Abstract
Genetic interactions or epistasis could make a substantial contribution to variation in human complex traits including longevity. However, detecting epistatic interactions in high dimensional datasets is difficult due to various reasons including multiple testing of correlated tests. We introduce a novel permutation strategy to the case-only analysis of gene by gene interaction using multiple SNPs. The method is applied to genes coding for Forkhead box O transcription factors which recently have been associated with human longevity across different populations hypothesizing that epistatic interaction in the regulation and expression of the FOXO gene family could contribute to the human longevity phenotype. Genotype data was collected from 1088 individuals from the Danish 1905 birth cohort aged over 92/93 years with 12 SNPs in the FOXO1a and 15 SNPs in the FOXO3a genes. Our analysis detected a joint effect between rs9486902 in FOXO3a and rs2701858 in FOXO1a that highly significantly contributes to human longevity (OR=3.23, 95% CI: 2.93–3.53) which is consistent in both males and females. Our results were compared with published studies and importance of our novel method and findings discussed.
Keywords: case-only analysis, epistatic effect, permutation, longevity, FOXO genes
Introduction
Complex diseases or phenotypes like longevity may have multiple genetic and environmental causes. The complexity arises from the fact that many genetic and environmental factors may interact with each other such that the expression of the phenotype may not be accurately predictable based on knowledge of the individual effects for each of the component factors considered alone. Genetic interactions or epistasis could hold the key to understanding of complex conditions such as Alzheimer’s disease and diabetes (Moore 2005; Carlborg and Haley, 2004). Recently, epistatic interactions have been used to explain the “missing heritability” in genome-wide association studies (GWAS) (Zuk et al. 2012) which assume that genetic variants act in an additive and independent manner. However, detecting epistatic interactions in high dimensional datasets is difficult because of the computational complexity due to all possible combinations of genetic variations across loci and problem of multiple testing under high dependence. As one solution, a recently proposed approach employs biological knowledge, such as functional pathway, to narrow down the evaluation to gene combinations with biologically concise reasons (Ma et al. 2012).
The FOXO (Forkhead box O) transcription factors, characterized by a conserved DNA binding domain, are essential in both development and adult physiology. Members of the FOXO family are believed to be evolutionarily conserved post-translational mediators of insulin and growth factor signaling. In C. elegans, the FOXO orthologue DAF-16 had been shown to regulate life span. In humans, genetic variations in genes coding for FOXO1a and FOXO3a have been associated with longevity in different populations (Soerensen et al. 2010; Li et al. 2009; Willcox et al. 2008; Kojima et al. 2004; Bonafe et al. 2003). Recently, Zeng et al. (2010) found evidence of gene by gene and gene by environment interactions that affects longevity in a case-control study of middle aged Chinese and Chinese centenarians.
The case-only design is a powerful method for analyzing gene by gene interaction effects on longevity (Tan et al. 2002) provided that the two genetic variants are not in linkage disequilibrium (LD). The method is characterized by assessment of interaction effects without controls, an important feature in longevity studies as compared with the traditional case-control design (Tan et al. 2006) for which the longevity phenotype in the young controls are actually censored. Similar to all association analyses, current application of the case-only design encounters problem of multiple testing due to the popularity of the single nucleotide polymorphism (SNP) markers. The situation is further complicated by the correlated structure among SNPs typed in each of the interacting genes. Resampling-based methods have been applied in analyzing microarray gene expression data for p value adjustment accounting for multiplicity and dependence structure in the thousands of genes tested simultaneously in a microarray experiment using the case-control design (Dudoit et al. 2002). In the case-only analysis of gene-gene interactions (G×G), the popular phenotype-based permutation test for multiple comparisons is inapplicable as all samples are of the same phenotype, i.e. cases. This paper aims at first introducing a novel genotype-based permutation scheme to the case-only analysis of G×G using multiple SNPs genotyped in each of the interacting genes and second, applying the method to SNPs data on FOXO1a and FOXO3a genes in the Danish 1905 birth cohort who survived over 93 years of age.
Materials and Methods
Longevity cases
Our longevity cases consist of 1088 subjects who were born in 1905 and who were first interviewed in 1998 at ages 92 to 93 years. All cases were collected by the Danish 1905 Cohort Study, a study that covers all Danes born in 1905 (Nybo et al. 2001). Blood samples were taken at the first interview for DNA extraction. The Danish 1905 Cohort Study was approved by the Danish National Committee on Medical Research Ethics.
Genotyping
The procedures for DNA extraction and genotyping were the same as described by Soerensen et al. (2010). In brief, DNA was purified from dry blood spots using QIAamp DNA Mini and Micro Kits (Qiagen, Dusseldorf, Germany) and genotyped by the Illumina GoldenGate assay (Illumina Inc, San Diego, USA). Genotyping was done for 12 SNPs in the FOXO1a gene on chromosome 13 (rs10507486, rs12854161, rs12876443, rs2180961, rs2701858, rs2721068, rs2755209, rs2755213, rs2951787, rs2984121, rs4581585, and rs9603776) and 15 SNPs in the FOXO3a gene on chromosome 6 (rs10499051, rs12206094, rs12207868, rs12212067, rs13217795, rs13220810, rs2764264, rs2802292, rs3800231, rs3800232, rs479744, rs7762395, rs9398172, rs9400239, and rs9486902). SNPs for genotyping were selected to cover the majority of known common genetic variations in the two genes. Selection of chromosome regions and tagging SNPs was described in detail elsewhere (Soerensen et al. 2010).
The case-only analysis
The case-only analysis is an efficient design in comparison with the traditional case-control design for detecting gene-gene (Yang et al. 1999) and gene-environment (Khoury and Flanders, 1996; Piegorsch, et al., 1994) interactions under the assumption that the interacting factors are independent. The design requires far fewer study samples (Yang, et al., 1997) and avoids difficulties in selecting appropriate controls (Khoury and Flanders, 1996) with the trade-off that no main effects other than interaction can be assessed. By treating longevity individuals (such as centenarians or nonagenarians) as cases, Tan et al. (2002) introduced and validated the method for detecting gene-gene interaction in longevity studies. Table 1 is a typical contingency table for case-only analysis of genes M and N with 0 and 1 for non-carrying and carrying of the minor allele of a SNP. The individual counts in the four cells are for samples that are non-carriers (a00), carriers (a11) of both minor alleles, carriers of minor allele in gene M and major allele in gene N (a01), and carriers of minor allele in gene N and major allele in gene M (a10). In brief, the case-only design detects an interaction effect on longevity as an association between two interacting genes in longevity samples with an observed positive association suggesting synergistic interaction while an inverse association indicating antagonistic interaction (Ottman, 1996). The null hypothesis is thus that the genotype distribution of one gene is independent of the other gene for which a standard χ2 statistic can be calculated as to test the null hypothesis with one degree of freedom, where n1․, n0․, n․1, n․0 are marginal sum of genotype counts and n the overall sum. The odds ratio for the interaction effect can be calculated as which has been shown to measure departure from the multiplicative joint effect on longevity from the two genes (Tan et al. 2002). The standard error for the natural log of OR can be calculated as . A confidence interval for ln(OR) can then be constructed and converted to the original scale.
Table 1.
A contingency table for calculating the test statistic in a case-only study
| Gene M | ||||
|---|---|---|---|---|
| 0 | 1 | Sum | ||
| Gene N | 0 | a00 | a01 | n0․ |
| 1 | a10 | a11 | n1․ | |
| Sum | n․0 | n․1 | N | |
Correction for multiple testing
As mentioned before, the case-only design for assessing G×G using SNPs data creates multiplicity of correlated nature due to LD in multiple SNPs genotyped in each of the interacting genes. Resampling-based methods have designed for correction of multiple comparisons in the situation of correlated test statistics (or p values). For example, the maxT and the minP approaches introduced by Westfall & Young (1993). These methods have been applied in analyzing genome-wide microarray gene expression data (Dudoit et al. 2002). The maxT method is also implemented in the popular Plink package for genetic association analysis (http://pngu.mgh.harvard.edu/~purcell/plink/). Given the special situation of case-only analysis, we need to introduce a novel permutation strategy in order to make use of the resampling-based methods for dealing with multiple testing.
Permutation strategy
In a traditional case-control study, a permutation test can be performed by shuffling the phenotype, i.e. randomly assign phenotype labels to the samples. The procedure produces a null distribution of the test statistic which can be used for obtaining an empirical significance level for the statistic of the original data.
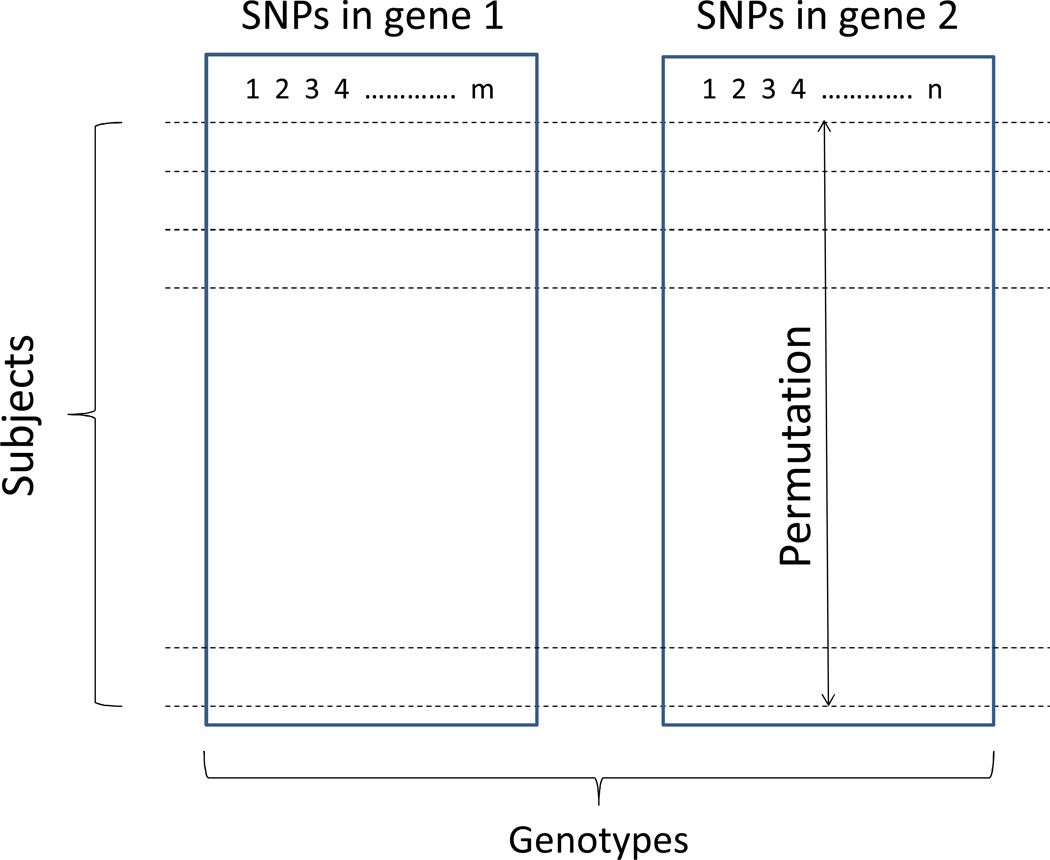
In the case-only design, phenotype-based permutation is inapplicable because all samples are of the same phenotype, i.e. cases. Instead of permuting the phenotypes, we introduce a genotype-based permutation procedure for assessing statistical significance of the test statistics and for correction of multiple testing due to multiple SNPs typed in the interacting genes. In Figure 1, we illustrate the procedure with two interacting genes typed for m SNPs in gene 1 and n SNPs in gene 2. The permutation is conducted by shuffling the genotypes among all subjects for one of the two interacting genes (for example, gene 2 in Figure 1). The idea is, through permutation, to destroy the genotype dependence between the two genes due to gene-gene interaction in each of the cases. The permuted samples are random samples in term of the epistatic interaction which can be used to generate the distribution of the χ2 statistic calculated from the case-only analysis of each random sample upon a large number of iterations. Note that, the χ2 statistics from all pairs of interacting SNPs are uniformly distributed with one degree of freedom, a situation that meets the assumption of “subset pivotality” in resampling-based multiple testing (Westfall & Troendle, 2008).
Figure 1.
A diagram showing the genotype-based permutation test for case-only design for assessing interaction between two genes typed for multiple SNPs (m in gene 1 and n in gene 2). Permutation can be performed on genotypes of one of the two genes (here exemplified on gene 2) or both genes.
P value correction
In order to correct for multiple testing, we adopt the step-down minP procedure introduced by Westfall & Young (1993) which consists of the following steps:
Order the raw p values from the original data for the m×n pairs of interacting SNPs: pr1 ≤ pr2 ≤ ⋯ ≤ prm×n
Permuting the genotypes as described above.
Compute m×n p values from the χ2 test applied to the bth permuted replicate.
Compute the successive minima of p values for the bth replicate, qi,b, with qm×n,b = prm×n,b, qi,b = min(qi+1,b, pri,b) for i = m × n − 1,⋯,1.
- Repeat 2–4 B times, and estimate the adjusted p values as
Here, I(·) is an indicator of value 1 for true and 0 for false conditions. Constraints on monotonicity is obtained by setting
Results
The combination of 12 SNPs in FOXO1a and 15 SNPs in FOXO3a genes created 180 pairs of interacting SNPs. The above described case-only analysis was applied to each combination in males and females separately and in the whole sample of cases as well. In order to correct for multiple testing, we performed the genotype-based permutation test to each of the three analyses with B=10,000 iterations. After adjustment, only one pair of interacting SNPs (rs2701858 in FOXO1a and rs9486902 in FOXO3a) remained significant (permutation p-value<0.0001, OR=3.23, 95% CI: 2.93–3.53). The same analysis was applied to males and females separately and interestingly both showed the same pair of SNPs as the only significant interacting SNPs (permutation p-value<0.0001, OR=4.42, 95% CI: 3.85–5 in males; permutation p-value<0.0001, OR=2.83, 95% CI: 2.47–3.18 in females).
Table 2 is a contingency table for the frequency counts by genotype for the two interacting SNPs in male, female and the whole samples. A total of 49 individuals were dropped due to missing genotypes in either of the two SNPs. Based on Table 2, we calculated frequencies of carriers of minor alleles of one SNP conditional on genotypes of the other SNP for males (Figure 1A, 1B), females (Figure 1C, 1D) and the whole samples (Figure 1E, 1F) together with 95% CIs. As shown by the figures, frequencies of the minor allele in one SNP is significantly higher in carriers of the minor allele than in homozygotes for the common allele of the other SNP suggesting high dependency between genotypes of the two SNPs as a result of interaction.
Table 2.
Sample counts by genotypes (0 for non-carriers and 1 for carriers of the minor alleles) at SNPs rs9486902 in FOXO3 and rs2701858 in FOXO1 genes and case-only test results.
| Minor allele rs2701858 |
Minor allele rs9486902 | ||
|---|---|---|---|
| 0 | 1 | Sum | |
| Males | |||
| 0 | 145 (0.54) | 73 (0.27) | 218 (0.81) |
| 1 | 22 (0.08) | 49 (0.11) | 71 (0.19) |
| Sum | 167 (0.62) | 122 (0.38) | 289 |
| Case-only test | χ2=26.28, p=2.96e-7, OR=4.42, 95% CI: 3.85–5.00 | ||
| Females | |||
| 0 | 413 (0.57) | 176 (0.24) | 589 (0.81) |
| 1 | 73 (0.09) | 88 (0.10) | 161 (0.19) |
| Sum | 486 (0.66) | 264 (0.34) | 750 |
| Case-only test | χ2=32.95, p=9.44e-9, OR=2.83, 95% CI: 2.47–3.18 | ||
| Sex combined | |||
| 0 | 558 (0.56) | 249 (0.25) | 807 (0.81) |
| 1 | 95 (0.08) | 137 (0.11) | 232 (0.19) |
| Sum | 653 (0.64) | 386 (0.36) | 1039 |
| Case-only test | χ2=60.16, p=8.76e-15, OR=3.23, 95% CI: 2.93–3.53 | ||
Discussion
We have introduced a novel resampling-based approach to enable case-only analysis of gene-gene interactions using multiple SNPs data. Application of our method to genotype data on FOXO genes in the Danish 1905 birth cohort surviving over ages 92 and 93 detected highly significant interaction effect by one pair of SNPs (rs9486902 in FOXO3a and rs2701858 in FOXO1a) that is beneficial for longevity in both males and females. Our application of the method is characterized by a novel genotype permutation strategy that enables resampling-based case-only analysis of high dimensional SNPs data for their interaction effects to handle multiple testing with correlated structure due to LD.
Detection of epistatic effects in the current genetic association studies is a challenge in genetic epidemiology due to issues created by the high dimensional analysis (Carlborg and Haley, 2004). Proper methods for dealing with multiple testing are demanding because of dependency of the tests, and conservative methods such as Bonferroni correction can only detect large interaction effects but will ignore subtle epistasis. Through example application to the FOXO genes, we have shown that our novel genotype-based permutation strategy can be applied to handle multiple comparisons in correlated tests on epistatic interaction between two genes each with multiple typed SNPs. Besides longevity, the strategy can be applied to case-only analysis of gene by gene interaction on disease phenotypes as well provided that the main interest is epistatic effects.
Soerensen et al. (2010) performed an association analysis of FOXO3a gene variants with human longevity using a case-control design with the same samples from the Danish 1905 birth cohort as used in the present study, and middle aged Danes as controls. The study identified SNP rs9486902 as benefiting longevity in a recessive model with higher statistical significance in males. Taking the same case-control data as in their study, we fitted a logistic regression model to the genotype data of SNPs rs2701858 in FOXO1a and rs9486902 in FOXO3a with an interaction term assuming dominant effects similar to that in our case-only analysis. The results only showed a significant interaction effect in male samples (OR=2.36, 95% CI: 1.04–5.35, p=0.04). Compared with our result for males, the interaction effect, although in the same direction, is relatively underestimated and the confidence interval is obviously large. This phenomenon exemplifies, in empirical data, the high efficiency of the case-only model in detecting interaction as compared with the traditional case-control design.
Although useful, the case-only model is unable to handle interactions between genes each with an additive mode of inheritance. Moreover, the permutation test can be computer intensive when applied to very high dimensional data such as the GWAS data. We emphasize that our permutation procedure is more suitable to case-only analysis of gene-gene interaction in candidate gene studies using multiple SNP markers. Recently, Pierce and Ahsan (2010) introduced a case-only genome-wide interaction analysis and suggested a screening procedure that searches the genome for variants that interact with a candidate polymorphism. Although interesting, the selection of candidates and managing the potential interactions can still be challenging issues. More work need to be done to introduce the powerful case-only model to genome-wide studies.
Zeng et al. (2010) reported epistatic effects on longevity in the FOXO gene family (SNPs rs2755209 and rs2755213 in FOXO1a; SNPs rs2802292 and rs2253310 in FOXO3a) in a Chinese study. Although their risk estimates did not reach statistical significance, consistent joint effects for carriers of the minor alleles of the two genes were found that increase chance of survival from middle age to over 100 years. Of the three SNPs also covered in our Danish data (rs2755209, rs2755213 in FOXO1a and rs2802292 in FOXO3a), neither significant interactions nor a consistent trend were found. Nevertheless, our two SNPs not covered in the Chinese study did exhibit a joint effect in favour of longevity in carriers of their minor alleles as indicated by the ORs in our case-only analysis. More replication studies are needed in order to establish the epistatic interactions on human longevity in the FOXO gene family.
Figure 2.
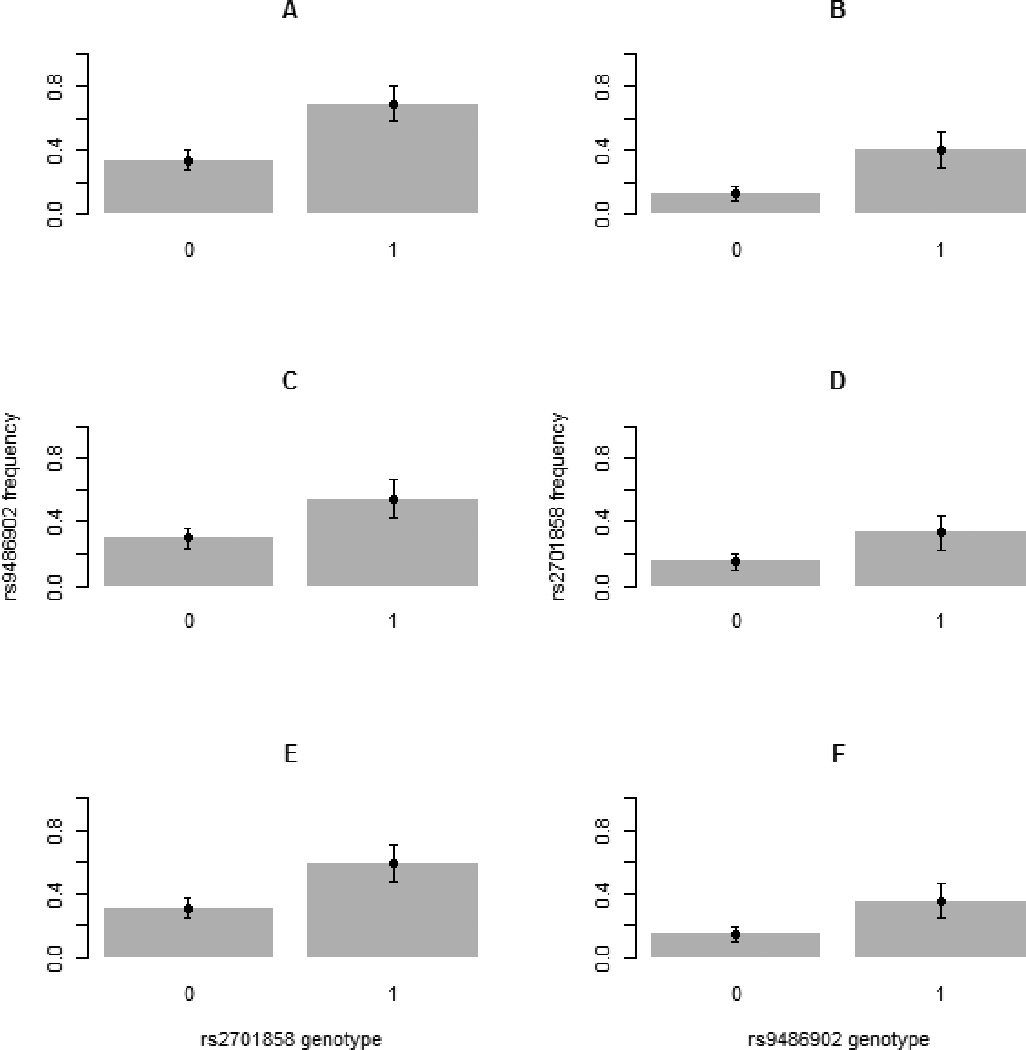
Frequencies of carriers of minor alleles of one SNP conditional on genotypes of the other SNP for males (Figure 1A, 1B), females (Figure 1C, 1D) and the whole samples (Figure 1E, 1F) together with 95% CIs. Frequencies of the minor allele carriers of one SNP is significantly higher in carriers of the minor allele than in homozygotes for the common allele of the other SNP indicting high correlation between genotypes of the two SNPs resulted from the interaction between FOXO1a and FOXO3a genes.
Acknowledgements
This work was partially supported by the EU Seventh Framework Programme (FP7/2007-2011) under grant agreement n° 259679 and NIH/NIA grant P01 AG08761.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Bonafè M, Barbieri M, Marchegiani F, Olivieri F, Ragno E, Giampieri C, Mugianesi E, Centurelli M, Franceschi C, Paolisso G. Polymorphic variants of insulin-like growth factor I (IGF-I) receptor and phosphoinositide 3-kinase genes affect IGF-I plasma levels and human longevity: cues for an evolutionarily conserved mechanism of life span control. J Clin Endocrinol Metab. 2003;88:3299–3304. doi: 10.1210/jc.2002-021810. [DOI] [PubMed] [Google Scholar]
- Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nat Rev Genet. 2004;5:618–625. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Yang YH, Callow MJ, Speed TP. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statistica Sinica. 2002;12:111–139. [Google Scholar]
- Khoury MJ, Flanders WD. Nontraditional epidemiologic approaches in the analysis of gene-environment interaction: case-control studies with no controls! Am J Epidemiol. 1996;144:207–213. doi: 10.1093/oxfordjournals.aje.a008915. [DOI] [PubMed] [Google Scholar]
- Kojima T, Kamei H, Aizu T, Arai Y, Takayama M, Nakazawa S, Ebihara Y, Inagaki H, Masui Y, Gondo Y, Sakaki Y, Hirose N. Association analysis between longevity in the Japanese population and polymorphic variants of genes involved in insulin and insulin-like growth factor 1 signaling pathways. Exp Gerontol. 2004;39:1595–1598. doi: 10.1016/j.exger.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang WJ, Cao H, Lu J, Wu C, Hu FY, Guo J, Zhao L, Yang F, Zhang YX, Li W, Zheng GY, Cui H, Chen X, Zhu Z, He H, Dong B, Mo X, Zeng Y, Tian XL. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18:4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Brautbar A, Boerwinkle E, Sing CF, Clark AG, Keinan A. Knowledge-driven analysis identifies a gene-gene interaction affecting high-density lipoprotein cholesterol levels in multi-ethnic populations. PLoS Genet. 2012;8:e1002714. doi: 10.1371/journal.pgen.1002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH. A global view of epistasis. Nature Genetics. 2005;37:13–14. doi: 10.1038/ng0105-13. [DOI] [PubMed] [Google Scholar]
- Nybo H, Gaist D, Jeune B, Bathum L, McGue M, Vaupel JW, Christensen K. The Danish 1905 cohort: a genetic-epidemiological nationwide survey. J Aging Health. 2001;13:32–46. doi: 10.1177/089826430101300102. [DOI] [PubMed] [Google Scholar]
- Ottman R. Gene-environment interaction: definitions and study designs. Prev Med. 1996;25:764–770. doi: 10.1006/pmed.1996.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piegorsch WW. Statistical models for genetic susceptibility in toxicological and epidemiological investigations. Environ Health Perspect. 1994;102:77–82. doi: 10.1289/ehp.94102s177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BL, Ahsan H. Case-only genome-wide interaction study of disease risk, prognosis and treatment. Genet Epidemiol. 2010;34:7–15. doi: 10.1002/gepi.20427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soerensen M, Dato S, Christensen K, McGue M, Stevnsner T, Bohr VA, Christiansen L. Replication of an association of variation in the FOXO3A gene with human longevity using both case-control and longitudinal data. Aging Cell. 2010;9:1010–1017. doi: 10.1111/j.1474-9726.2010.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, De Benedictis G, Ukraintseva SV, Franceschi C, Vaupel JW, Yashin AI. A centenarian-only approach for assessing gene-gene interaction in human longevity. Eur J Hum Genet. 2002;10:119–124. doi: 10.1038/sj.ejhg.5200770. [DOI] [PubMed] [Google Scholar]
- Tan Q, Kruse TA, Christensen K. Design and analysis in genetic studies of human ageing and longevity. Ageing Res Rev. 2006;5:371–387. doi: 10.1016/j.arr.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Westfall PH, Troendle JF. Multiple testing with minimal assumptions. Biometrical Journal. 2008;5:745–755. doi: 10.1002/bimj.200710456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PH, Young SS. Resampling-based Multiple Testing: Examples and methods for p-value adjustment. John Wiley & Sons; 1993. [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Khoury MJ, Sun F, Flanders WD. Case-only design to measure gene-gene interaction. Epidemiology. 1999;10:167–170. [PubMed] [Google Scholar]
- Yang Q, Khoury MJ, Flanders WD. Sample size requirements in case-only designs to detect gene-environment interaction. Am J Epidemiol. 1997;146:713–720. doi: 10.1093/oxfordjournals.aje.a009346. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Cheng L, Chen H, Cao H, Hauser ER, Liu Y, Xiao Z, Tan Q, Tian XL, Vaupel JW. Effects of FOXO genotypes on longevity: a biodemographic analysis. J Gerontol A Biol Sci Med Sci. 2010;65:1285–1299. doi: 10.1093/gerona/glq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]