiple sclerosis (MS), the Experimental Allergic Encephalomyelitis (EAE), in pancreatic islets in NOD mice, the pre-clinical model of T1D and at the joint level in animal models of Rheumatoid Arthritis (RA).
Several studies have investigated the role of the microbiota in shaping the Th17/Treg cell ratio thus influencing development of EAE, encouraged by the observation that oral antibiotic treatment protected mice from the autoimmune disease by reducing the burden and altering the composition of the gut commensal flora [44]. In those experiments, the beneficial effect of oral antibiotic treatment was dependent on enhanced percentages of FoxP3+ Treg cells in the MLNs (GALT) as well as in the cervical lymph nodes (CLNs) that drain the CNS and are distant from the GALT. Oral antibiotics also enhanced the frequency in the CLNs of regulatory IL-10-producing CD1dhigh CD5+ B cells that inhibit EAE initiation in mice [45,46]. Similarly to oral antibiotic-treated mice, germ-free (GF) mice are resistant to EAE [4]. Autoimmune protection in GF mice was associated with significantly decreased levels of the pro-inflammatory cytokine IL-17A and of the transcription factor RORγt, responsible for lineage differentiation of Th17 cells compared to control mice hosted in Specific Pathogen Free facilities (SPF). Moreover, DCs from MLNs of GF mice were defective in promoting both IL-17A and IFN-γ expression in co-cultured T cells, compared to SPF mice. Intestinal mono-colonization of GF mice with SFB restored EAE by promoting pro-inflammatory mechanisms, i.e., effector Th1 and Th17 cell differentiation. Re-colonization with conventional commensal microbiota restored not only pro-inflammatory T cell responses but also MOG-specific autoantibody production [4]. Altogether, these data reveal that the microbiota is able to modulate autoimmune disease in the CNS by altering pro-inflammatory Th1 and Th17 responses as well as autoimmune B cell proliferation and function.
The ability of gut microbiota to modulate autoimmune disease was also demonstrated in pre-clinical models of RA. The first connection between joint inflammation and the intestinal microbiota dates back to 1979, when germ-free but not conventional or specific pathogen-free rats developed severe arthritis with 100% incidence after intradermal adjuvant injection [47]. This observation suggested a protective role of the microbiota on the development of autoimmune arthritis. Mono-colonization of those rats with Escherichia coli suppressed disease development, favoring the hypothesis that this and maybe other commensal microbial species inhibit the pathologic process leading to arthritis in this model [48]. Other reports in different rat and mouse models confirmed the role of the microbiota in modulating autoimmune arthritis, either with protective or exacerbating properties. For instance, HLA-B27 rats, which are transgenic for the human major histocompatibility complex class I allele that confers susceptibility to spondyloarthropathies, did not develop gut and joint inflammation under germ-free conditions [49]. Also SKG mice, in which a mutation in the gene encoding ZAP-70 leads to spontaneous T cell-mediated chronic autoimmune arthritis, failed to develop the disease when housed in a strictly controlled specific pathogen-free environment. In that model, arthritis was triggered by fungal components such as β-glucans, recognized by DCs via the C-type lectin receptor Dectin-1 [50].
IL-1 receptor antagonist-knockout (Il1rn -/-) mice spontaneously develop autoimmune arthritis due to excessive IL-1 signaling and enhanced Th17 cell function. In those mice, onset of arthritis was abrogated in the absence of commensal microbiota, while reintroduction of Lactobacillus bifidus alone was sufficient to activate the autoimmune process. In particular, TLR4 deficiency protected SPF Il1rn -/- mice from severe autoimmune arthritis by negatively regulating IL-23 and IL-1 production by APCs and consequently number and function of Th17 cells [51]. An even stronger connection between commensals, Th17 cells and arthritis was recently outlined in the K/BxN T-cell receptor transgenic mouse model, that showed diminished systemic Th1 and Th17 cell signatures and attenuated arthritis in germ-free versus SPF conditions. Strikingly, mono-colonization of those mice with SFB accelerated arthritis onset by promoting Th17 cell differentiation in the small intestinal lamina propria and their migration to the spleen [52]. Although this study suggests the existence of a gut-joint inflammation axis, there is an open issue on how microbial components and/or immune cells that have been primed in the GALT are able to reach the site of inflammation at organs distant from the intestine such as the joint but also the CNS. Another related question concerns antigen-specificity of those immune cells primed in the gut, i.e., which mechanisms make effector T cells trafficking out of the LP able to modulate tissue specific-damage outside the intestines. A suggestive hypothesis derives from recent discoveries on the initiation phases of RA and EAE. In those murine models of autoimmune diseases, local accumulation of Th17 cells that were not specific for CNS or joint auto-antigens and expressed the gut homing receptor CCR6 occured in the early phases of the disease and was sufficient to trigger the initiation of tissue-specific autoimmunity [53,54]. Of note, expression of the CCR6 chemokine receptor also mediated the homing of CD4+ T cells to the GALT [55,56], thus suggesting that Th17 cells trafficking out of the lamina propria are directly responsible for triggering pathogenesis of EAE and RA. According to this hypothesis, the balance between effector Th1/Th17 and Treg cell differentiation in the gut, which is regulated by several factors such as the microbiota composition, can finely modulate the autoimmune T cell response outside the intestine and be involved in the early phases of autoimmune diseases such as EAE and RA (Figure 1).
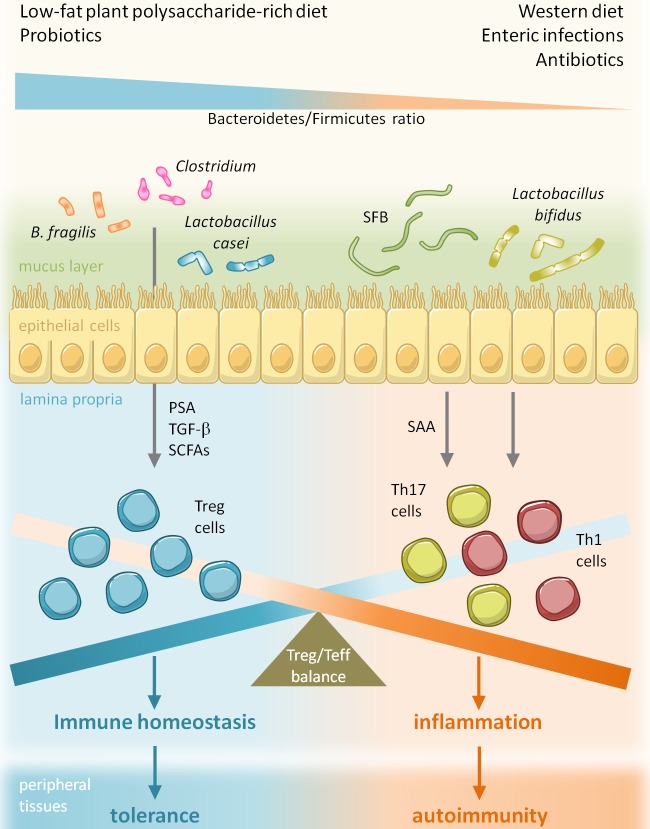
Figure 1.
Environmental factors modify pathogenesis of autoimmune diabetes by altering gut microbiota composition. Several environmental factors such as diet, enteric infections and drugs (antibiotics) can affect the pathogenesis of autoimmune diseases like T1D by altering the gut microbiota and, specifically, by changing the Bacteroidetes/Firmicutes ratio thus modifying the Treg/Teff balance in the GALT and systemically.
The gut environment and autoimmune Type 1 diabetes
Multiple studies both in patients with T1D and animal models revealed a strong association between T1D development and intestinal alterations that often precede clinical signs of the disease. Major evidence supporting such connection is represented by the high prevalence of clinical and subclinical celiac disease (CD) in diabetic patients. T1D patients often present with chronic inflammation of the intestinal mucosa even in the absence of clinical symptoms of CD [57]. Several human studies detected ultra-structural mucosal alterations and increased intestinal permeability in non-celiac diabetic patients in the preclinical phase of the disease [58]. Gut mucosal alterations in T1D patients could be due to microbiota composition. In fact, a recent metagenomic analysis of the gut microbiome in four pairs of age and sex-matched new-onset T1D case and control subjects revealed significant differences at the taxonomic and functional level [43]. Specifically, Actinobacteria, Bacteroidetes and Proteobacteria were more represented in T1D patients than in controls, while Firmicutes were less present. Importantly, butyrate-producer genera were scarcely represented in T1D patients. Since those species are responsible for butyrate-dependent increase of mucin synthesis and tight junction assembly, the absence of those bacteria could explain the mucosal abnormalities, whereas the concomitant deficiency of mucin-degrading genera such as Prevotella in T1D patients could be responsible for increased gut permeability. Interestingly, microbiota of T1D patients was more efficient at producing other SCFAs, namely propionate, succinate and acetate, which affect neutrophils and could explain the impaired neutrophil functionality observed in T1D patients [59,60]. The latter observation suggests that microbiota can also affect autoimmune diabetes by altering gut immunity and immune regulation. In fact, there is clear indication that T1D patients have excessive activation of the gut immune system, documented by increased expression levels of class II MHC molecules and high frequency of IL-1α and IL-4-secreting cells in the LP even in T1D patients lacking evident mucosal alterations [59,61]. Gut immune activation in basal conditions could be explained by defective immune regulatory mechanisms in the intestine of T1D patients. Indeed, a defect of FoxP3+ Treg cells was recently found in the small intestinal mucosa of T1D patients compared to healthy subjects, due to the inability of intestinal DCs to induce FoxP3+ Treg cell conversion [62].
Although a clear correlation between alterations of gut immunity and immune regulation and the pathogenesis of autoimmune diabetes in patients is still missing, studies in pre-clinical models of T1D indicated that the microbiota can directly modulate T cell autoimmunity against pancreatic islets. Studies in gnotobiotic animal models of spontaneous T1D helped defining the immunological influence of the microbiota on the pathogenesis of autoimmune diabetes. Analysis of microbiota composition in young pre-diabetic rats revealed that animals that subsequently developed diabetes had a significantly higher amount of Bacteroides species and abundant Lactobacillus and Bifidobacterium compared to those that remained diabetes-free [63,64]. Furthermore, antibiotic treatment decreased diabetes incidence in BB rats [63] and in NOD mice. It is well known that the characteristics of the animal housing facility deeply influence the incidence of spontaneous diabetes in NOD mice, the most used pre-clinical model of T1D, most likely by altering microbiota composition [65]. The definitive evidence that the gut microbiota influences autoimmune diabetes in NOD mice was provided by studies in MyD88-deficient NOD mice. MyD88 is an adaptor molecule involved in signaling of many TLRs and its absence completely modifies the gut microbiota composition and protected NOD mice from autoimmune diabetes. In that model, protection from T1D in MyD88KO was integrally dependent on the presence of a specific gut microbiota, characterized by a low Firmicutes/Bacteroidetes ratio and rich in Lactobacillaceae, since re-derivation of MyD88KO mice as germ-free restored autoimmune diabetes.
The protective effect of Lactobacillaceae species in T1D was also proven by the protection obtained in NOD mice treated with the VSL#3 probiotic preparation [66]. Also, changes of the gut microbiota obtained by feeding NOD mice with a gluten-free diet significantly reduced incidence of autoimmune diabetes [67]. Some studies have associated single bacterial species with protection or exacerbation of T1D. Spontaneous monoculture gut colonization with an aerobic spore-forming bacterium, Bacillus cereus, delayed onset and reduced incidence of diabetes in NOD mice [68]. In a different animal facility, protection of female NOD mice from T1D segregated with SFB colonization, and coincided with significant increase of Th17 cell immunity in the small intestinal LP, although no differences were observed at the level of PLN [69]. This observation supports the hypothesis that in T1D, unlike other autoimmune diseases such as MS and RA, Th17 cells do not act as effector pathogenic lymphocytes but rather play an immuneregulatory, suppressive role against autoimmunity. However, the role of Th17 cells in T1D is still debated, given the detection of enhanced circulating Th17 cells, possibly of intestinal origin (expressing the mucosal-homing chemokine receptor CCR6) in new-onset T1D patients [70,71] and the increased Th17 immunity in the PLNs of T1D patients [72].
Those data clearly demonstrate the impact of the microbiota on gut immunity and on the pathogenesis of autoimmune T1D. However, as discussed in the previous paragraph for pre-clinical models of MS and RA, it is still unclear how gut immunity can affect autoimmune processes at site distal from the intestine such as the pancreatic islets. It must be considered that pancreatic lymph nodes that drain pancreatic tissues are mesenteric lymph nodes. Hence, an immunological axis between the gut mucosa, GALT and pancreatic islets exists as indicated by the ability of orally administered antigens to prime T cell responses in the PLNs [41]. Studies in pre-clinical models of T1D helped to better define the anatomical association between autoimmune diabetes and intestinal immunity. The transfer of lymphocytes isolated from MLNs of 3-week-old pre-diabetic NOD mice into NOD. Scid recipients induced diabetes, thus revealing that diabetogenic T cells are primed in the GALT at early stages of T1D pathogenesis [73]. Indeed, the constitutive expression of the adhesion molecule MAdCAM-1 by pancreatic islets allows homing of T cells of intestinal origin that up-regulated α4β7 integrin on their surface following activation by gut associated DCs [74].
New therapeutic approaches in autoimmune diabetes: shaping autoimmunity through gut microbiota modification
The assembly of the symbiotic species that constitute the intestinal microbiota is a complex process that starts at birth and continues throughout life, with many host and environmental factors actively shaping its composition. Neonates normally acquire their microbiota at the time of delivery, when they are exposed to bacteria of maternal origin. The mode of delivery deeply influences the species that first colonize the intestines of newborns, as demonstrated by the observation that vaginally delivered infants harbor bacterial communities that are similar to those found in the maternal vagina, whereas microbiota from infants delivered via caesarean section resembles the skin communities of the mother [75]. Community assembly continues during infancy in a nonrandom fashion, with a gradual increase in diversity over time and abrupt shifts in the abundance of some taxa, strictly dependent on life events such as infections, antibiotic treatment and dietary changes [76]. Several studies reported that introduction of formula and solid food coincided with increased Bacteroidetes and production of SCFAs, which marked the transition toward an adult-like microbiota [76,77]. After weaning, the body encounters several new sources of bacteria that are able to colonize the gastrointestinal tract depending on a number of factors, including body temperature, availability of nutrients and competing symbionts and pathogens. As we mentioned above, the microbiota deeply affects the development of the immune system and its functions, but the crosstalk also acts in the reverse direction, i.e., the GALT plays a fundamental role in containment and selection of bacterial species that inhabit mucosal surfaces, mainly by secretion of antibacterial products [78,79]. Given the importance of commensals for the education of the immune response and prevention of autoimmune diseases, we now recapitulate the major environmental factors that are known to influence the composition of the microbiota in the adult and can be potentially modified for prevention and/or treatment of autoimmune diseases and T1D (Figure 1).
Diet
While postnatal acquisition of the primary microbiome relies on delivery mode, diversification of the bacterial populations that inhabit the gastrointestinal tract from those present in other anatomical sites occurs at later stages, and is deeply influenced by weaning timing of, introduction of solid food and dietary habits [76]. In fact, bacterial species inhabiting our gastrointestinal tract require distinct nutrients and metabolic pathways according to the hydrolytic enzymes encoded by the microbiota genome (microbiome). Many complex polysaccharides that are ingested with human diet and cannot undergo host-mediated degradation often represent a major source of carbon and energy for commensal microbes [80]. Studies in gnotobiotic mice demonstrated that those prebiotics play a crucial role for proliferation and competition between different bacterial species. For instance, Bacteroides thetaiotaomicron, an obligate anaerobe found abundantly within the human microbiota, is able to harvest and metabolize polysaccharides from host mucus in the absence of dietary fructans [81]. On the other hand, an inulin-restricted diet favored the growth of Bacteroides caccae over B. thetaiotaomicron’s, due to differences in the gene content of their respective polysaccharide utilization loci (PULs). Another useful model for the study of the effects of diet on bacterial communities was obtained by transplanting adult human fecal microbiota into germ-free mice. The humanized gut microbiota murine model not only provided interesting insights into the mechanisms of postnatal assembly and spatial distribution of the bacterial community, but also revealed the abrupt effect of a change in diet on the composition of the gut microflora. Indeed, switching those mice from a low-fat, plant polysaccharide-rich diet to a high-fat, high-sugar Western-like diet rapidly affected the relative abundances of bacterial taxa, epitomized by the increased Firmicutes/Bacteroidetes ratio [82]. Similarly, a recent study demonstrated that children living in a rural village of Burkina Faso and fed on a traditional rural African diet rich in fiber and plant polysaccharides have a microbiota profile rich in Actinobacteria and Bacteroidetes while Firmicutes and Proteobacteria dominated the microbiota composition of European children, fed on a typical high-fat Western diet [83]. Furthermore, African children had significantly higher amounts of SCFAs compared to European children, in particular propionate and butyrate. Since those microbial species and metabolites can have a pivotal role in maintenance of gut immune homeostasis and prevention of autoimmunity in the gut and at distal organs, it is evident that the dietary habits of African children can explain the low incidence of autoimmune diseases in those populations [83]. The important implications of those findings is that a diet rich in plant polysaccharides and low in fat and animal proteins able to favor pro-tolerogenic microbial species and promote gut immune regulation can be proposed to individuals genetically at risk for autoimmune diseases such as T1D.
Enteric infections
Enteric infections either result from the encounter of the host with a pathogen or follow pathological overgrowth of an opportunistic bacterium normally present at low abundances in the microbiota [84]. In both cases, the invasive agent needs to find or create a favorable environment rich of necessary nutrients and without antimicrobial activities. The healthy microbiota represents an obstacle to enteric infections, not only because it competes for space and nutrients but also because it promotes protective immunity and secretion of antimicrobial peptides by the gut mucosa [85]. This pathogen colonization resistance can be overcome by inflammation, which can precede or be caused by the enteric pathogen. For example, wild-type Salmonella enterica Serovar Typhimurium, but not a strain deficient in virulence-associated type III secretion systems, triggered colitis that was sufficient to alter the cecal microbiota composition thus creating a favorable niche for pathogen colonization [86]. Accordingly, specific Bifidobacteria strains present in the healthy microbiota protected from lethal enterohaemorragic Escherichia coli infection by secreting acetate that inhibited the inflammatory and pro-apoptotic effects of that pathogen [87]. The ability of a pathogen to compete with the gut microbiota by regulating its virulence over time was recently confirmed in another model of infection. Citrobacter rondentium significantly but transiently decreased the number of commensals in the colon of infected mice, due to early expression of virulence factors [88]. These studies clearly indicate the capacity of invading pathogens to modify the microbiota and suggest that repeated enteric infections can also play a critical role in shaping the commensal microbiota and altering gut immunity and immune regulation.
Antibiotics
In the absence of a pathogen-induced inflammatory response, iatrogenic causes are often responsible for alteration of the healthy microbiota [89-91] and can also provide conditions that favor outgrow of pathogens. Indeed, antibiotic treatment currently represents the major cause of disturbance for the diversity and composition of the microbiota. A study in healthy humans revealed that ciprofloxacin treatment significantly decreased the taxonomic richness, diversity and evenness of the gut microbiota, which returned to the pretreatment state by 4 days after the last antibiotic administration. However, it must be noted that some taxa did not recover during a six-months follow up [92]. Clindamycin had similar effects in another cohort of human volunteers, causing in particular a reduction in the diversity of Bacteroides species that persisted over a two-year period [93]. Treatment with vancomycin, an antibiotic that specifically kills Gram-positive bacteria, caused dramatic expansion of Enterobacteriaceae in the ileum and cecum of mice, which was not contained but rather increased in the two-week period after the last antibiotic administration [91]. These studies indicate that, despite the existence of some mechanisms that are able to promote community recovery after disturbance, antibiotic treatments cause long-term effects on the composition of the gut microbiota, and therefore their use should be carefully considered, given their effects on the intestinal immune system and, indirectly on the pathogenesis of autoimmune diseases like T1D. One example of this profound interconnection is represented by the ability of vancomycin to reduce the frequency of Treg cells in the colon of mice, by specifically depleting Gram-positive Clostridium species [38].
Probiotics
Probiotics are beneficial live microbes that can be administered to the host in many different formulations as drugs or dietary supplements [94]. Unlike the long-term effects on microbiota composition recorded after antibiotic treatment, probiotic administration can trigger only transient modifications of commensal bacterial communities that lasted less than 15 days after treatment interruption [95]. However, continuous administration of high concentrations of probiotics can reduce gut immunity and maintain remission from inflammatory bowel disease [95,96]. Interestingly, some probiotics were found to protect both human and mice from autoimmune diseases. In a preclinical model of rheumatoid arthritis, one strain of Lactobacillus casei inhibited inflammation and suppressed the progression of the disease [97]. Oral probiotic administration also prevented spontaneous autoimmune diabetes in the NOD mouse [66]. Given the beneficial effects of some orally administered probiotic species in the aforementioned inflammatory and autoimmune diseases, the problem of short-term microbiota modification in humans may be overcome by combination strategies with antibiotics that are known to reduce colonization resistance. Alternatively, in order to restore a healthy and protective gut microbiota for prevention of autoimmune diseases, it is possible to perform fecal bacteriotherapy, i.e., administration of a suspension of feces from a healthy subject to individuals depleted of their own microbiota by antibiotics. This approach that is already experimentally used in some forms of enterocolitis and IBD [98] can be also proposed to patients affected by organ-specific autoimmune diseases such as T1D who have a demonstrated alteration of the microbiota composition and documented increased of pro-inflammatory species.
Another possible strategy to overcome the problems of limited viability and colonization of probiotics for human therapy may be the administration of the soluble bacterial factors that mediate their beneficial effects. As mentioned above, PSA from B. fragilis proved to be as efficient as the entire bacterium at inducing FoxP3+ Treg cell conversion in the intestine of mice [13,37]. Germ-free mice fed acetate in drinking water were protected from DSS-induced colitis and experimental arthritis [12]. Although these and other molecules may not fully reproduce the complex immune-modulatory effects of living microorganisms, they represent a good alternative to modify gut immunity and immune regulation in controlled experimental and clinical settings.
Concluding remarks
It is now clear that the environment plays a crucial role in shaping the immune system in both physiologic and pathologic conditions and modulates the pathogenesis of autoimmune diseases such as T1D. The aforementioned studies indicate that the gut microbiota can have a fundamental role as intermediary between the high number of environmental triggers that alter autoimmune processes and the diverse immune cells, possibly including autoimmune T cells that patrol our mucosal surfaces. Furthermore, direct links between microbiota alterations and autoimmunity have been found. The discovery that specific nutrients and dietary supplements can selectively affect colonization capacity of either beneficial or detrimental species of the microbiota open to the possibility of their therapeutic exploitation in the prevention or treatment of autoimmune diseases. Genetic predisposition still accounts for a large part of the etiology of these diseases, however environmental modifications and, specifically, interventions that can modulate gut microbiota can be proposed to genetically at-risk individuals to prevent or delay the onset of autoimmune diseases like T1D.
References
- 1.Bogdanos DP, Smyk DS, Rigopoulou EI, Mytilinaiou MG, Heneghan MA, Selmi C, Gershwin ME. Twin studies in autoimmune disease: genetics, gender and environment. J Autoimmun. 2012;38:J156–69. doi: 10.1016/j.jaut.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol. 2011;7:569–578. doi: 10.1038/nrrheum.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, Wekerle H, Krishnamoorthy G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 5.McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA, Newberry RD, Miller MJ. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483:345–349. doi: 10.1038/nature10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 7.Williams A, Flavell RA, Eisenbarth SC. The role of NOD-like Receptors in shaping adaptive immunity. Curr Opin Immunol. 2010;22:34–40. doi: 10.1016/j.coi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol. 2009;9:465–479. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 10.Biswas A, Wilmanski J, Forsman H, Hrncir T, Hao L, Tlaskalova-Hogenova H, Kobayashi KS. Negative regulation of Toll-like receptor signaling plays an essential role in homeostasis of the intestine. Eur J Immunol. 2011;41:182–194. doi: 10.1002/eji.201040479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng T, Wang L, Schoeb TR, Elson CO, Cong Y. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J Exp Med. 2010;207:1321–1332. doi: 10.1084/jem.20092253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12:5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 15.Semmrich M, Plantinga M, Svensson-Frej M, Uronen-Hansson H, Gustafsson T, Mowat AM, Yrlid U, Lambrecht BN, Agace WW. Directed antigen targeting in vivo identifies a role for CD103(+) dendritic cells in both tolerogenic and immunogenic T-cell responses. Mucosal Immunol. 2012;5:150–160. doi: 10.1038/mi.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, Flavell RA, Littman DR, Pamer EG. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uematsu S, Fujimoto K, Jang MH, Yang BG, Jung YJ, Nishiyama M, Sato S, Tsujimura T, Yamamoto M, Yokota Y, Kiyono H, Miyasaka M, Ishii KJ, Akira S. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat Immunol. 2008;9:769–776. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- 20.Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med. 2012;209:139–155. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu W, Troutman TD, Edukulla R, Pasare C. Priming microenvironments dictate cytokine requirements for T helper 17 cell lineage commitment. Immunity. 2011;35:1010–1022. doi: 10.1016/j.immuni.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber S, Gagliani N, Esplugues E, O’Connor W Jr, Huber FJ, Chaudhry A, Kamanaka M, Kobayashi Y, Booth CJ, Rudensky AY, Roncarolo MG, Battaglia M, Flavell RA. Th17 cells express interleukin-10 receptor and are controlled by Foxp3(-) and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity. 2011;34:554–565. doi: 10.1016/j.immuni.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, Pan F. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 25.Raz E. Mucosal immunity: aliment and ailments. Mucosal Immunol. 2010;3:4–7. doi: 10.1038/mi.2009.123. [DOI] [PubMed] [Google Scholar]
- 26.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 29.Bauer H, Horowitz RE, Levenson SM, Popper H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am J Pathol. 1963;42:471–483. [PMC free article] [PubMed] [Google Scholar]
- 30.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 33.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuwahara T, Ogura Y, Oshima K, Kurokawa K, Ooka T, Hirakawa H, Itoh T, Nakayama-Imaohji H, Ichimura M, Itoh K, Ishifune C, Maekawa Y, Yasutomo K, Hattori M, Hayashi T. The lifestyle of the segmented filamentous bacterium: a non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res. 2011;18:291–303. doi: 10.1093/dnares/dsr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baba N, Samson S, Bourdet-Sicard R, Rubio M, Sarfati M. Selected commensal-related bacteria and Toll-like receptor 3 agonist combinatorial codes synergistically induce interleukin-12 production by dendritic cells to trigger a T helper type 1 polarizing programme. Immunology. 2009;128:e523–31. doi: 10.1111/j.1365-2567.2008.03022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 37.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 41.Turley SJ, Lee JW, Dutton-Swain N, Mathis D, Benoist C. Endocrine self and gut non-self intersect in the pancreatic lymph nodes. Proc Natl Acad Sci U S A. 2005;102:17729–17733. doi: 10.1073/pnas.0509006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip M, Hyoty H, Veijola R, Simell T, Simell O, Neu J, Wasserfall CH, Schatz D, Atkinson MA, Triplett EW. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Foureau DM, Haque-Begum S, Kasper LH. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J Immunol. 2009;183:6041–6050. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- 45.Matsushita T, Yanaba K, Bouaziz JD, Fujimoto M, Tedder TF. Regulatory B cells inhibit EAE initiation in mice while other B cells promote disease progression. J Clin Invest. 2008;118:3420–3430. doi: 10.1172/JCI36030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ochoa-Reparaz J, Mielcarz DW, Haque-Begum S, Kasper LH. Induction of a regulatory B cell population in experimental allergic encephalomyelitis by alteration of the gut commensal microflora. Gut Microbes. 2010;1:103–108. doi: 10.4161/gmic.1.2.11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohashi O, Kuwata J, Umehara K, Uemura F, Takahashi T, Ozawa A. Susceptibility to adjuvant-induced arthritis among germfree, specific-pathogen-free, and conventional rats. Infect Immun. 1979;26:791–794. doi: 10.1128/iai.26.3.791-794.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohashi O, Kohashi Y, Takahashi T, Ozawa A, Shigematsu N. Suppressive effect of Escherichia coli on adjuvant-induced arthritis in germ-free rats. Arthritis Rheum. 1986;29:547–553. doi: 10.1002/art.1780290413. [DOI] [PubMed] [Google Scholar]
- 49.Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernandez-Sueiro JL, Balish E, Hammer RE. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180:2359–2364. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshitomi H, Sakaguchi N, Kobayashi K, Brown GD, Tagami T, Sakihama T, Hirota K, Tanaka S, Nomura T, Miki I, Gordon S, Akira S, Nakamura T, Sakaguchi S. A role for fungal {beta}-glucans and their receptor Dectin-1 in the induction of autoimmune arthritis in genetically susceptible mice. J Exp Med. 2005;201:949–960. doi: 10.1084/jem.20041758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abdollahi-Roodsaz S, Joosten LA, Koenders MI, Devesa I, Roelofs MF, Radstake TR, Heuvelmans-Jacobs M, Akira S, Nicklin MJ, Ribeiro-Dias F, van den Berg WB. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest. 2008;118:205–216. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murakami M, Okuyama Y, Ogura H, Asano S, Arima Y, Tsuruoka M, Harada M, Kanamoto M, Sawa Y, Iwakura Y, Takatsu K, Kamimura D, Hirano T. Local microbleeding facilitates IL-6- and IL-17-dependent arthritis in the absence of tissue antigen recognition by activated T cells. J Exp Med. 2011;208:103–114. doi: 10.1084/jem.20100900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arima Y, Harada M, Kamimura D, Park JH, Kawano F, Yull FE, Kawamoto T, Iwakura Y, Betz UA, Marquez G, Blackwell TS, Ohira Y, Hirano T, Murakami M. Regional neural activation defines a gateway for autoreactive T cells to cross the blood-brain barrier. Cell. 2012;148:447–457. doi: 10.1016/j.cell.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 55.Williams IR. CCR6 and CCL20: partners in intestinal immunity and lymphorganogenesis. Ann N Y Acad Sci. 2006;1072:52–61. doi: 10.1196/annals.1326.036. [DOI] [PubMed] [Google Scholar]
- 56.Ito T, Carson WF 4th, Cavassani KA, Connett JM, Kunkel SL. CCR6 as a mediator of immunity in the lung and gut. Exp Cell Res. 2011;317:613–619. doi: 10.1016/j.yexcr.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bosi E, Molteni L, Radaelli MG, Folini L, Fermo I, Bazzigaluppi E, Piemonti L, Pastore MR, Paroni R. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49:2824–2827. doi: 10.1007/s00125-006-0465-3. [DOI] [PubMed] [Google Scholar]
- 58.Secondulfo M, Iafusco D, Carratu R, deMagistris L, Sapone A, Generoso M, Mezzogiomo A, Sasso FC, Carteni M, De Rosa R, Prisco F, Esposito V. Ultrastructural mucosal alterations and increased intestinal permeability in non-celiac, type I diabetic patients. Dig Liver Dis. 2004;36:35–45. doi: 10.1016/j.dld.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 59.Bilgic S, Aktas E, Salman F, Ersahin G, Erten G, Yilmaz MT, Deniz G. Intracytoplasmic cytokine levels and neutrophil functions in early clinical stage of type 1 diabetes. Diabetes Res Clin Pract. 2008;79:31–36. doi: 10.1016/j.diabres.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 60.Valle A, Giamporcaro GM, Scavini M, Stabilini A, Grogan P, Bianconi E, Sebastiani G, Masini M, Maugeri N, Porretti L, Bonfanti R, Meschi F, De Pellegrin M, Lesma A, Rossini S, Piemonti L, Marchetti P, Dotta F, Bosi E, Battaglia M. Reduced circulating neutrophils precedes and accompanies type 1 diabetes. Diabetes. 2013;62:2072–7. doi: 10.2337/db12-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westerholm-Ormio M, Vaarala O, Pihkala P, Ilonen J, Savilahti E. Immunologic activity in the small intestinal mucosa of pediatric patients with type 1 diabetes. Diabetes. 2003;52:2287–2295. doi: 10.2337/diabetes.52.9.2287. [DOI] [PubMed] [Google Scholar]
- 62.Badami E, Sorini C, Coccia M, Usuelli V, Molteni L, Bolla AM, Scavini M, Mariani A, King C, Bosi E, Falcone M. Defective differentiation of regulatory FoxP3+ T cells by small-intestinal dendritic cells in patients with type 1 diabetes. Diabetes. 2011;60:2120–2124. doi: 10.2337/db10-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brugman S, Klatter FA, Visser JT, Wildeboer-Veloo AC, Harmsen HJ, Rozing J, Bos NA. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia. 2006;49:2105–2108. doi: 10.1007/s00125-006-0334-0. [DOI] [PubMed] [Google Scholar]
- 64.Roesch LF, Lorca GL, Casella G, Giongo A, Naranjo A, Pionzio AM, Li N, Mai V, Wasserfall CH, Schatz D, Atkinson MA, Neu J, Triplett EW. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J. 2009;3:536–548. doi: 10.1038/ismej.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pozzilli P, Signore A, Williams AJ, Beales PE. NOD mouse colonies around the world--recent facts and figures. Immunol Today. 1993;14:193–196. doi: 10.1016/0167-5699(93)90160-M. [DOI] [PubMed] [Google Scholar]
- 66.Calcinaro F, Dionisi S, Marinaro M, Candeloro P, Bonato V, Marzotti S, Corneli RB, Ferretti E, Gulino A, Grasso F, De Simone C, Di Mario U, Falorni A, Boirivant M, Dotta F. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia. 2005;48:1565–1575. doi: 10.1007/s00125-005-1831-2. [DOI] [PubMed] [Google Scholar]
- 67.Hansen AK, Ling F, Kaas A, Funda DP, Farlov H, Buschard K. Diabetes preventive gluten-free diet decreases the number of caecal bacteria in non-obese diabetic mice. Diabetes Metab Res Rev. 2006;22:220–225. doi: 10.1002/dmrr.609. [DOI] [PubMed] [Google Scholar]
- 68.King C, Sarvetnick N. The incidence of type-1 diabetes in NOD mice is modulated by restricted flora not germ-free conditions. PLoS One. 2011;6:e17049. doi: 10.1371/journal.pone.0017049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Honkanen J, Nieminen JK, Gao R, Luopajarvi K, Salo HM, Ilonen J, Knip M, Otonkoski T, Vaarala O. IL-17 immunity in human type 1 diabetes. J Immunol. 2010;185:1959–1967. doi: 10.4049/jimmunol.1000788. [DOI] [PubMed] [Google Scholar]
- 71.Marwaha AK, Crome SQ, Panagiotopoulos C, Berg KB, Qin H, Ouyang Q, Xu L, Priatel JJ, Levings MK, Tan R. Cutting edge: Increased IL-17-secreting T cells in children with new-onset type 1 diabetes. J Immunol. 2010;185:3814–3818. doi: 10.4049/jimmunol.1001860. [DOI] [PubMed] [Google Scholar]
- 72.Ferraro A, Socci C, Stabilini A, Valle A, Monti P, Piemonti L, Nano R, Olek S, Maffi P, Scavini M, Secchi A, Staudacher C, Bonifacio E, Battaglia M. Expansion of Th17 cells and functional defects in T regulatory cells are key features of the pancreatic lymph nodes in patients with type 1 diabetes. Diabetes. 2011;60:2903–2913. doi: 10.2337/db11-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jaakkola I, Jalkanen S, Hanninen A. Diabetogenic T cells are primed both in pancreatic and gut-associated lymph nodes in NOD mice. Eur J Immunol. 2003;33:3255–3264. doi: 10.1002/eji.200324405. [DOI] [PubMed] [Google Scholar]
- 74.Hanninen A, Nurmela R, Maksimow M, Heino J, Jalkanen S, Kurts C. Islet beta-cell-specific T cells can use different homing mechanisms to infiltrate and destroy pancreatic islets. Am J Pathol. 2007;170:240–250. doi: 10.2353/ajpath.2007.060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 79.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 82.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sekirov I, Finlay BB. The role of the intestinal microbiota in enteric infection. J Physiol. 2009;587:4159–4167. doi: 10.1113/jphysiol.2009.172742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schauber J, Svanholm C, Termen S, Iffland K, Menzel T, Scheppach W, Melcher R, Agerberth B, Luhrs H, Gudmundsson GH. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut. 2003;52:735–741. doi: 10.1136/gut.52.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 88.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 89.Woo H, Okamoto S, Guiney D, Gunn JS, Fierer J. A model of Salmonella colitis with features of diarrhea in SLC11A1 wild-type mice. PLoS One. 2008;3:e1603. doi: 10.1371/journal.pone.0001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, Pamer EG. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 94.Guarner F, Khan AG, Garisch J, Eliakim R, Gangl A, Thomson A, Krabshuis J, Lemair T, Kaufmann P, de Paula JA, Fedorak R, Shanahan F, Sanders ME, Szajewska H, Ramakrishna BS, Karakan T, Kim N. World gastroenterology organisation global guidelines: probiotics and prebiotics october 2011. J Clin Gastroenterol. 2012;46:468–481. doi: 10.1097/MCG.0b013e3182549092. [DOI] [PubMed] [Google Scholar]
- 95.Venturi A, Gionchetti P, Rizzello F, Johansson R, Zucconi E, Brigidi P, Matteuzzi D, Campieri M. Impact on the composition of the faecal flora by a new probiotic preparation: preliminary data on maintenance treatment of patients with ulcerative colitis. Aliment Pharmacol Ther. 1999;13:1103–1108. doi: 10.1046/j.1365-2036.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 96.Gionchetti P, Rizzello F, Helwig U, Venturi A, Lammers KM, Brigidi P, Vitali B, Poggioli G, Miglioli M, Campieri M. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124:1202–1209. doi: 10.1016/s0016-5085(03)00171-9. [DOI] [PubMed] [Google Scholar]
- 97.So JS, Kwon HK, Lee CG, Yi HJ, Park JA, Lim SY, Hwang KC, Jeon YH, Im SH. Lactobacillus casei suppresses experimental arthritis by down-regulating T helper 1 effector functions. Mol Immunol. 2008;45:2690–2699. doi: 10.1016/j.molimm.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 98.Grehan MJ, Borody TJ, Leis SM, Campbell J, Mitchell H, Wettstein A. Durable alteration of the colonic microbiota by the administration of donor fecal flora. J Clin Gastroenterol. 2010;44:551–561. doi: 10.1097/MCG.0b013e3181e5d06b. [DOI] [PubMed] [Google Scholar]