Abstract
Aicardi-Goutières syndrome (AGS) is an inflammatory disorder resulting from mutations in TREX1, RNASEH2A/2B/2C, SAMHD1 or ADAR1. Here we provide molecular, biochemical and cellular evidence for the pathogenicity of two synonymous variants in RNASEH2A. Firstly, the c.69G>A (p.Val23Val) mutation causes the formation of a splice donor site within exon 1, resulting in an out of frame deletion at the end of exon 1, leading to reduced RNase H2 protein levels. The second mutation, c.75C>T (p.Arg25Arg), also introduces a splice donor site within exon 1, and the internal deletion of 18 amino acids. The truncated protein still forms a heterotrimeric RNase H2 complex, but lacks catalytic activity. However, as a likely result of leaky splicing, a small amount of full-length active protein is apparently produced in an individual homozygous for this mutation. Recognition of the disease causing status of these variants allows for diagnostic testing in relevant families.
Keywords: Aicardi-Goutières syndrome, AGS, RNASEH2A, Synonymous mutations, splicing
First described in 1984 by Jean Aicardi and Françoise Goutières (Aicardi and Goutieres, 1984), Aicardi-Goutières syndrome (AGS) is a rare genetic encephalopathy that can mimic the clinical, radiological and biochemical features of acquired in utero viral infection (Goutieres, et al., 1998). An elevation of interferon alpha in serum and cerebrospinal fluid is frequently documented in AGS patients, and some affected individuals show immunological features overlapping those seen in the autoimmune disease systemic lupus erythematosus. Type I interferon mediated inflammation is thought to be induced by accumulating nucleic acid substrates that are normally degraded by cellular nucleases. AGS is a genetically heterogeneous disorder with six disease causing genes identified to date: TREX1, a 3’ to 5’ DNA exonuclease (Crow, et al., 2006a), RNASEH2A (MIM# 606034), RNASEH2B and RNASEH2C encoding the three components of the RNase H2 complex (Crow, et al., 2006b), SAMHD1, a deoxynucleoside triphosphate triphosphohydrolase (Goldstone, et al., 2011; Powell, et al., 2011; Rice, et al., 2009), and the double stranded RNA editing enzyme ADAR1 (Rice, et al., 2012). Null mutations are common in TREX1 and SAMHD1 (Rice, et al., 2007; Rice, et al., 2009), but biallelic null mutations in RNASEH2A, RNASEH2B, RNASEH2C and ADAR1 have not been definitively confirmed (Rice, et al., 2007; Rice, et al., 2012). Thus, for TREX1 and SAMHD1, truncated, inactive or, in the case of SAMHD1, complete absence of protein is still compatible with life, albeit often in the context of profound disability (Abdel-Salam, et al., 2010; Leshinsky-Silver, et al., 2010; Ramesh, et al., 2010). In the case of RNASEH2A/B/C, mutations are apparently hypomorphic, suggesting that biallelic null alleles in any of these three genes may be lethal during embryonic development (Crow, et al., 2006b; Rice, et al., 2007). Indeed, complete absence of RNaseH2 activity was recently shown to be incompatible with mouse development post-gastrulation (Reijns, et al., 2012).
We previously identified two individuals with single heterozygous missense mutations in RNASEH2A, and no additional recognised pathogenic lesions (Rice, et al., 2007). However, each of these individuals carried a synonymous transition on the other RNASEH2A allele (GenBank reference sequence: NM_006397.2), c.69G>A (p.Val23Val) (hereafter referred to as c.69G>A) or c.75C>T (p.Arg25Arg) (hereafter referred to as c.75C>T) (nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence. The initiation codon is codon 1). These changes have not been annotated as single nucleotide polymorphisms (SNPs), nor did we find them in any of our controls. These variants have been submitted to the LOVD RNASEH2A database (www.lovd.nl/RNASEH2A). This study was instigated to determine if these synonymous changes might be pathogenic.
Five patients with synonymous mutations in RNASEH2A were identified in four out of 251 families with a clinical diagnosis of AGS, that had been recruited internationally through collaborating physicians. The clinical characteristics of the patients studied are presented in Table 1.
Table 1.
Patient clinical data
| Family / patient |
Sex | Consanguinity | Ancestry | Age at presentation |
Features at presentation |
CSF white cell count* |
CSF IFN (IU/ml) (age)† |
CSF pterins [age]†† |
Status (age) | Calcification | Leukodystrophy | Developmental level |
Nucleotide alteration# |
Amino Acid alteration |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1_P1 | F | No | Spanish | 4wk | SP, PF, DD | “Raised” | 18 (18mo) | nr | Alive (12yr) | Yes | Yes | Severe delay | c.75C>T het + c.704G>A het |
p.Arg25Arg + p.Arg235Gln |
| 1_P2** | F | No | Spanish | 2wk | SP, PF, DD | “Raised” | nr | nr | Alive (12yr) | nr | Yes | Severe delay | c.75C>T het + c.704G>A het |
p.Arg25Arg + p.Arg235Gln |
| 2 | F | No | Belgian | 4mo | DD | 38 (4mo) | nr | nr | Alive (7yr) | Yes | Yes | Severe delay | c.75C>T hom | p.Arg25Arg (Hom) |
| 3 | M | No | Italian | Birth | PF, DD | 86 (4mo) | 200 (4mo) | T 110 (20–70) N 2000 (15–35)[4mo] |
Died (3yr 6mo) | Yes | Yes | Severe delay | c.69G>A het + c.556C>T het |
p.Val23Val + p.Arg186Trp |
| 4 | M | No | White US | Birth | PF, DD | 5 (17mo) | nr | T 66 (20–58) N 170 (7–65)[17mo] |
Alive (11yr) | Yes | Yes | Severe delay | c.69G>A het + c.635A>T het |
p.Val23Val + p.Asn212Ile |
SP Spasticity, PF Poor feeding, DD Developmental delay, nr Not recorded, wk Week, mo Month,
Abnormal >5 cells/mm3,
Normal levels <2 IU/ml,
T Tetrahydrobiopterin, N Neopterin values (normal range),
this sibling was not analysed in the present study.
GenBank reference sequence: NM_006397.2. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence. The initiation codon is codon 1.
In addition to the synonymous mutations, sequencing of RNASEH2A identified the following single heterozygous missense mutations in these patients: c.704G>A (p.Arg235Gln) in patient P1, c.556C>T (p.Arg186Trp) in patient 3, and c.635A>T (p.Asn212Ile) in patient 4 (Coffin, et al., 2011; Rice, et al., 2007). All of these mutations affect evolutionary conserved residues, with the arginine residues at positions 186 and 235 conserved in all eukaryotes, and the asparagine at position 212 conserved in amniota (Supp. Figure S1A). Furthermore, both Arg186 and Arg235 are thought to have roles in stabilizing the conformation of a highly conserved GRG motif (Supp. Figure S1B and C) responsible for recognition of the substrate RNA-DNA junction (Figiel, et al., 2010), likely explaining why the Arg186Trp and Arg235Gln mutations dramatically reduce substrate binding and enzyme activity in vitro (Coffin, et al., 2011). The Asn212Ile substitution has no detectable effect on the catalytic activity of recombinant RNase H2, although it does cause a small reduction in substrate binding affinity (Coffin, et al., 2011). No other variants were identified from sequencing the coding exons and splice junctions of RNASEH2A (aside from a common SNP, c.128 -61A>T (rs55668927)). Likewise, no pathogenic mutations were identified on sequencing TREX1, RNASEH2B, RNASEH2C and SAMHD1 in these patients (see Supp. Materials and Methods).
Synonymous mutations, c.75C>T in patient P1 and c.69G>A in patients 3 and 4 were present in combination with one of the heterozygous missense mutations described above, while patient 2 was homozygous for the c.75C>T variant. In each case, the synonymous variants were inherited from one parent and segregated with the disease phenotype (Supp. Table S1). We therefore hypothesized that the synonymous variants were in fact pathogenic mutations. Notably, splice site predictions using the SplicePort (http://spliceport.cs.umd.edu/) (Dogan, et al., 2007) and NetGene2 (http://www.cbs.dtu.dk/) (Hebsgaard, et al., 1996) algorithms suggested that both the c.75C>T and c.69G>A variants created new donor splice sites within exon 1. Indeed the computational scores for the new donor splice sites were higher than those for the canonical donor site in intron 1 (SplicePort): 0.77 vs 0.10 for c.69G>A; and 0.89 vs 0.10 for c.75C>T. We therefore predicted that the newly created donor splice sites would be used in preference over the original site.
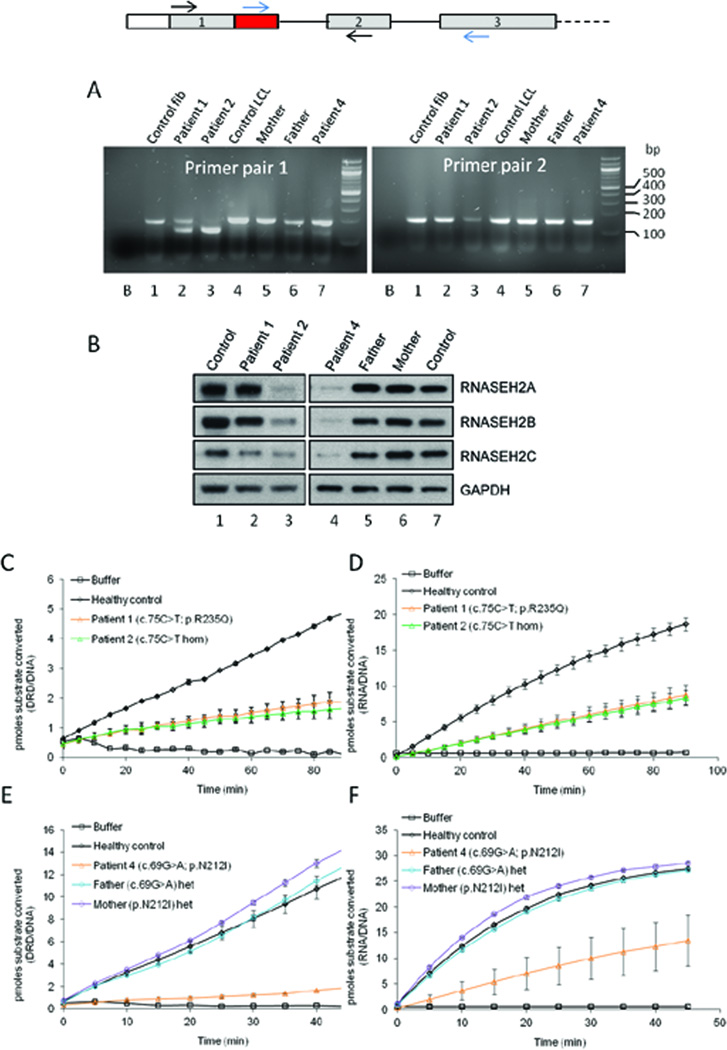
To examine this possibility further, RT-PCR of exons 1–2 of the RNASEH2A gene was performed on patient cell lines (Supp. Materials and Methods, Supp. Table S2). A smaller sized PCR product was observed in all the patients carrying heterozygous or homozygous (patient 2) synonymous mutations (Figure 1A, Supp. Table S3). The PCR product size was of a size expected from the use of these new splice sites.
Figure 1. The c.75C>T and c.69G>A synonymous variants result in internal deletion and premature truncation of RNASEH2A transcript respectively, causing a reduction in RNase H2 protein levels and enzyme activity.
A: RT-PCR confirms splicing defects in AGS patients with the c.75C>T and c.69G>A variants and demonstrates leaky splicing of the c.75C>T transcript. Schematic of exons 1 to 3 of the RNASEH2A gene, with red box highlighting the region of exon 1 that is excluded during pre-mRNA splicing as a result of the c.75C>T mutation. Primer pair 1 (RNASEH2A1F and RNASEH2AdelR2) is situated on either side of the skipped region (black arrows in schematic above); primer pair 2 (RNASEH2AdelFnew and RNASEH2AdelRnew) has the forward primer in the skipped region and the reverse primer in exon 3 (blue arrows in schematic above). Lane 1: healthy control fibroblasts; lane 2: patient 1 fibroblasts (c.75C>T + p.Arg235Gln); lane 3: patient 2 fibroblasts (c.75C>T hom); lane 4: healthy control lymphoblastoid cells; lane 5: patient 4 Mother lymphoblastoid cells (p.Asn212Ile het); lane 6: patient 4 Father lymphoblastoid cells (c.69G>A het); lane 7: patient 4 lymphoblastoid cells (c.69G>A + p.Asn212Ile). Additional lower bands in lanes 2, 6 and 7 with primer pair 1 are consistent with internal deletions due to aberrant splicing. The faint band in lane 3 with primer pair 2 is indicative of the presence of a small amount of full-length mRNA. ‘B’ on both gels represents the non-template control for each reaction. B: Reduced RNase H2 protein levels in AGS patient cells. RNASEH2A, B and C proteins were detected by immunoblotting of lysates from patient fibroblasts (left panel) and lymphoblastoid cells (right panel). Loading control, GAPDH. Lane 1: healthy control; lane 2: patient 1 (c.75C>T + p.Arg235Gln); lane 3: patient 2 (c.75C>T hom); lane 4: patient 4 (c.69G>A + p.Asn212Ile); lane 5: patient 4 Father (c.69G>A het); lane 6: patient 4 Mother (p.Asn212Ile het); lane 7: healthy control. C-F: Reduced RNase H2 activity in patient cells. Whole cell lysates from AGS patient fibroblasts (C, D) and lymphoblastoid cells (E, F) were used to determine RNase H type 2-specific activity (cleavage of double stranded DNA substrate with a single ribonucleotide in one of the strands (DRD/DNA); C and E) and overall RNase H enzyme activity (cleavage of RNA/DNA hybrid substrate; D and F). Lysates from healthy control cells were included for comparison. Data points and error bars shown represent averages and standard deviations respectively for six measurements from two independent experiments.
Sequencing of the RT-PCR product from patient 2 (homozygous for the c.75C>T mutation) confirmed the preferential usage of the new donor splice site, with loss of sequence corresponding to the end of exon 1 (Supp. Figure S2). The aberrant transcript has a 54 base pairs in-frame deletion, resulting in an 18 amino acids (residues 26 to 43) internal deletion in the RNASEH2A protein (Supp. Figures S1 and S2). A small peak corresponding to full-length cDNA was observed in patient 2 using fluorescent primers flanking the skipped 3’ end of exon 1 (Supp. Figure S3 panel C). When a primer within the 3’ end of exon 1 was used in conjunction with a reverse primer in exon 3, a small amount of full-length RNASEH2A transcript (Figure 1A, Supp. Figure S3 panel K) was also detected. These data were corroborated by expression analysis using quantitative RT-PCR (qPCR) in patient cells and peripheral whole blood (Supp. Figure S4).
We next determined the consequences of the c.75C>T variant on the RNase H2 enzyme. The variant RNASEH2A protein was co-expressed with the RNASEH2B and C subunits in E.coli, and the heterotrimeric complex was purified to determine the effect of the 18-amino acid deletion on in vitro activity (see Supp. Materials and Methods). The truncated RNASEH2A protein was expressed at levels similar to those seen for full length RNASEH2A, but with a noticeable difference in migration, consistent with the internal deletion (Supp. Figure S5). Although a mutant trimeric complex was formed, it exhibited no detectible catalytic activity on any of three different RNA/DNA hybrid substrates (Supp. Figure S6). The complete lack of catalytic activity is consistent with loss of key metal binding residues Asp34 and Glu35 in the mutant RNase H2 enzyme (Figiel, et al., 2010; Reijns, et al., 2011; Shaban, et al., 2010) (Supp. Figure S1B).
Finally, to determine the cellular effects of the c.75C>T variant, we analysed RNase H2 levels and enzyme activity in available patient cell lines. Proteins were separated by SDS-PAGE after cell lysis, allowing detection of all three RNase H2 subunits by western blot analysis (Supp. Materials and Methods). To determine levels of cellular RNase H2 activity, enzyme assays were performed using cell lysates and a fluorometric assay as previously described (Reijns, et al., 2011) (Supp. Materials and Methods). The levels of all three subunits were reduced in patient 2 (homozygous for the c.75C>T variant, lane 3 in Figure 1B), showing that ‘ex vivo’ this mutation also has consequences for RNase H2 complex stability. Normal electrophoretic mobility of RNASEH2A for this patient suggests that it is produced from the remaining full-length transcript. In contrast, RNase H2 protein levels were only modestly affected in patient P1, carrying the c.75C>T variant in combination with a p.Arg235Gln mutation (lane 2 in Figure 1B). Despite this, cellular enzyme activity in patient P1 was reduced to a level similar to that seen in patient 2, both for RNA/DNA and DRD/DNA substrates (Figure 1C and D). As enzyme activity was not completely lost in patient 2, sufficient full-length transcript must still be produced and translated to generate residual enzymatic activity.
The consequences of the c.69G>A variant on splicing and enzyme activity were similarly examined. Firstly, RT-PCR product from patient 4, heterozygous for c.69G>A and p.Asn212Ile, was cloned and inserts were sequenced, demonstrating the loss of the end of exon 1 (Supp. Figure S2). This results in an out of frame deletion and the insertion of 20 amino acids followed by a premature stop codon at amino acid 42. As would be predicted, patient 4 and his father both had two different sized RT-PCR products consistent with the presence of a full-length as well as a truncated transcript (Figure 1A lanes 6 and 7). In contrast, his mother (lane 5), who is heterozygous for the Asn212Ile mutation, had a single band equivalent in size to the control (lane 1). These data were corroborated by denaturing capillary electrophoresis of fluorescent RT-PCR products (Supp. Figure S3, Supp. Table S4), and by qPCR (Supp. Figure S4). Western blot analysis demonstrated a reduction of all three RNase H2 subunits in patient 4, to a fraction of that seen in control cells (lane 4 in Figure 1B). RNase H2 enzyme activity was strongly reduced in cell lysates from this patient, whilst enzyme activity in parental cells was similar to control levels (Figure 1 E and F).
RNase H2 is the major nuclear enzyme with the ability to degrade the RNA strand of RNA/DNA hybrids and to hydrolyse the 5’ phosphodiester bond of ribonucleotides embedded in a DNA duplex (Arudchandran, et al., 2000; Rydberg and Game, 2002). These activities may be enhanced by the binding of the PIP-box (PCNA-interacting-peptide) of RNASEH2B to PCNA (proliferating cell nuclear antigen) (Bubeck, et al., 2011). Mis-incorporation of single ribonucleotides into genomic DNA by DNA polymerases may occur frequently in eukaryotic cells (Nick McElhinny, et al., 2010), which, if not removed, likely contributes to genome instability (Reijns, et al., 2012). In addition, RNase H2 can cleave R-loops, a thermodynamically stable form of RNA/DNA hybrid formed during transcription (Chon, et al., 2013).
Of the three RNase H2 subunits, RNASEH2A contains the active site residues, whilst the B and C subunits are thought to serve as a platform for complex assembly and to modulate its interaction with other proteins. The crystal structures of the mouse (Shaban, et al., 2010) and human (Figiel, et al., 2010; Reijns, et al., 2011) RNase H2 complexes demonstrate that the RNASEH2B and C subunits adopt a tightly intertwined dimer, whereas the C-terminal extension of the catalytic RNASEH2A subunit interacts with this dimer and is essential to the formation of an active RNase H2 trimer.
In silico predictions suggested that the synonymous c.69G>A and c.75C>T variants might activate cryptic splice donor sites within exon 1 of RNASEH2A. Using RT-PCR and cDNA sequencing we confirmed that this is indeed the case. The c.75C>T variant causes the formation of a 5’ cryptic splice site in exon 1, resulting in a protein product missing amino acids 26 to 43. In vitro analysis demonstrated that this truncated protein is stable and allows formation of the trimeric complex, but that this is catalytically inactive. We were therefore surprised to identify an individual (patient 2) homozygous for the c.75C>T transition, since RNase H2 is apparently essential for mammalian development (Reijns, et al., 2012). We hypothesised that the splicing defect might be leaky, allowing a small amount of active protein to be formed. Indeed, RT-PCR analysis, qPCR and RNase H2 activity assays revealed the presence of some remaining RNase H2 activity in patient cells, suggesting that the c.75C>T variant is not a true null mutation.
Furthermore, we show that the c.69G>A mutation leads to an additional incorrectly spliced transcript with an out of frame deletion at the end of exon 1, and the consequent introduction of a premature stop codon. We observed reduced cellular RNase H2 protein levels and activity in a patient compound heterozygous for this variant. A reduction in the level of all three RNase H2 subunits is in agreement with a requirement for the three subunits in complex stability, and possibly suggests that excess RNASEH2B and RNASEH2C may be degraded in the absence of RNASEH2A.
Recently, a novel RNASEH2B splice site mutation (c.322-3C>G) was described in patients with AGS from the Faroe Islands (Ostergaard, et al., 2012). PCR analysis of cDNA from fibroblasts demonstrated skipping of exon 5 leading to an out-of-frame transcript. It is suggested that the relatively severe phenotype seen in this population isolate might be explained by homozygosity for the presumed null mutation. However, no protein or enzyme activity data were presented so that, as the authors state, the possibility of some residual normal product cannot be excluded.
AGS is a debilitating disorder, with the majority of affected subjects showing severe motor and cognitive impairment. Treatment of AGS is currently only symptomatic. As a result, for many families with an affected child, the possibility of prenatal diagnosis is important, and the identification of disease causing mutations in AGS families is of high clinical significance. Moreover, elucidation of the spectrum of disease causing mutations may help in understanding the biology of the enzymes involved. In this study, we identified two synonymous variants in RNASEH2A that result in loss of RNase H2 enzyme function and the consequent AGS phenotype. In our cohort of 265 genetically confirmed AGS pedigrees, four of thirteen families with RNASEH2A mutations harbour one of the two splicing variants described here. Clarification of the disease causing status of these variants opens up the possibility of carrier and prenatal testing in these families.
Supplementary Material
Acknowledgments
We would like to thank the families for their cooperation in the research presented here. YJC acknowledges the Manchester NIHR Biomedical Research Centre. The research leading to these results has received funding from the European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement number 241779 (YJC, GIR, HG, DG, MS, BA, GMAF). APJ, MAMR, AL and KRA are funded by an MRC Senior Clinical Fellowship and the Lister Institute for Preventative Medicine. FWP and SRC are supported by National Institutes of Health Grant GM069962 (FWP), by the Alliance for Lupus Research 179222 (FWP), and by National Institutes of Health Postdoctoral Fellowship Award F32GM095290 (SRC).
References
- Abdel-Salam GM, El-Kamah GY, Rice GI, El-Darouti M, Gornall H, Szynkiewicz M, Aymard F, Zaki MS, Abdel-Aleem AK, Lebon P, Crow YJ. Chilblains as a diagnostic sign of aicardi-goutieres syndrome. Neuropediatrics. 2010;41:18–23. doi: 10.1055/s-0030-1255059. [DOI] [PubMed] [Google Scholar]
- Aicardi J, Goutieres F. A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann Neurol. 1984;15:49–54. doi: 10.1002/ana.410150109. [DOI] [PubMed] [Google Scholar]
- Arudchandran A, Cerritelli S, Narimatsu S, Itaya M, Shin DY, Shimada Y, Crouch RJ. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells. 2000;5:789–802. doi: 10.1046/j.1365-2443.2000.00373.x. [DOI] [PubMed] [Google Scholar]
- Bubeck D, Reijns MA, Graham SC, Astell KR, Jones EY, Jackson AP. PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 2011;39:3652–3666. doi: 10.1093/nar/gkq980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H, Sparks JL, Rychlik M, Nowotny M, Burgers PM, Crouch RJ, Cerritelli SM. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 2013;41:3130–3143. doi: 10.1093/nar/gkt027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin SR, Hollis T, Perrino FW. Functional Consequences of the RNase H2A Subunit Mutations That Cause Aicardi-Goutieres Syndrome. J Biol Chem. 2011;286:16984–16991. doi: 10.1074/jbc.M111.228833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, Corry PC, Cowan FM, et al. Mutations in the gene encoding the 3'-5' DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006a;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006b;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- Dogan RI, Getoor L, Wilbur WJ, Mount SM. SplicePort--an interactive splice-site analysis tool. Nucleic Acids Res. 2007;35:W285–W291. doi: 10.1093/nar/gkm407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figiel M, Chon H, Cerritelli SM, Cybulska M, Crouch RJ, Nowotny M. The Structural and Biochemical Characterization of Human RNase H2 Complex Reveals the Molecular Basis for Substrate Recognition and Aicardi-Goutieres Syndrome Defects. J Biol Chem. 2010;286:10540–10550. doi: 10.1074/jbc.M110.181974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- Goutieres F, Aicardi J, Barth PG, Lebon P. Aicardi-Goutieres syndrome: an update and results of interferon-alpha studies. Ann Neurol. 1998;44:900–907. doi: 10.1002/ana.410440608. [DOI] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshinsky-Silver E, Malinger G, Ben-Sira L, Kidron D, Cohen S, Inbar S, Bezaleli T, Levine A, Vinkler C, Lev D, Lerman-Sagie T. A large homozygous deletion in the SAMHD1 gene causes atypical Aicardi-Goutieres syndrome associated with mtDNA deletions. Eur J Hum Genet. 2010;19:287–292. doi: 10.1038/ejhg.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard E, Joensen F, Sundberg K, Duno M, Hansen FJ, Batbayli M, Sorensen N, Born AP. A novel RNASEH2B splice site mutation responsible for Aicardi-Goutieres syndrome in the Faroe Islands. Acta Paediatr. 2012;101:e509–e513. doi: 10.1111/j.1651-2227.2012.02807.x. [DOI] [PubMed] [Google Scholar]
- Powell RD, Holland PJ, Hollis T, Perrino FW. Aicardi-Goutieres syndrome gene and HIV- 1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem. 2011;286:43596–43600. doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh V, Bernardi B, Stafa A, Garone C, Franzoni E, Abinun M, Mitchell P, Mitra D, Friswell M, Nelson J, Shalev SA, Rice GI, et al. Intracerebral large artery disease in Aicardi- Goutieres syndrome implicates SAMHD1 in vascular homeostasis. Dev Med Child Neurol. 2010;52:725–732. doi: 10.1111/j.1469-8749.2010.03727.x. [DOI] [PubMed] [Google Scholar]
- Reijns MA, Bubeck D, Gibson LC, Graham SC, Baillie GS, Jones EY, Jackson AP. The Structure of the Human RNase H2 Complex Defines Key Interaction Interfaces Relevant to Enzyme Function and Human Disease. J Biol Chem. 2011;286:10530–10539. doi: 10.1074/jbc.M110.177394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, et al. Enzymatic removal of ribonucleotides from DNA is essential for Mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, Baxter P, Benko WS, et al. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am J Hum Genet. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, et al. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, Jenkinson EM, Bacino CA, et al. Mutations in 16 ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat Genet. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Natl Acad Sci U S A. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaban NM, Harvey S, Perrino FW, Hollis T. The structure of the mammalian RNase H2 complex provides insight into RNA.NA hybrid processing to prevent immune dysfunction. J Biol Chem. 2010;285:3617–3624. doi: 10.1074/jbc.M109.059048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.