Abstract
Antiretroviral therapy can inhibit HIV replication in patients and prevent progression to AIDS. However, it is not curative. Here we provide an overview of what antiretroviral drugs do and how the virus persists during therapy in rare reservoirs, such as latently infected CD4+ T cells. We also outline several innovative methods that are currently under development to eradicate HIV from infected individuals. These strategies include gene therapy approaches intended to create an HIV-resistant immune system, and activation/elimination approaches directed towards flushing out latent virus. This latter approach could involve the use of novel chemically synthesized analogs of natural activating agents.
Keywords: HIV, cure, latency, gene therapy, reservoir, antiretroviral
Beginning with its first transmission into humans from chimpanzees approximately 100 years ago1, human immunodeficiency virus (HIV) has had a devastating effect throughout the world. At the end of 2011 an estimated 34 million people were living with HIV, and in that year approximately 1.7 million people died of acquired immunodeficiency syndrome (AIDS)2. There is currently no vaccine to prevent HIV infection, and while efforts to develop such a prophylactic vaccine are beginning to show promise3, it is a complex challenge which is unlikely to be achieved in the near future. Significant advances for treating HIV have been made in the area of antiviral therapy. However, these treatments are expensive and can result in side-effects, adherence issues, and the development of drug-resistant virus. Most importantly, they are not curative, and must instead be taken for the remainder of the patient’s life to effectively contain the virus and prevent progression to AIDS. Furthermore, access to such drugs is limited for individuals in many parts of the world2. Developing a cure for HIV to eliminate the virus from people who are already infected is therefore an important area of research. The purpose of this review is to describe what currently available HIV therapies do, outline our understanding of why they do not cure the infection, and discuss several novel approaches that are currently under development for eliminating HIV from infected individuals.
HIV infects and kills cells of the immune system, including CD4+ T cells and macrophages4. These cells are critical for mounting effective immune responses against invading pathogens. Over time, HIV replication causes depletion of these cells, leading to lower total CD4+ T cell numbers, damage to the architecture of lymph nodes and other lymphoid tissues, immune activation, and general dysregulation of immune function4. After an average infection time of around 10 years, the immune system is damaged to the point that the infected individual progresses to AIDS. At this stage the individual becomes highly susceptible to both common and unusual infections and cancers, which ultimately result in death4.
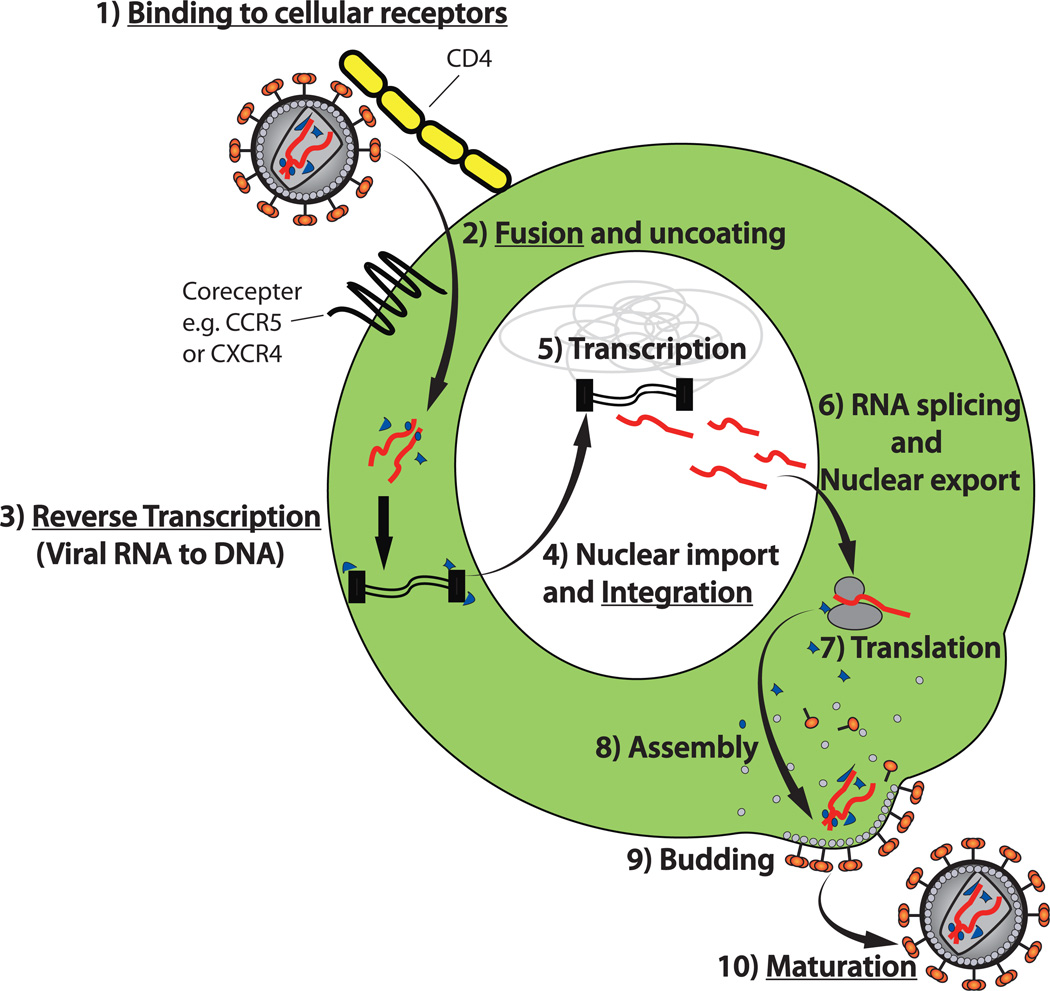
Since HIV replication is required for the development of disease, antiretroviral drugs have been developed to prevent this replication and stop progression to AIDS5. A diagram outlining the major steps in the HIV life cycle and the targets of clinically approved antiretroviral drugs is shown (Fig 1). As a retrovirus, the genetic material in HIV virions (virus particles) is RNA, but the virus replicates through a DNA intermediate that is integrated into the DNA of the host cell. During infection, the virus first binds to the CD4 protein and a coreceptor protein at the cell surface. The most commonly utilized coreceptors are the chemokine receptors CCR5 and CXCR4, with CCR5 usage generally predominating in early infection, and often maintained throughout infection6. The viral and host cell membranes then fuse, and the virion-borne reverse transcriptase enzyme catalyzes conversion of the viral RNA into DNA. This DNA is transported into the nucleus as part of a pre-integration complex. The viral integrase enzyme then mediates the integration process, whereby the viral DNA is inserted into the host cell’s chromosomes. At this point the resultant proviral DNA is permanently integrated and will be maintained for the lifespan of the host cell. HIV RNA is then transcribed from the integrated provirus, and is either translated into proteins (following RNA splicing for certain viral proteins), or directly incorporated into new virions. The virions assemble and bud from the plasma membrane. Finally, the viral protease enzyme cleaves polyproteins within the virion to produce mature infectious virus particles that are ready to infect a new cell.
Fig. 1. Essential steps in the HIV life cycle and targets of currently available antiretroviral drugs.
1) HIV virus particles (virions) bind to CD4 and a coreceptor (generally CCR5 or CXCR4) on target cells. 2) The viral envelope proteins mediate fusion of the viral and host cell membranes, allowing the viral RNA to be released into the host cell cytoplasm. 3) The viral RNA is reverse transcribed into double stranded DNA by the HIV reverse transcriptase enzyme. 4) Double stranded viral DNA is translocated into the nucleus and the HIV integrase enzyme catalyzes integration of this DNA into the host cell’s chromosomes. At this point the HIV genome is referred to as “proviral DNA” or an “integrated provirus”. 5) Transcription of the HIV genome is mediated by host cell polymerases. 6) and 7) HIV RNA is exported to the cytoplasm for translation or incorporation into new virions. For expression of some proteins, the RNA is spliced prior to nuclear export. 8 and 9) New virions assemble and bud from the plasma membrane. 10) As virions bud, the viral protease enzyme cleaves HIV polyproteins into individual subunits, producing infectious, mature virions. Underscored steps represent those that are targeted by clinically-approved antiretroviral drugs.
Over 20 antiretroviral drugs have been approved for use in HIV infected patients5. These drugs variously inhibit virus entry (fusion/entry inhibitors), reverse transcription (reverse transcriptase inhibitors), integration (integrase inhibitors), or maturation (protease inhibitors) (Fig 1). Modern antiretroviral therapy regimes typically consist of specific combinations of three antiretroviral drugs termed combination antiretroviral therapy (cART) or highly active antiretroviral therapy (HAART). The rationale behind using multiple drugs with non-overlapping resistance profiles is to increase the suppression of virus replication achieved by the therapy while also reducing the likelihood of the virus becoming resistant to the drugs. Importantly, these antiretroviral drugs only inhibit virus replication. Therefore they can stop the virus from spreading to new cells but have no direct effect on an integrated HIV provirus.
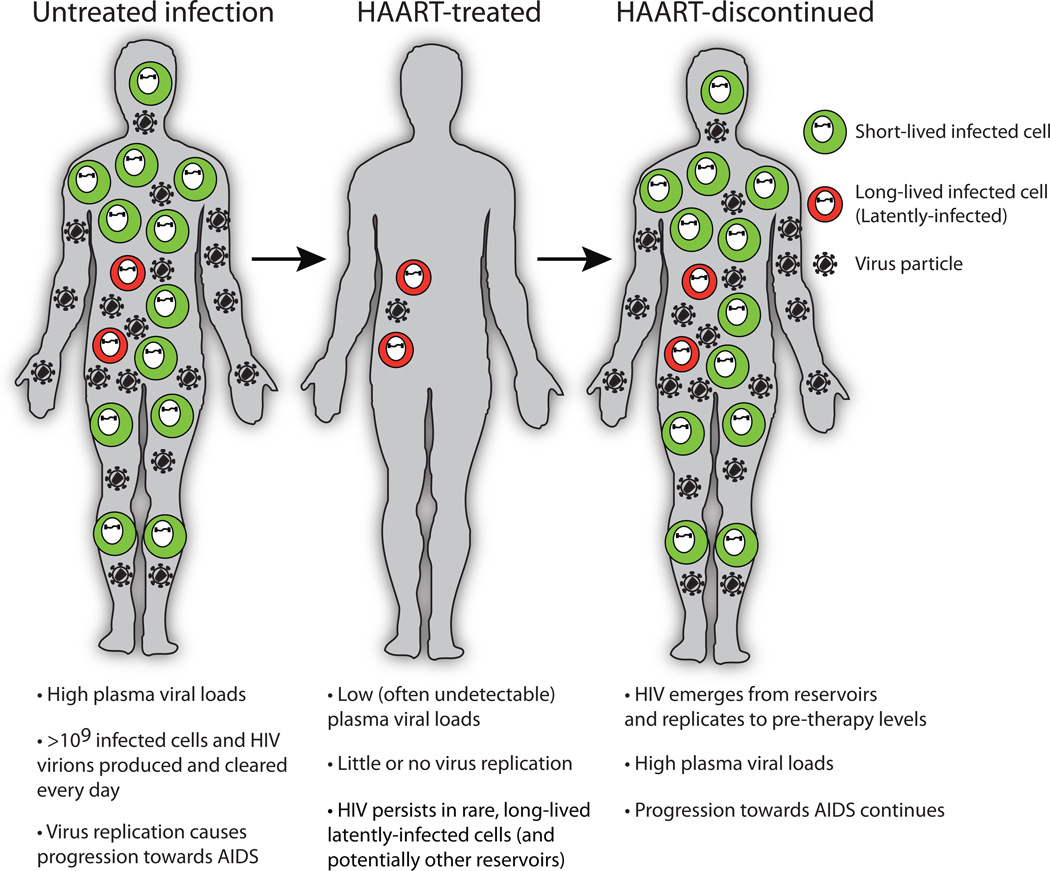
Untreated HIV infection is generally characterized by a continuous battle between the virus and the host immune response, with billions of new virions and infected cells produced and cleared every day (Fig 2)7. The fact that the adaptive immune response maintains much of its function for years during this onslaught is a testament to its strength and regenerative capacity. Treatment with HAART eliminates the vast majority of (or potentially all) HIV replication, and plasma viral loads often fall to levels that are undetectable with standard clinical assays (such assay limits are typically 50 copies of virion RNA/ml of plasma)4. However, certain reservoirs of replication-competent virus persist during therapy. Therefore if HAART is stopped then virus can emerge from these reservoirs and rapidly spread8, causing renewed progression towards AIDS. The resultant rebound in plasma viral loads typically occurs within several weeks of stopping therapy, indicating that virus is being released from reservoirs with regularity9.
Fig. 2. Suppression of HIV replication during therapy.
The natural course of HIV infection is associated with extremely high levels of virus replication. Treatment with highly active antiretroviral therapy (HAART) inhibits the vast majority of this replication, but HIV persists at low levels in very rare cellular reservoirs in these individuals. If HAART is stopped for any reason, the virus can emerge from these reservoirs, allowing replication to continue at pretherapy levels.
The best understood reservoir of HIV during HAART consists of latently-infected CD4+ T cells. These cells are resting (primarily central and transitional memory) CD4+ T cells that harbor an integrated HIV provirus and express little or no viral RNA and no viral proteins, but can be induced to produce infectious virus if the cell becomes activated10–13. In this non-expressing state the latent provirus cannot be recognized by the immune system, and consequently the infected cell is not eliminated by immune effector mechanisms. Latently-infected CD4+ T cells are relatively rare in HAART-treated patients, with approximately 1 latently-infected cell per million total resting CD4+ T cells, translating into about one million latently-infected cells per patient14. These memory CD4+ T cells are a key component of immunological memory responses, and it is believed that they can survive for decades even while harboring latent HIV proviruses. The stability of this reservoir during HAART is formidable. It has been estimated that depletion of the latent reservoir using HAART alone would take over 70 years15.
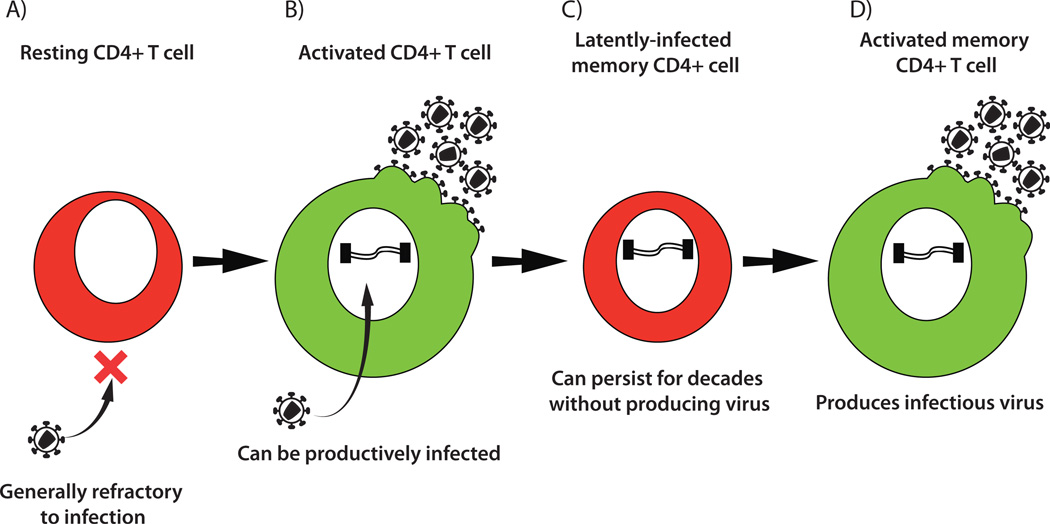
Studies of latently-infected cells from infected individuals as well as in vitro experiments using both cell lines and primary CD4+ cells have provided insights into how latency can be established (Fig 3)16, 17. Resting CD4+ T cells cannot be efficiently infected by HIV, with incoming virus facing multiple blocks to infection. The most pronounced block is a reduced efficiency of reverse transcription18. However, if the cell is activated, it then becomes susceptible to productive infection by HIV. This activation generally happens when the cell encounters an antigen-presenting cell displaying a foreign peptide that the T cell recognizes, resulting in signaling through its T cell receptor. Partial activation with cytokines can also shift the cell from a G0 resting phenotype into a state that can support HIV infection19. The majority of activated T cells that are infected by HIV will die, but a small subset of activated T cells naturally transition into resting memory cells. This transition is associated with a reduction in cellular transcriptional activity and a variety of other changes that together are capable of preventing expression of HIV. Thus if an infected cell undergoes this transition before it can be killed by the virus or the immune response, then it can become a latently-infected cell harboring a non-expressing HIV provirus. If the cell subsequently becomes activated, for example by recognition of its cognate antigen or exposure to a proinflammatory cytokine environment, then new infectious virus can be produced. Additional mechanisms for establishment of latency may also contribute to the pool of latently-infected cells observed in vivo, such as rare direct infection of resting CD4+ T cells20, or infection during certain stages of T cell development (thymopoiesis)21.
Fig. 3. Generation of HIV latency.
A) Resting CD4+ T cells cannot generally support HIV replication. B) However, if the cells are stimulated by cytokines or recognition of their cognate antigen then they become activated and susceptible to HIV infection. Infection will generally result in death of the host cell, but a small subset of these cells will transition back to a resting state before they can be killed by the virus. C) The result is a shut-down of HIV expression and production of a long-lived latently-infected cell that harbors an HIV provirus that is not producing viral proteins. D) Subsequent activation of this latently infected cell (perhaps many years later) results in re-initiation of virus expression and production of new infectious virions.
In addition to CD4+ T cells, other cell types such as hematopoietic stem cells22 and monocyte/macrophage cells23 might also become latently-infected. However, the regularity in which this occurs and the contribution of these cell types to persistence of HIV over the course of many years of therapy is unclear. Macrophages can also become chronically infected in some cases, whereby they continuously produce low levels of virus over extended periods of time without being killed by the virus24. There is also evidence that HAART does not prevent all HIV replication in some individuals. Ongoing virus replication due to incomplete HAART suppression in these individuals would clearly provide a source of HIV to seed viral rebound upon cessation of therapy. Therefore, the latent reservoir in CD4+ T cells might not be the only source of replication-competent HIV that persists during HAART. In spite of this possibility, latently-infected CD4+ T cells are a major focus of HIV cure research because this reservoir is the only one that has been convincingly demonstrated to be capable of maintaining replication-competent HIV over many years of effective HAART15, 25. Hence, even if other reservoirs are also present, the latent reservoir still represents a key barrier to eradication that will need to be surmounted before a cure for HIV is achieved.
Several approaches for HIV eradication are currently being investigated. Most of these can be broadly categorized as gene therapy/transplantation approaches that are generally intended to protect cells of the immune system from HIV infection, and activation/elimination approaches directed towards flushing out the latent virus. These methodologies are not necessarily mutually exclusive, but each is intended to either cure the infection by eliminating all replication competent virus from the individual, or produce a “functional cure” whereby the levels of HIV are substantially reduced in a durable manner without ongoing therapy, such that disease progression does not occur and transmission of virus to new hosts is highly unlikely.
The idea of using allogeneic (non-self) transplants to cure HIV is supported by a remarkable single case which has galvanized the field26, 27. This individual, sometimes referred to as the “Berlin Patient”, was HIV-positive with suppressed viral loads through effective HAART. He then developed a life-threatening acute myeloid leukemia. Treatment for the leukemia involved the use of extremely aggressive chemotherapy to eliminate the cancerous cells, which also depleted the majority of his HIV host cells (CD4+ T cells and macrophages). He then received two bone marrow transplants to provide CD34+ hematopoietic stem cells that could differentiate into new blood cells. Matched donor cells were identified that were homozygous for a CCR5 delta-32 mutation. This is a naturally-occurring mutation that results in cells which cannot express the CCR5 HIV coreceptor, but because of overlapping functionality and redundancy in the immune signaling network, it does not appear to affect the health of individuals carrying it.
Approximately 1% of Caucasians are homozygous for this mutation, and are therefore naturally resistant to infection by the CCR5-tropic strains of HIV that generally predominate in most infected individuals6. During the transplant process the patient stopped HAART, and HIV viral loads did not increase. Testing of blood and tissues over the subsequent four years has failed to find evidence for replicating HIV. It therefore appears that in this particular case, the combination of chemotherapy to deplete endogenous HIV host cells followed by repopulation with HIV-resistant cells was sufficient to clear the infection. Additional contributing factors such as graft versus host disease, which occurred during the course of treatment, could also have contributed towards the elimination of pre-existing cellular reservoirs of virus26. This case is tentatively being hailed as the first instance of an individual being cured of HIV27.
However, there are numerous reasons that this approach cannot be extended to all infected individuals. The aggressive chemotherapy alone carries significant risks. In addition, locating an appropriate bone marrow donor match that is also CCR5 delta-32 homozygous is not possible for many individuals, and use of imperfectly-matched allogeneic donor cells can result in problems associated with immune incompatibilities. Finally, the expense and complexity of the process require well-funded and sophisticated medical facilities that are not available for most people infected with HIV. Nevertheless, this case has provided a proof-of-concept that HIV can be cured. Therefore alternative approaches based on similar principles are being developed that could be more broadly applied.
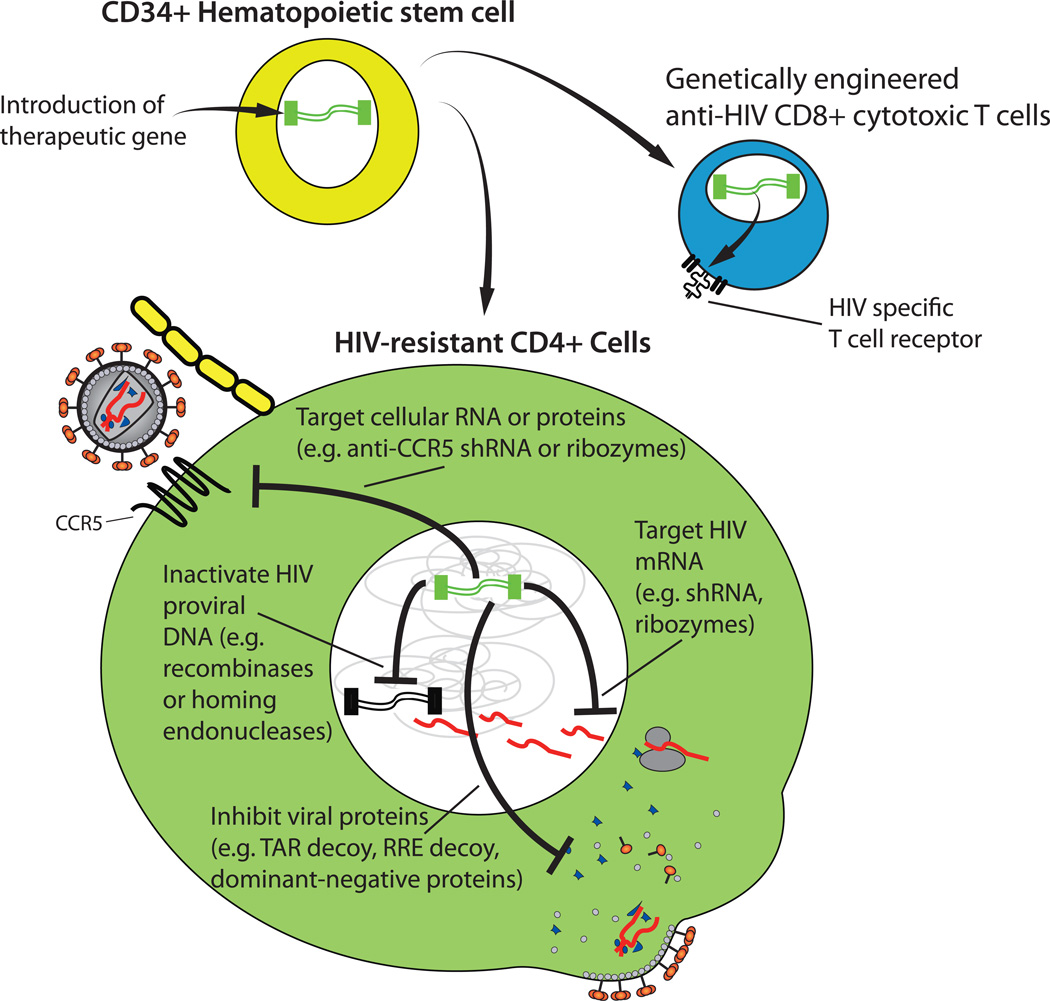
Several gene therapy approaches for treating HIV are under investigation. These are generally directed towards introducing an anti-HIV gene of some type into cells obtained from the infected patient, then reinfusing these cells back into the same patient. The use of autologous cells from the same patient should reduce or eliminate the problems with immunological incompatibility that are characteristic of allogeneic cell transfers. The genes could be introduced into differentiated peripheral cells such as CD4+ T cells. However, most of these cells do not persist for long periods of time in vivo. Therefore an alternative approach is to introduce the therapeutic gene into CD34+ hematopoietic stem cells (Fig 4), which are capable of self-renewal, can differentiate into all blood cell types, and are theoretically able to produce differentiated cells expressing the anti-HIV genes indefinitely28. Under ideal circumstances, such cells would be refractory to HIV infection and would thus be able to maintain an effective immune response and prevent progression to AIDS.
Fig 4. Gene therapy approaches for eliminating HIV.
Therapeutic genes can be introduced into CD34+ hematopoietic stem cells. As these cells differentiate, the introduced genes will also be present and expressed in progeny cells. In this way CD4+ T cells can be produced that are resistant to HIV infection or encode genes for anti-HIV factors. Examples shown here include: genes encoding T cell receptors against HIV, that could be used to produce cytotoxic T lymphocytes that target HIV-infected cells; genes encoding short-hairpin RNAs (shRNAs) and ribozymes, that can cleave viral RNA or the RNA for the CCR5 receptor for HIV, making the cell resistant to infection; genes encoding recombinases or homing endonucleases, that can inactivate integrated HIV DNA, and various RNAs and proteins that can interfere with HIV protein functions.
A diverse armamentarium of anti-HIV approaches have been developed and tested for gene therapy applications29–31. These include antisense RNAs, short peptides, intrabodies, short-hairpin RNAs (shRNAs), short-interfering RNAs (siRNAs), RNA decoys, zinc-finger nucleases and ribozymes. Some of these gene-based therapeutics directly target HIV proteins or RNA to prevent productive infection altogether or to reduce the amount of virus expression in an infected cell. For example shRNAs are capable of causing mRNA degradation and inhibition of protein translation in a sequence-specific manner. Therefore shRNAs have been designed that target mRNA regions across the HIV genome, including coding sequences for the essential HIV regulatory proteins Tat and Rev, accessory proteins such as Nef, and the major open reading frames encoding the structural (Gag), polymerase (Pol), and envelope (Env) proteins32. The HIV Rev protein is required for exporting partially spliced and unspliced HIV transcripts from the nucleus. Early studies have shown that the function of this protein can be disrupted by expression of a dominant negative version of Rev, and this fact has been exploited in exploratory gene therapy approaches33, 34. HIV Tat protein is critical for producing high levels of HIV transcription. Both Tat and Rev function by binding to specific RNA secondary structures on the HIV genome, termed the transactivation response element (TAR) and Rev response element (RRE), respectively. RNA decoys that resemble these natural structures have been produced, which bind to the viral proteins and interfere with their function35, 36. A ribozyme (catalytic RNA) that sequence-specifically cleaves HIV RNA has also been developed and tested through Phase I and Phase II clinical trials37. Moreover, a membrane-anchored peptide termed C46 has also been shown to prevent fusion of the viral and host cell membranes when expressed in potential HIV host cells, by binding to and inhibiting the gp41 region of HIV Env38.
Given that latent HIV consists of an integrated provirus that is not directly affected by antiviral drugs, several interesting methods to inactivate such non-expressing proviruses are also under investigation. One method involves introducing a gene encoding for an evolved recombinase that is capable of excising the integrated provirus39. A second involves the use of homing endonucleases that can cleave the proviral DNA, leading to disruption of its coding sequence following DNA repair by the host cell40.
Gene-based strategies that affect cellular proteins are also under investigation. One such approach involves knock-down or knock-out of CCR5 to protect the host cells from infection by CCR5-tropic HIV variants and hopefully achieve similar results to those observed with the Berlin Patient. Successful methods for reducing levels of CCR5 expression in cells have included the use of shRNA41, 42 and ribozyme43 technologies that specifically target CCR5 mRNA.
Another promising approach for eliminating CCR5 expression is the use of zinc finger nucleases44 that can irreversibly inactivate the CCR5 gene following transient expression in hematopoietic stem cells, and thus do not require a therapeutic gene to be permanently maintained in the cells. This CCR5-specific zinc finger nuclease approach has been successfully applied to a humanized mouse model of HIV infection45.
Utilizing a combination of genes with different mechanisms of action would probably provide better protection to the cells than use of a single protective gene, and it would also make selection of resistant virus less likely. To this end, combinatorial gene therapy approaches are being developed. For example, a recent clinical trial involved the introduction of a vector encoding a TAR decoy along with an shRNA targeting the tat/rev region of the HIV genome and an anti-CCR5 ribozyme46.
Another way that gene therapy may be utilized in treating HIV infection is via the introduction of genes that generate more effective immune responses against the virus. One example of this is the introduction of an HIV-specific T cell receptor, which results in the production of cytotoxic T lymphocytes that can target and kill cells expressing particular HIV proteins31.
As with the development of all new therapeutics, safety considerations are of paramount importance. To date, the majority of clinical studies using gene therapy for HIV treatment have been focused on determining the safety and feasibility of the procedures rather than optimizing efficacy. These trials have demonstrated that stem cell-based gene-transfer clinical trials can be conducted safely with a relatively large number of study participants (in one case, 74 HIV-infected individuals37). Challenges that these trials have highlighted are currently being addressed, including the need to increase the frequency of cells harboring the newly-introduced transgene in vivo. Further limitations to current gene therapy approaches include the expense and complexity of the procedures, the potential for long-term complications associated with introduction of foreign genes into cells (for example insertional mutagenesis or off-target DNA damage), and side effects of chemotherapy that may be required to deplete some endogenous cells in order to “make space” for the newly-introduced cells. Nevertheless, if optimized, these gene therapy approaches would offer the possibility for HIV-infected individuals to receive a single treatment, or collection of treatments, that would provide lifelong protection against development of AIDS without the continual need for antiretroviral drugs.
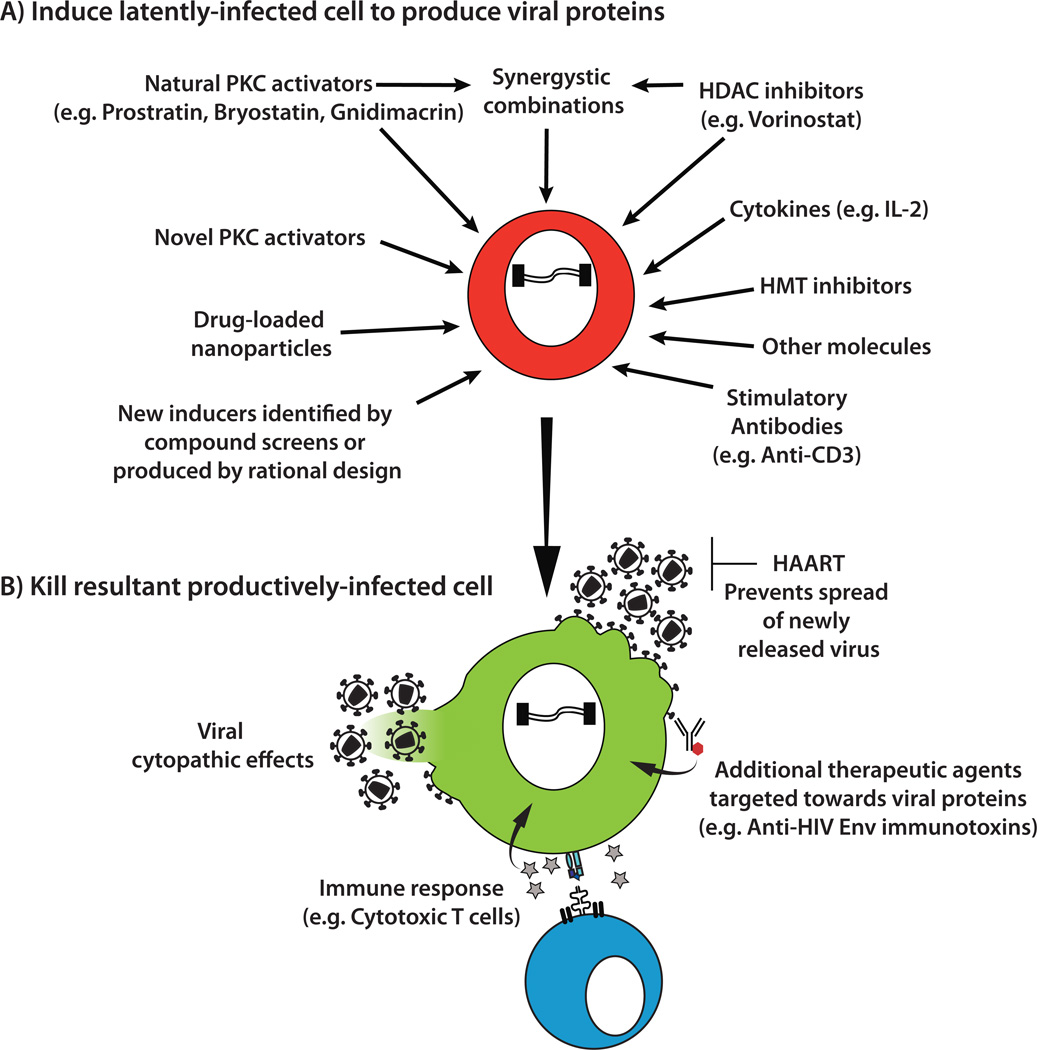
The second major strategy for purging latently-infected cells is often referred to as an “activation/elimination” approach (Fig 5). The goal of this strategy is to flush out the latent virus by inducing it to express viral proteins. If high levels of HIV expression are generated then the cell may be killed by the damaging effects of virus production itself (viral cytopathic effects) or apoptosis. Once proteins are expressed, the infected cell should also become visible to the immune system and can be targeted by effector cells such as CD8+ cytotoxic T lymphocytes or other therapeutic agents directed towards viral proteins. In order to prevent the activated virus from initiating new rounds of virus replication, activation/elimination approaches would be performed during continuous HAART.
Fig. 5. Activation/elimination approaches to purge the latent HIV reservoir.
A) A variety of different stimuli have been shown to be capable of inducing a latent HIV provirus to express new viral proteins. In some cases combinations of inducers (such as PKC activators and HDAC inhibitors) can act synergistically in this process. B) Once virus expression has been activated in the latently-infected cell, HIV spread to new cells will be inhibited by the continued presence of HAART. The host cell may be killed directly by cytopathic effects associated with virus replication. Alternatively, the cell could be killed by immune effector mechanisms or novel therapeutic approaches targeted towards viral proteins. HDAC (histone deacetylase), HMT (histone methyltransferase), PKC (protein kinase C), IL (interleukin).
Early clinical attempts to purge latent virus in vivo relied on stimulation of T cells with the cytokine interleukin (IL)-2. A significant effect on latently-infected cell numbers was identified in these studies, but viral rebound was still observed following cessation of antiretroviral therapy8, 9, 47. Administration of IL-2 in conjunction with an anti-CD3 monoclonal antibody that stimulates T cells through the T cell receptor was also attempted, but the treatment was associated with toxic side-effects48. These important exploratory studies demonstrated that the “blunt force” approach of inducing global T cell activation would not be sufficient to achieve the goal of HIV eradication. Consequently more selective methods based on our growing understanding of latency have been investigated. Though a detailed description of the molecular mechanisms underlying HIV latency and strategies for activating latent virus is provided elsewhere16, 17, examples of several areas of current interest are outlined below.
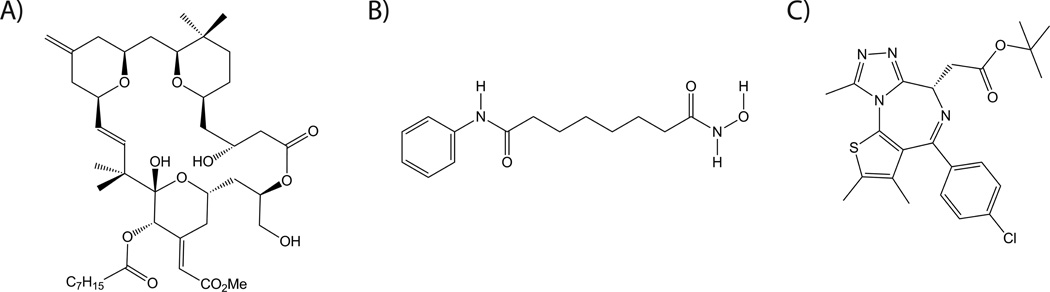
Protein kinase C (PKC) activators such prostratin, a non-tumor-inducing phorbol ester, and bryostatin 1, a marine macrolide, can induce HIV from latency via activation of the transcription factor NF-κB17. Bryostatin 1 has been tested in over 30 clinical trials as a cancer therapeutic, but isolation from natural sources is expensive and problematic. In collaboration with the Wender lab at Stanford University, we recently described the synthesis of bryostatin analogs that are inexpensive to produce and display improved HIV latency activation properties in a cell line model49 (for example, Fig 6A). The Wender laboratory has also succeeded in synthesizing analogues of prostratin50. The in vivo safety and efficacy of these new compounds has yet to be determined, but since these analogs can in principle be tuned for performance, there is now greater latitude for optimization during preclinical testing.
Fig. 6. Examples of HIV latency activating compounds currently under investigation.
A) Bryostatin analog (PKC activator) B) Vorinostat (HDAC inhibitor) C) (+)-JQ1 (bromodomain inhibitor).
Histone deacetylases (HDACs) can produce hypoacetylated nucleosomes at the HIV promoter, which reduces access of transcription factors and contributes to the maintenance of HIV latency. Conversely, HDAC inhibitors can induce latent HIV expression. One such compound termed “vorinostat” (suberoylanilide hydroxamic acid, Fig 6B), is already approved for clinical use for cancer applications. Initial results from a recent clinical trial using vorinostat to activate HIV from latency have shown that it can increase HIV transcription in resting CD4+ T cells from HAART-treated patients, demonstrating that the latent reservoir can be perturbed in vivo using HDAC inhibitors51. Additional HDAC inhibitors including romidepsin, panobinostat and others have also shown promise as HIV latency activating compounds and are currently being further studied. Moreover, HDAC inhibitors and PKC activators have also been found to induce HIV from latency in a synergistic manner. Therefore combinations of different latency activators may prove more effective than either one individually.
In addition, a variety of other mechanisms may contribute to latency, such as the actions of histone methyltransferases, DNA methyltransferases, bromodomain proteins, and the lack of positive transcription elongation factor b (pTEFb) in latently-infected cells. Hence, compounds that affect these factors are also under investigation17. For example the transcriptional regulator bromodomain containing 4 (BRD4) competes with HIV Tat for pTEFb, thereby reducing HIV transcription and potentially contributing to maintenance of HIV latency52. Consistent with this concept, the BRD4 inhibitor JQ1 (Fig 6C) has been shown to be capable of activating HIV from latency52, 53. Consequently this and other similar bromodomain inhibitors54 may prove useful in HIV eradication efforts.
Since expression of HIV is closely associated with the activation state of the host cell, there is concern that the most effective latency-activating agents may cause generalized immune activation, for example via induction of abundant proinflammatory cytokines (sometimes referred to as a cytokine storm). This has the potential to be directly toxic to the patient, and it also would create a large number of activated T cells at the time of latency induction, making virus spread to these potential new host cells more difficult to contain with HAART. One potential approach for enhancing efficacy while reducing off target effects is to direct the latency activating compounds more selectively to the cell type of interest. For example, we recently described the use of lipid nanoparticles for introducing bryostatin 2 specifically to CD4+ cells55. Furthermore, the copackaging of the HIV protease inhibitor nelfinavir into these particles allowed the same particle to simultaneously activate HIV from latency while inactivating any virions released from the cell.
There is also some evidence that activation of the virus from latency may not be sufficient to kill the cell, particularly if only a small amount of virus expression is induced56. Hence, additional interventions such as therapeutic vaccination to enhance anti-HIV immune responses, use of genetically modified HIV-specific cytotoxic T lymphocytes (Fig 4), or immunotoxins that can bind to and kill cells expressing HIV Env on the surface (Fig 5) may prove useful in killing the recently activated latently-infected cells57. Furthermore, our understanding of HIV latency has focused primarily on the 2–3% of T cells that are circulating in the peripheral blood, and we have limited information regarding how latently-infected cells residing in tissues will respond to potential therapies. The use of relevant small animal models may allow a more thorough investigation of this important question58, 59.
Eliminating HIV from infected individuals is a complex problem. However, there is increasing optimism within the field that curing HIV is an achievable goal. Some of the approaches that are under investigation are expensive, labor intensive, and complicated. If a cure is developed then the next challenge will be to make it available to infected individuals throughout the world, including those who currently have no access to available HIV therapies.
Acknowledgments
We thank Dr. Sherry M. Tsai for the illustration of molecular structures shown in Fig 6. The authors gratefully acknowledge support from the National Institutes of Health (AI70010 and U19AI096113, project 3.4 to J.Z.) and a Grand Challenges Explorations grant (OPP1032668) from the Bill and Melinda Gates Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, Muyembe JJ, Kabongo JM, Kalengayi RM, Van Marck E, Gilbert MT, Wolinsky SM. Nature. 2008:455–661. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.UNAIDS. UNAIDS Report on the Global AIDS Epidemic. 2012 [Google Scholar]
- 3.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. N Engl J Med. 2009:361–2209. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 4.Moir S, Chun TW, Fauci AS. Annu Rev Pathol. 2011:6–223. doi: 10.1146/annurev-pathol-011110-130254. [DOI] [PubMed] [Google Scholar]
- 5.Thompson MA, Aberg JA, Hoy JF, Telenti A, Benson C, Cahn P, Eron JJ, Gunthard HF, Hammer SM, Reiss P, Richman DD, Rizzardini G, Thomas DL, Jacobsen DM, Volberding PA. JAMA. 2012:308–387. doi: 10.1001/jama.2012.7961. [DOI] [PubMed] [Google Scholar]
- 6.Moore JP, Kitchen SG, Pugach P, Zack JA. AIDS Res Hum Retroviruses. 2004:20–111. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- 7.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Nature. 1995;373:123. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 8.Chun T-W, Davey RT, Engel D, Lane HC, Fauci AS. Nature. 1999;401:874. doi: 10.1038/44755. [DOI] [PubMed] [Google Scholar]
- 9.Davey RT, Jr, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, Kovacs JA, Polis MA, Walker RE, Falloon J, Masur H, Gee D, Baseler M, Dimitrov DS, Fauci AS, Lane HC. Proc Natl Acad Sci U S A. 1999;96:15109. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. Nat Med. 1995;1:1284. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 11.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. Science. 1997;278:1295. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 12.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. Science. 1997;278:1291. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 13.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. Nat Med. 2009;15:893. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. Nature. 1997;387:183. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 15.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. Nat Med. 1999;5:512. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 16.Marsden MD, Zack JA. Future Virology. 2010;5:97. doi: 10.2217/fvl.09.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing S, Siliciano RF. Drug Discov Today. 2012 doi: 10.1016/j.drudis.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. Cell. 1990;61:213. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 19.Unutmaz D, KewalRamani VN, Marmon S, Littman DR. J Exp Med. 1999;189:1735. doi: 10.1084/jem.189.11.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swiggard WJ, Baytop C, Yu JJ, Dai J, Li C, Schretzenmair R, Theodosopoulos T, O'Doherty U. J Virol. 2005;79:14179. doi: 10.1128/JVI.79.22.14179-14188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks DG, Kitchen SG, Kitchen CM, Scripture-Adams DD, Zack JA. Nature medicine. 2001;7:459. doi: 10.1038/86531. [DOI] [PubMed] [Google Scholar]
- 22.Carter CC, Onafuwa-Nuga A, McNamara LA, Riddell Jt, Bixby D, Savona MR, Collins KL. Nat Med. 2010;16:446. doi: 10.1038/nm.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coleman CM, Wu L. Retrovirology. 2009;6:51. doi: 10.1186/1742-4690-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gendelman HE, Orenstein JM, Martin MA, Ferrua C, Mitra R, Phipps T, Wahl LA, Lane HC, Fauci AS, Burke DS, et al. J Exp Med. 1988;167:1428. doi: 10.1084/jem.167.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisele E, Siliciano RF. Immunity. 2012;37:377. doi: 10.1016/j.immuni.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. N Engl J Med. 2009;360:692. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 27.Allers K, Hutter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, Schneider T. Blood. 2011;117:2791. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 28.Kiem HP, Jerome KR, Deeks SG, McCune JM. Cell Stem Cell. 2012;10:137. doi: 10.1016/j.stem.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scherer LJ, Rossi JJ. Hum Mol Genet. 2011;20:R100. doi: 10.1093/hmg/ddr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson CW, Younan P, Jerome KR, Kiem HP. Gene Ther. 2013 doi: 10.1038/gt.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitchen SG, Shimizu S, An DS. Virology. 2011;411:260. doi: 10.1016/j.virol.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Mol Ther. 2006;14:883. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Malim MH, Freimuth WW, Liu J, Boyle TJ, Lyerly HK, Cullen BR, Nabel GJ. J Exp Med. 1992;176:1197. doi: 10.1084/jem.176.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Podsakoff GM, Engel BC, Carbonaro DA, Choi C, Smogorzewska EM, Bauer G, Selander D, Csik S, Wilson K, Betts MR, Koup RA, Nabel GJ, Bishop K, King S, Schmidt M, von Kalle C, Church JA, Kohn DB. Mol Ther. 2005;12:77. doi: 10.1016/j.ymthe.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 35.Lee SW, Gallardo HF, Gilboa E, Smith C. J Virol. 1994;68:8254. doi: 10.1128/jvi.68.12.8254-8264.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullenger BA, Gallardo HF, Ungers GE, Gilboa E. Cell. 1990;63:601. doi: 10.1016/0092-8674(90)90455-n. [DOI] [PubMed] [Google Scholar]
- 37.Mitsuyasu RT, Merigan TC, Carr A, Zack JA, Winters MA, Workman C, Bloch M, Lalezari J, Becker S, Thornton L, Akil B, Khanlou H, Finlayson R, McFarlane R, Smith DE, Garsia R, Ma D, Law M, Murray JM, von Kalle C, Ely JA, Patino SM, Knop AE, Wong P, Todd AV, Haughton M, Fuery C, Macpherson JL, Symonds GP, Evans LA, Pond SM, Cooper DA. Nat Med. 2009;15:285. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egelhofer M, Brandenburg G, Martinius H, Schult-Dietrich P, Melikyan G, Kunert R, Baum C, Choi I, Alexandrov A, von Laer D. J Virol. 2004;78:568. doi: 10.1128/JVI.78.2.568-575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar I, Hauber I, Hauber J, Buchholz F. Science. 2007;316:1912. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 40.Aubert M, Ryu BY, Banks L, Rawlings DJ, Scharenberg AM, Jerome KR. PLoS One. 2011;6:e16825. doi: 10.1371/journal.pone.0016825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin XF, An DS, Chen IS, Baltimore D. Proc Natl Acad Sci U S A. 2003;100:183. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimizu S, Hong P, Arumugam B, Pokomo L, Boyer J, Koizumi N, Kittipongdaja P, Chen A, Bristol G, Galic Z, Zack JA, Yang O, Chen IS, Lee B, An DS. Blood. 2010;115:1534. doi: 10.1182/blood-2009-04-215855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng Y, Leavitt M, Tritz R, Duarte E, Kang D, Mamounas M, Gilles P, Wong-Staal F, Kennedy S, Merson J, Yu M, Barber JR. Virology. 2000;276:271. doi: 10.1006/viro.2000.0536. [DOI] [PubMed] [Google Scholar]
- 44.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH. Nat Biotechnol. 2008;26:808. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. Nat Biotechnol. 2010;28:839. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiGiusto DL, Krishnan A, Li L, Li H, Li S, Rao A, Mi S, Yam P, Stinson S, Kalos M, Alvarnas J, Lacey SF, Yee JK, Li M, Couture L, Hsu D, Forman SJ, Rossi JJ, Zaia JA. Sci Transl Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chun TW, Engel D, Mizell SB, Hallahan CW, Fischette M, Park S, Davey RT, Jr, Dybul M, Kovacs JA, Metcalf JA, Mican JM, Berrey MM, Corey L, Lane HC, Fauci AS. Nat Med. 1999;5:651. doi: 10.1038/9498. [DOI] [PubMed] [Google Scholar]
- 48.Prins JM, Jurriaans S, van Praag RM, Blaak H, van Rij R, Schellekens PT, ten Berge IJ, Yong SL, Fox CH, Roos MT, de Wolf F, Goudsmit J, Schuitemaker H, Lange JM. Aids. 1999;13:2405. doi: 10.1097/00002030-199912030-00012. [DOI] [PubMed] [Google Scholar]
- 49.DeChristopher BA, Loy BA, Marsden MD, Schrier AJ, Zack JA, Wender PA. Nat Chem. 2012;4:705. doi: 10.1038/nchem.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wender PA, Kee JM, Warrington JM. Science. 2008;320:649. doi: 10.1126/science.1154690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. Nature. 2012;487:482. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J, Gaiha GD, John SP, Pertel T, Chin CR, Gao G, Qu H, Walker BD, Elledge SJ, Brass AL. Cell Rep. 2012;2:807. doi: 10.1016/j.celrep.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Banerjee C, Archin N, Michaels D, Belkina AC, Denis GV, Bradner J, Sebastiani P, Margolis DM, Montano M. J Leukoc Biol. 2012;92:1147. doi: 10.1189/jlb.0312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bamborough P, Diallo H, Goodacre JD, Gordon L, Lewis A, Seal JT, Wilson DM, Woodrow MD, Chung CW. J Med Chem. 2012;55:587. doi: 10.1021/jm201283q. [DOI] [PubMed] [Google Scholar]
- 55.Kovochich M, Marsden MD, Zack JA. PLoS One. 2011;6:e18270. doi: 10.1371/journal.pone.0018270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang HC, Zhang H, Margolick JB, Blankson JN, Siliciano RF. Immunity. 2012;36:491. doi: 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brooks DG, Hamer DH, Arlen PA, Gao L, Bristol G, Kitchen CM, Berger EA, Zack JA. Immunity. 2003;19:413. doi: 10.1016/s1074-7613(03)00236-x. [DOI] [PubMed] [Google Scholar]
- 58.Marsden MD, Kovochich M, Suree N, Shimizu S, Mehta R, Cortado R, Bristol G, An DS, Zack JA. J Virol. 2012;86:339. doi: 10.1128/JVI.06366-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Denton PW, Olesen R, Choudhary SK, Archin NM, Wahl A, Swanson MD, Chateau M, Nochi T, Krisko JF, Spagnuolo RA, Margolis DM, Garcia JV. J Virol. 2012;86:630. doi: 10.1128/JVI.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]