Abstract
Burkholderia sp. C3, an efficient polycyclic aromatic hydrocarbon (PAH) degrader, can utilize 9 of the 10 N-methylcarbamate insecticides including carbaryl as a sole source of carbon. Rapid hydrolysis of carbaryl in C3 is followed by slow catabolism of the resulting 1-naphthol. This study focused on metabolomes and proteomes in C3 cells utilizing carbaryl in comparison to those using glucose or nutrient broth. Sixty of the 867 detected proteins were involved in primary metabolism, adaptive sensing and regulation, transport, stress response, and detoxification. Among the 41 proteins expressed in response to carbaryl were formate dehydrogenase, aldehyde-alcohol dehydrogenase and ethanolamine utilization protein involved in one carbon metabolism. Acetate kinase and phasin were 2 of the 19 proteins that were not detected in carbaryl-supported C3 cells, but detected in glucose-supported C3 cells. Down-production of phasin and polyhydroxyalkanoates in carbaryl-supported C3 cells suggests insufficient carbon sources and lower levels of primary metabolites to maintain an ordinary level of metabolism. Differential metabolomes (approximately 196 identified polar metabolites) showed up-production of metabolites in pentose phosphate pathways and metabolisms of cysteine, cystine and some other amino acids, disaccharides and nicotinate, in contract to down-production of most of the other amino acids and hexoses. The proteomic and metabolomic analyses showed that carbaryl-supported C3 cells experienced strong toxic effects, oxidative stresses, DNA/RNA damages and carbon nutrient deficiency.
Keywords: Proteomics, Metabolomics, Biodegradation, Bioremediation, Pesticide, Catabolism
Introduction
N-Methylcarbamate insecticides (N-methylcarbamates) are a class of the most widely used insecticides in agricultural fields, gardens and households. They inhibit acetylcholine esterase catalyzing rapid hydrolysis of the neurotransmitter acetylcholine in cholinergic synapses preventing overstimulation of the post-synaptic membrane (Wilson 2010). N-Methylcarbamates generally undergo biological or chemical cleavage of the carbamyl ester followed by further degradation of the resulting phenolic metabolites. Various microorganisms capable of degrading N-methylcarbamates have been isolated from soil (Topp et al. 1993; Qing et al. 2003; Swetha and Phale 2005). Being similar to the other N-methylcarbamates, carbaryl is hydrolyzed into 1-naphthol, methylamine and CO2 (Topp et al. 1993). Carbaryl hydrolase is characterized from several bacterial species (Singh et al. 1999; Hashimoto et al. 2002; Swetha and Phale 2005). 1-Naphthol is metabolized via salicylate to either gentisate or catechol (Larkin and Day 1986).
Burkholderia sp. C3 was recently isolated from petroleum-contaminated soil in Hawaii (Seo et al. 2007a). The strain C3 can rapidly degrade polycyclic aromatic hydrocarbons (PAHs) via initial 1,2- and 3,4-dioxygenations and meta- and ortho-cleavages of naphthalene-1,2-diol (Seo et al. 2007b; Tittabutr et al. 2011). It is known that substituted 1-naphthols and salicylate are common metabolites of PAHs. The strain C3, therefore, may degrade carbaryl into 1-naphthol and then follow the PAH catabolism pathways for 1-naphthol degradation. Therefore, 10 N-methylcarbamates were tested for biodegradation by the strain C3, while carbaryl was selected for detailed catabolic studies in the present work.
Rapid accumulation of genomic data provides basic information for studying proteins involved in biochemical metabolic pathways. Currently, genomes of more than 200 microorganisms have been fully sequenced, including xenobiotics-degrading bacteria (e.g., Arthrobacter aurescens TC1, Burkholderia xenovorans LB400, several Mycobacterium species, Novosphingobium aromaticivorans DSM1244 and Sphingomonas wittichii RW1). Proteomes of these bacteria have also been studied (Denef et al. 2004 and 2005; Ishii et al. 2007; Kweon et al. 2007). Studies via polyomic approaches offer promise in providing comprehensive overview of the bacterial catabolism of pesticides. Proteomes and metabolomes are dynamic and respond to xenobiotics exposure (Kweon et al. 2007; Lee et al. 2007; Keum et al. 2008 and 2010; Qi and Li 2010). Most noticeable differential proteomes include stress-related proteins alleviating the toxic effects of xenobiotics and metabolites on the host cells. Genomic and proteomic responses to benzoate were studied for Burkholderia xenovorans LB400 (Denef et al. 2004). Limited studies, however, have been reported on comparative proteomes and metabolomes.
Phn and Nag-like dioxygenases metabolize PAHs in Burkholderia sp. C3 (Tittabutr et al. 2011). Both phn and nag-like genes in C3 were cloned and identified. The two types of dioxygenases degrade naphthalene, phenanthrene and dibenzothiophene in varying efficiencies. Phenanthrene is metabolized to 1-hydroxy-2-naphthanoic acid and 2-hydroxy-1-naphthanoic acid via 3,4- and 1,2-dioxygenation, respectively. The dioxygenases encoded by phn genes play a major role in 1,2-/3,4-dioxygenation and 3,4-dioxygenation dominates, whereas the dioxygenases encoded by nag-like genes mainly govern the lower catabolic pathways in C3. Understanding of metabolic pathways and enzyme involvements may lead to develop better bioremediation technologies for cleaning up PAHs contaminated sites.
The objectives of this study were to determine the degradation ability of Burkholderia sp. C3 to ten N-methylcarbamate insecticides, metabolome and proteome in C3 cells responding to carbaryl as a substrate in comparison with glucose and nutrient broth, and study degradation pathways of carbaryl. The experiments were designed to understand comprehensive networks of proteins and metabolites in the C3 cells.
Materials and Methods
Chemicals
Authentic standards (purity 96–99%) of the N-methylcarbamate insecticides aminocarb, bendiocarb, bufencarb, carbaryl, carbofuran, methiocarb, mexacarbate, pirimicarb, propoxur, and xylylcarb were obtained from the US EPA. Methyl 2-methoxydroxybenzal pyruvate was previously prepared (Keum et al. 2005). Phenolic metabolites were prepared via alkaline hydrolysis from the N-methylcarbamates and purified with thin layer chromatography. Sodium pyruvate, methoxylamine hydrochloride, pyridine, N,O-bis(trimethylsilyl) trifluoroacetamide with trimethylchlorosilane (BSTFA-TMCS), and poly(3-hydroxybutyric acid-co-3-hydroxyvaleric acid) (PHA) were obtained from Aldrich (Milwaukee, WI, USA). Standards of the primary metabolites were obtained from Aldrich or TCI (Tokyo, Japan). Methanol and other solvents were high performance liquid chromatography (HPLC) grade or higher.
Growth of bacterium and extraction of N-methylcarbamates and their metabolites
Burkholderia sp. C3 was cultured in 15 ml of minimal salt medium (MSM) (Bastiaens et al. 2000) supplemented with an N-methylcarbamate (50 mg/l) with shaking (120 rpm/min) at 28 °C for 5 days. After centrifugation of the culture medium (6000 g, 20 min), the supernatant was extracted with ethyl acetate (20 ml × 2). After removal of the organic solvent, the residue was reconstituted in methanol (1 ml) and analyzed on a Hewlett Packard (HP) 1100 series high performance liquid chromatograph (Agilent Korea Ltd., Korea) that was equipped with a Luna 18 column (20 cm × 4.6 mm i.d., 5 µm; Phenomenex, Torrance, CA, USA). Mobile phase was 50% aqueous acetonitrile (ACN) solution set at a flow rate of 1 ml/min. Elution profile was monitored at 210 nm. A control sample was prepared with heat-sterilized cells through the same procedures. For analyses of metabolomes and proteomes, the strain C3 was grown in glucose-(100 mg/l, control) or carbaryl- (50 mg/l) supplemented MSM (1.0 l) or nutrient broth (NB) at 28 °C under constant shaking (120 rpm/min). After 5 days incubation at 28 °C, cells were harvested by centrifugation (6000 g, 20 min).
Neutral and acidic metabolites of carbaryl were extracted and, respectively, derivatized with n-butylboronic acid diazomethane according to the procedure previously published (Keum et al. 2006; Seo et al. 2011). The derivatized metabolites of the two fractions were analyzed on a Varian QP-5000 GC Saturn-2000 mass spectrometer system (GC-MS) (Varian, Palo Alto, CA, USA) as previously reported (Keum et al. 2006; Seo et al. 2011).
Protein extraction, one dimensional SDS polyacrylamide gel electrophoresis (1D-SDS PAGE) and protein in-gel digestion
Cells were harvested and proteins were extracted in triplicate at 0 °C according to the procedures previously described (Tittabutr et al. 2011). Protein content was measured with Coomassie Plus™ protein assay (Pierce, IL, USA). 1D-SDS PAGE, protein in-gel digestion and alkylation of cysteine with iodoacetamide were performed as previously described (Tittabutr et al. 2011). The digested peptides were extracted from each gel slice twice with 30 µl of water/ACN/trifluoroacetic acid (TFA) (93:5:2, v/v/v) by sonication for 10 min in an ice bath. Both extracts were combined for LC-MS/MS analysis.
LC-MS/MS analyses of digested peptides
The digested peptides were analyzed in triplicate with an Ultimate™ nano-electrospray LC system interfaced to an esquireHCTultra ion trap mass spectrometer (Bruker Daltonics, Billerica, MA, USA) (Lee et al. 2007; Tittabutr et al. 2011). MS/MS spectra were interpreted with Mascot (Matrix Science, London, UK) via Biotools 2.2 software (Bruker); and peptide mass fingerprint (PMF) searches were performed with the Swiss-Prot and MSDB databases through the Mascot server. Peptides were assumed to be monoisotopic, oxidized at methionine residues and carbamidomethylated at cysteine residues. Up to one missed trypsin cleavage was allowed, although matches that contained any missed cleavages were not noticed. Mass tolerance was set at 1.0 Da. Probability-based molecular weight search (MOWSE) scores were estimated by comparison of search results against estimated random match populations and were reported as: 10 × log10(p), where p is the absolute probability. Scores in Mascot greater than the MOWSE score at p = 0.05 were considered statistically significant, meaning that the probability of the match being a random event is lower than 0.05. The false-positive rate (FPR) was estimated (Elias et al. 2005) to be smaller than 2% [FPR = FP/(FP+TP), where FP is the number of FPR hits; TP is the number of true-positive hits]. Only proteins identified with at least two peptide hits (p ≤ 0.0025) in triplicate analyses, with each peptide containing two tryptic termini, were accepted. The MS/MS spectra of all positively identified peptides were also manually inspected for data accuracy.
Protein profiles of the treatment samples were compared with those of the appropriate control samples. A protein detected in the treatment sample but not in the control is referred to as up-expression/production of the protein, whereas a protein undetected in the treatment sample but detected in the control is referred to as down-expression/production of the protein.
Extraction and derivatization of intrinsic metabolites and GC-MS analyses
The cell pellet was washed with distilled water. Bacterial cells (200 mg) were suspended in 70% methanol (15 ml) and disrupted by sonication (550 Sonic Dismembrator, Fisher Scientific; 1 min pulse and 2 min cooling, 8 cycles). Cellular debris was removed by centrifugation (15000 g, 10 min) and the supernatant was dried under vacuum. The residue was re-suspended in 500 µl of dry pyridine, derivatized with 100 µl of methoxylamine (100 mg methoxylamine hydrochloride in 10 ml pyridine) for 1 h, and subsequently with 200 µl of BSTFA-TMCS for 2 h at 80 °C. Profiles of medium to long chain fatty acids and PHA content were analyzed according to the procedure previously described (Keum et al. 2008).
Metabolite profiles were analyzed on a Shimadzu GCMS QP-2010 system equipped with DB-1 column (60 m, 0.25 µm film thickness, 0.2 mm i.d.). Helium was a carrier gas at a flow rate of 1 ml/min. The column temperature for metabolites and PHA analyses was started at 95 °C for 10 min, raised to 295 °C at a rate of 2 °C /min and then held for 20 min. For fatty acid methyl ester analysis, the column temperature was started at 50 °C for 10 min, raised to 280 °C at a rate of 2.5 °C /min and then held for 20 min. Injector and interface temperatures were 270 and 280 °C, respectively. The mass spectrometer was operated at electron impact (EI) mode at 70 eV.
Identification of intrinsic metabolites and statistical analyses of the data
Chromatographic data from GC-MS were converted to netCDF files, deconvluted to single components with AMDIS software (http://chemdata.nist.gov), and aligned with SpectConnect (http://spectconnect.mit.edu). Chemical identities of each peak and peak areas were determined via GCMS Solution software with implemented Wiley mass spectral library (Dongil-Shimadzu Co., Korea). Prior to the statistical analyses, the peak area of each metabolite was normalized with an average total peak value of overall sample set. Descriptive statistics, principal component analysis (PCA) and the other statistics were performed with SAS package (SAS Korea, Korea) or SIMCA-P (Umetrics, Sweden). Notable, but not identified peaks are reported as unknowns. To visualize the overall changes of metabolomes, up- and down-productions of representative metabolites were located in common primary metabolic maps with KEGG atlas (Kyoto Encyclopedia of Genes and Genomes) (Fig. S1A–C).
Results
Catabolism of N-methylcarbamate insecticides
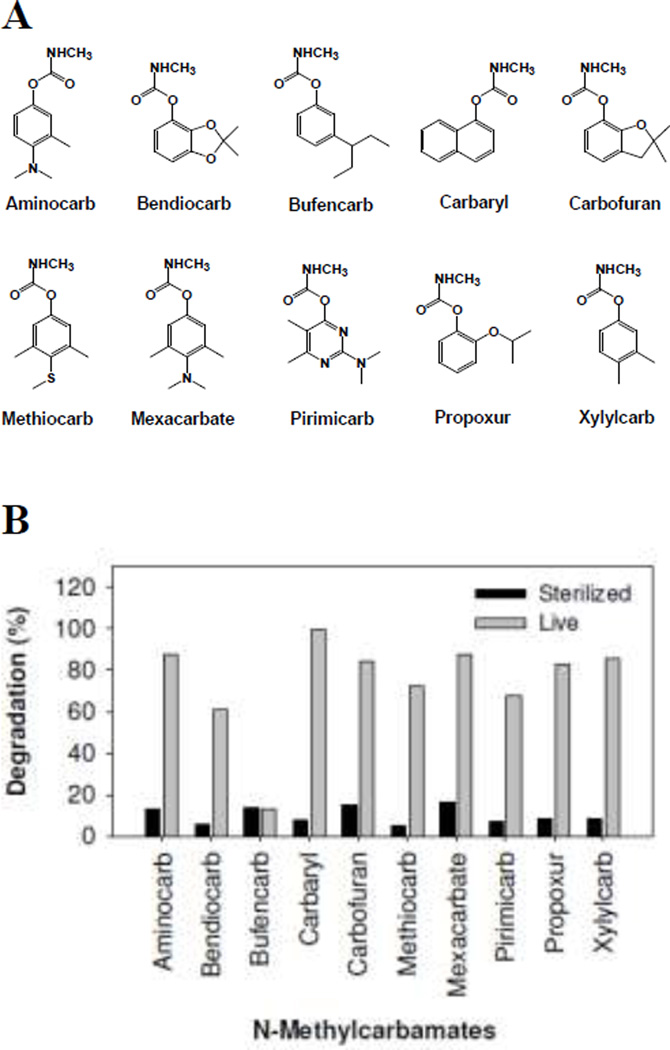
The strain C3 can utilize 9 out of the 10 N-methylcarbamates (Fig. 1). Approximately 60–100% of the insecticides were degraded within 5 days of C3 cultivation, while 7–15% was counted for abiotic degradation. In the C3 culture media, the most abundant metabolites were from hydrolytic cleavage of N-methylcarbamyl esters (Table S1. The common metabolite was N-methylcarbamic acid, which can be the nitrogen and carbon sources for bacterial growth. Low extent demethylation of the N,N-dimethylamino substituent was also observed from aminocarb and mexacarbate in the C3 cultures. Therefore, carbaryl was selected for further catabolic, proteomic and metabolic studies.
Fig. 1.
Degradation percentages (B) of N-methylcarbamate insecticides by Burkholderia sp. C3 after 5 days of incubation at an initial concentration of 50 mg/l. Data were averages of two independent experiments.
Catabolism of carbaryl
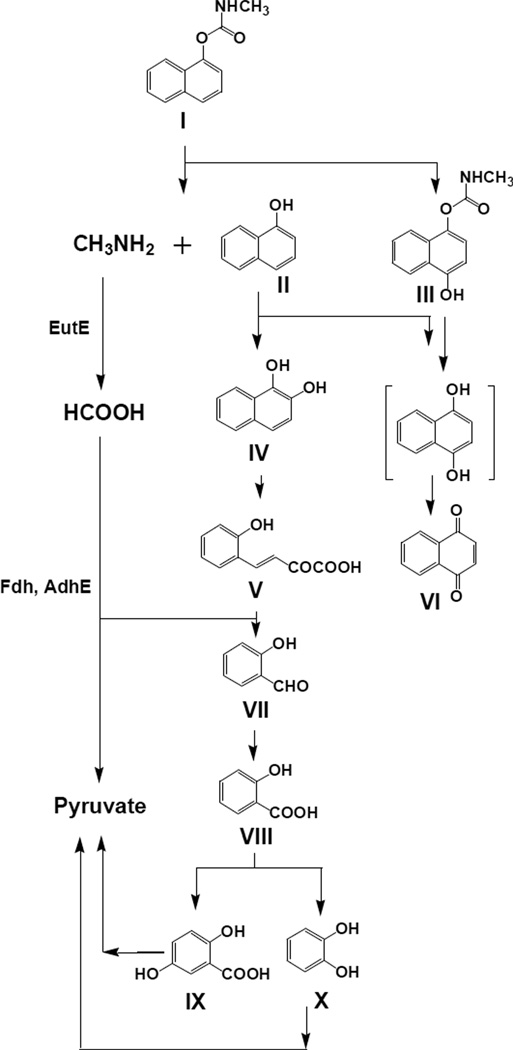
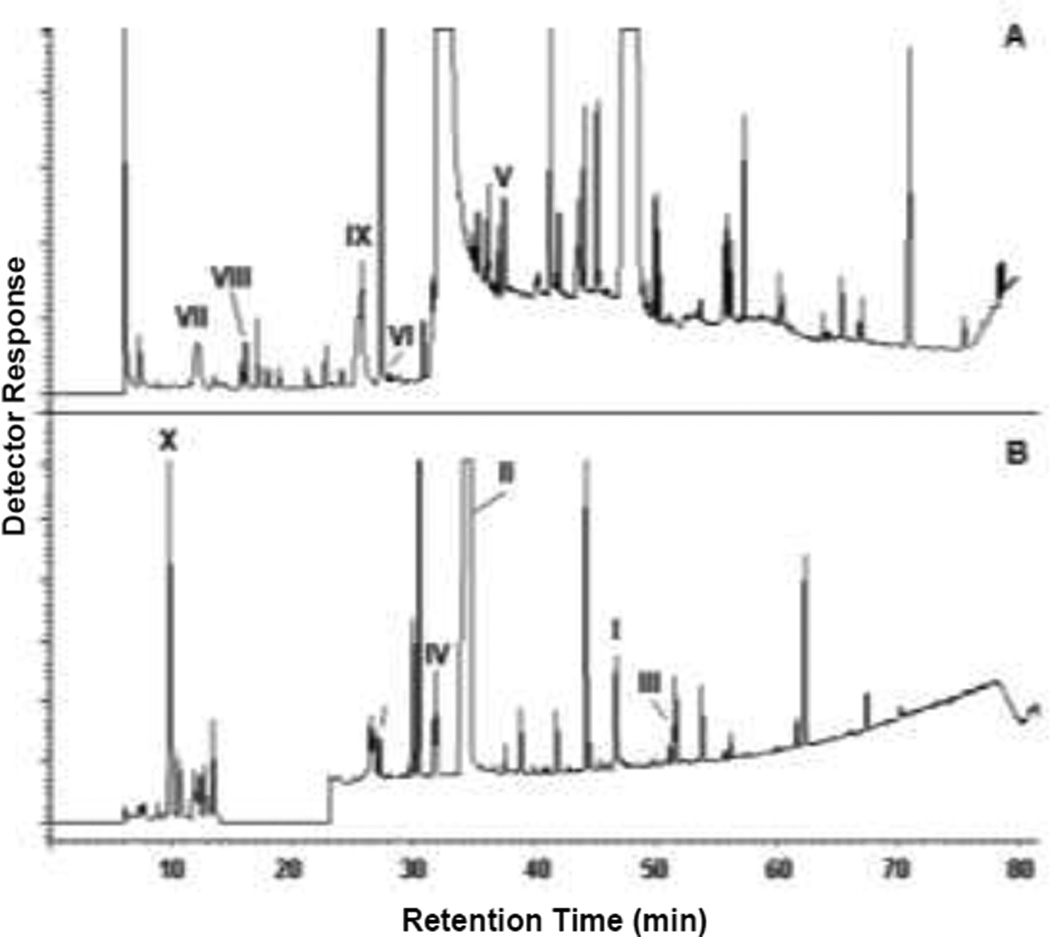
Nine carbaryl metabolites (II–IX in Fig. 2) were identified by GC-MS from the C3 culture medium. 1-Naphthol eluting at 32.7 min was the most abundant (II in Fig. 3B and Table 1). Metabolite III eluting at 50.3 min showed a similar MS fragmentation pattern as that of carbaryl with m/z 217 as a molecular ion (III in Fig. 3B and Table 1). This metabolite was tentatively identified as 4-hydroxycarbaryl. A trace amount of naphthoquinone (VI) was found at 27.3 min in the acidic fraction. However, its hydroquinone analogue was not observed in any samples. Metabolite IV eluting at 31.8 min showed a characteristic MS fragmentation pattern of PAH diol n-butylboronates (Keum et al. 2006; Seo et al. 2007b) and identified as naphthalene-1,2-diol (IV in Fig. 3B and Table 1). Metabolite V eluting at 36.2 min was identified as trans-2-hydroxybenzalpyruvate through the comparison with the authentic standard (methyl 2-methoxybenzalpyruvate). Early eluting metabolites at 14.4 min and 16.2 min were identified as salicylaldehyde (VII) and salicylic acid (VIII), respectively (Fig. 3A). Gentisic acid (IX) was eluted at 25.4 min. One of the most abundant peaks in the neutral fraction was n-butylboronate ester of catechol at 9.8 min (X in Fig. 3B).
Fig. 2.
Putative degradation pathways of carbaryl in Burkholderia sp. C3 and involvement of key enzymes differentially up-expressed. EutE, ethanolamine utilization protein; Fdh, formate dehydrogenase; AdhE, aldehyde-alcohol dehydrogenase. Roman numbers are ID numbers of the metabolites of which the names are given in Table 1.
Fig. 3.
GC-MS chromatograms of acidic (A) and neutral (B) metabolites of carbaryl in the cultural medium of Burkholderia sp. C3. Names of the Roman-numbered metabolites are are given in Table 1
Table 1.
GC retention times and MS fragmentation patterns of carbaryl and its metabolites isolated from the culture of Burkholderia sp. C3
| ID | Chemical namea | tRb (min) | Fragmentation pattern (intensity in %) |
|---|---|---|---|
| I | Carbaryl | 48.5 | 201 (M+, 28), 144 (100), 115 (26) |
| II | 1-Naphthol | 32.7 | 144 (M+, 100), 115 (25) |
| III | 4-Hydroxy carbaryl | 50.3 | 217 (M+, 6), 199 (1), 173 (2), 160 (100), 145 (7), 131 (10) |
| IV | 1,2-Dihydroxynaphthalene (nBuB) | 31.8 | 226 (M+, 100), 196 (5), 182 (2), 170 (76) |
| V | trans-2-Hydroxybenzalpyruvate | 36.2 | 220 (M+, 2), 205 (1), 189 (13), 161 (100), 145 (9), 129 (11) |
| VI | Naphthoquinone (diMe) | 27.3 | 158 (M+, 100), 130 (42), 102 (52) |
| VII | Salicylaldehyde | 14.4 | 122 (M+, 100), 93 (27) |
| VIII | Salicylic acid (Me) | 16.2 | 152 (M+, 70), 137 (1), 120 (100), 92 (78) |
| IX | Gentisic acid (Me) | 25.4 | 168 (M+, 100), 153 (1), 136 (90), 108 (74) |
| X | Catechol (nBuB) | 9.8 | 176 (M+, 100), 147 (5), 134 (4), 120 (74) |
Detected as derivatives: nBuB, n-butylboronate ester; diMe, dimethyl ester or methyl ester/ether; Me, methyl ester
tR, retention time
Proteomes in carbaryl-supported cells
A total of 876 proteins were identified (Table S2). In comparison with proteins in Burkholderia sp. C3 cells grown in the glucose-supplemented medium (i.e., glucose-supported cells), numerous proteins were differentially expressed in the cells grown in the carbaryl-supplemented medium (i.e., carbaryl-supported cells). Table 2 shows 41 up-expressed proteins in carbaryl-supported cells in comparison with glucose-supported cells. Up-expressed proteins in carbaryl-supported cells include formate dehydrogenase δ-subunit (FdhD) and aldehyde-alcohol dehydrogenase (AdhE) that are related to metabolism of formate and other C1 metabolism. Ethanolamine utilization protein (EutE) and spermidine synthase (SpeE) were up-expressed. Tryptophan 2-monooxygenase was found only in carbaryl-supported cells. Many proteins in gluconeogenesis and tricarboxylic acid (TCA) cycle were also up-expressed during carbaryl metabolism. Several regulatory or environmental sensing-related proteins were highly expressed in carbaryl-supported C3 cells, including methyl-accepting chemotaxis citrate transducer (Tcp) and HTH-type transcriptional regulator (PcaQ). Other sensory proteins, up-expressed by carbaryl treatment, were adaptive response regulator (Ada), GTP pyrophosphokinase (RelA) and starvation sensing protein (RspA). Stress-responsive proteins including chaperones, peroxidase and proteases were also differentially up-expressed. It is noteworthy that glutathione synthase (GshB) were strongly expressed in carbaryl-supported cells. Some metal ion-transporters were up-expressed. In relation to carbohydrate metabolism, several trehalose synthesis-related proteins (TreC, TreS, and TreY) were highly expressed during carbaryl metabolism.
Table 2.
Proteins differentially expressed in Burkholderia sp. C3 grown in minimal salt medium supplemented with carbaryl in comparison to those supplemented with glucose
| Protein name | Matcha | IDb | Function |
|---|---|---|---|
| Detected (up-expressed) proteins | |||
| Ethanolamine utilization protein (EutE) | 27/7 | P77445 | Primary metabolism |
| Spermidine synthase (SpeE) | 22/3 | Q7TY95 | Primary metabolism |
| Ketol-acid reductoisomerase | 30/4 | Q8UDV0 | Primary metabolism |
| Bifunctional putA protein | 35/5 | P09546 | Primary metabolism |
| Tryptophan 2-monooxygenase | 24/3 | P0A3V2 | Primary metabolism |
| UDP-N-acetylmuramate--L-alanine ligase | 32/4 | Q8UDM9 | Primary metabolism |
| UDP-N-acetylenolpyruvoylglucosamine reductase | 43/6 | Q9JV28 | Primary metabolism |
| Thiamine-phosphate pyrophosphorylase | 36/3 | Q9S2V2 | Primary metabolism |
| ATP synthase epsilon chain | 45/7 | Q9JW69 | Primary metabolism |
| Long-chain-fatty-acid--CoA ligase | 25/4 | P63521 | Primary metabolism |
| Phosphoenolpyruvate carboxykinase [ATP] | 46/5 | Q8UJ94 | Primary metabolism |
| L-Serine dehydratase 1 | 35/4 | P16095 | Primary metabolism |
| Glucose-6-phosphate isomerase | 31/4 | Q8NS31 | Primary metabolism |
| Adenosine deaminase | 19/3 | Q8UJ05 | Primary metabolism |
| Bifunctional purine biosynthesis protein | 45/5 | Q8Z335 | Primary metabolism |
| Succinate dehydrogenase flavoprotein | 19/4 | P0AC41 | Primary metabolism |
| Malate:quinone oxidoreductase | 48/4 | Q8FFQ5 | Primary metabolism |
| Trehalose-6-phosphate hydrolase (TreC) | 28/6 | P28904 | Primary metabolism, stress |
| Maltooligosyl trehalose synthase (TreY) | 21/4 | Q44315 | Primary metabolism, stress |
| Trehalose synthase (TreS) | 42/4 | P72235 | Primary metabolism, stress |
| Formate dehydrogenase, δ-subunit (FdhD) | 37/5 | Q8UC79 | Primary metabolism, stress |
| Aldehyde-alcohol dehydrogenase (AdhE) | 33/3 | P0A9Q7 | Primary metabolism, stress |
| Protein TolB precursor | 49/7 | Q5HX54 | Primary metabolism, transport |
| Major ferric ion-binding protein | 18/5 | P0A0Y3 | Primary metabolism, transport |
| Manganese transport protein (MntH) | 27/4 | O69443 | Primary metabolism, transport |
| Cell division inhibitor (SurA) | 12/2 | P0A241 | Sensors/regulators |
| Methyl-accepting chemotaxis citrate transducer (TcP) | 30/4 | Q02755 | Sensors/regulators |
| HTH-type transcriptional regulator (PcaQ) | 26/4 | P0A4T6 | Sensors/regulators |
| Starvation sensing protein (RspA) | 20/3 | P38104 | Sensors/regulators, stress |
| GTP pyrophosphokinase (RelA) | 35/3 | P0AG20 | Sensors/regulators, stress |
| ADA regulatory protein (Ada) | 51/5 | P26189 | Sensors/regulators, stress |
| 60 kDa chaperonin 1 (GroL) | 46/6 | Q00767 | Stress |
| Chaperone protein (DnaK) | 54/7 | Q9L7P1 | Stress |
| Protein SufB | 14/3 | P77522 | Stress, detoxification |
| Peroxidase/catalase HPI (KatG) | 47/7 | Q8Z303 | Stress, detoxification |
| ATPase ravA (RavA) | 19/4 | P31473 | Stress |
| UvrABC system protein A (UvrA) | 41/5 | Q8X5U9 | Stress, detoxification |
| Glutathione synthetase (GshB) | 31/5 | P58580 | Stress, detoxification |
| GTP-binding protein | 27/3 | Q9PIB6 | Unknown |
| Acyl carrier protein phosphodiesterase | 12/4 | Q8UG34 | Primary metabolism (xenobiotics) |
| Aliphatic nitrilase | 17/2 | Q02068 | Primary metabolism (xenobiotics) |
| Undetected (down-expressed) proteins | |||
| Chromosome partitioning protein (ParB) | 41/4 | Q9JW77 | Sensors/regulators |
| 3-Isopropylmalate dehydratase | 33/4 | Q8UBY9 | Primary metabolism |
| 3-Demethylubiquinone-9 3-methyltransferase | 31/4 | Q9JWE6 | Primary metabolism |
| Tryptophan synthase alpha chain | 15/3 | Q6AF66 | Primary metabolism |
| 5-Methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | 23/4 | Q82LG4 | Primary metabolism |
| 2-Isopropylmalate synthase | 39/5 | Q8FU05 | Primary metabolism |
| Dihydrodipicolinate reductase | 26/4 | Q9JX48 | Primary metabolism |
| Threonine-phosphate decarboxylase | 44/6 | Q8Z8H8 | Primary metabolism |
| L-Aspartate oxidase (NadB) | 48/7 | Q49617 | Primary metabolism |
| Enolase | 56/5 | Q9JZ53 | Primary metabolism |
| Acetate kinase (AckA) | 20/2 | P37877 | Primary metabolism |
| Glycerol-3-phosphate acyltransferase | 31/4 | P0A7A7 | Primary metabolism |
| Adenylate cyclase | 17/3 | P0A1A7 | Sensors/regulators |
| Phasin | 59/6 | Q7W4W5 | Primary metabolism |
| Adenylosuccinate synthetase | 33/2 | Q5HST3 | Primary metabolism |
| Malate dehydrogenase | 41/5 | Q8FN62 | Primary metabolism |
| Malate synthase G | 24/4 | O32913 | Primary metabolism |
| 2',3'-Cyclic-nucleotide 2'-phosphodiesterase | 35/3 | P08331 | Sensors/regulators |
| ATP-dependent Clp protease | 41/5 | P0A6H1 | Stress |
Mascot scores that were larger than the MOWSE scores at p = 0.05 / number of matched peptides
Protein ID in Swiss-Prot
Table 2 also shows 19 down-expressed proteins in carbaryl-supported cells in comparison with glucose-supported cells. Several enzymes for amino acid biosynthesis were down-expressed (i.e., not detected). An example was aspartate oxidase (NadB). Some respiratory proteins were also down-regulated. Among the metabolic enzymes, acetate kinase (AckA) was down-expressed in carbaryl-supported cells, but strongly expressed in glucose-supported C3 cells. Several enzymes, involved in cofactor biosynthesis were down- expressed during carbaryl metabolism. In relation to PHA biosynthesis, phasin was found in glucose-supported cells, but not in carbaryl-supported cells.
Metabolomes in carbaryl-supported cells
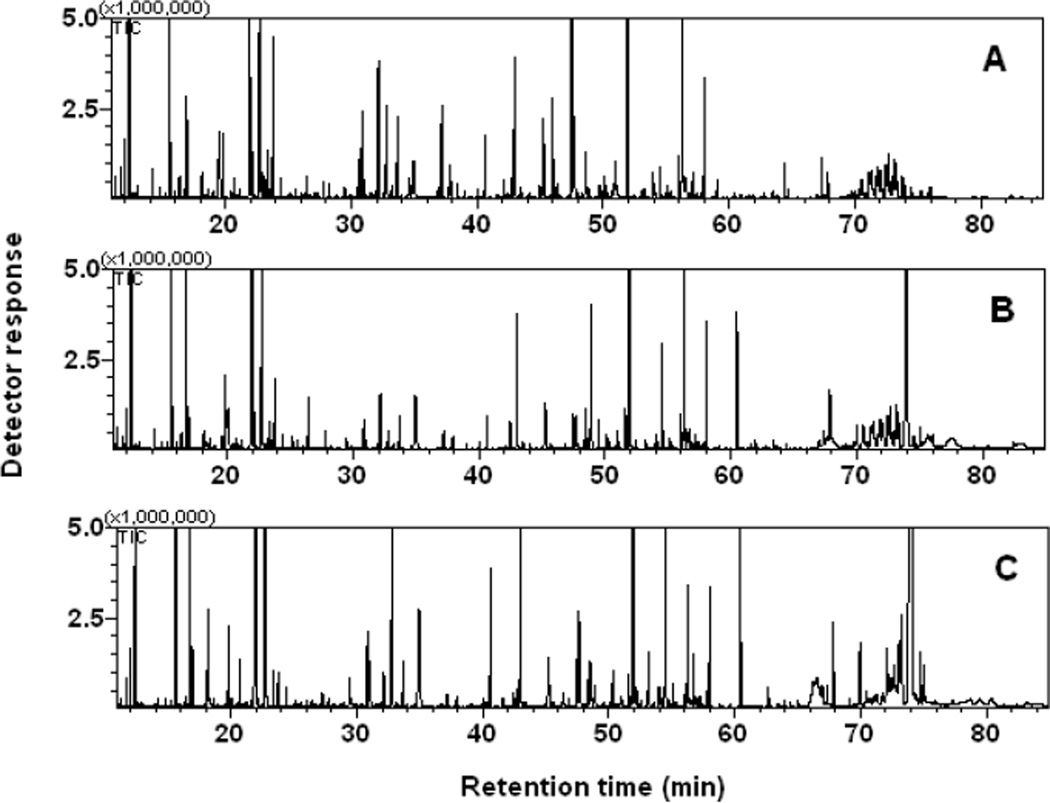
The metabolome in the C3 cells grown in the complex carbon nutrients (i.e., NB) was compared with that in single carbon sources (i.e., carbaryl or glucose). In general, the diversity and intensity of detected metabolites in NB-supported cells were intermediary between those in glucose and carbaryl-supported cells.
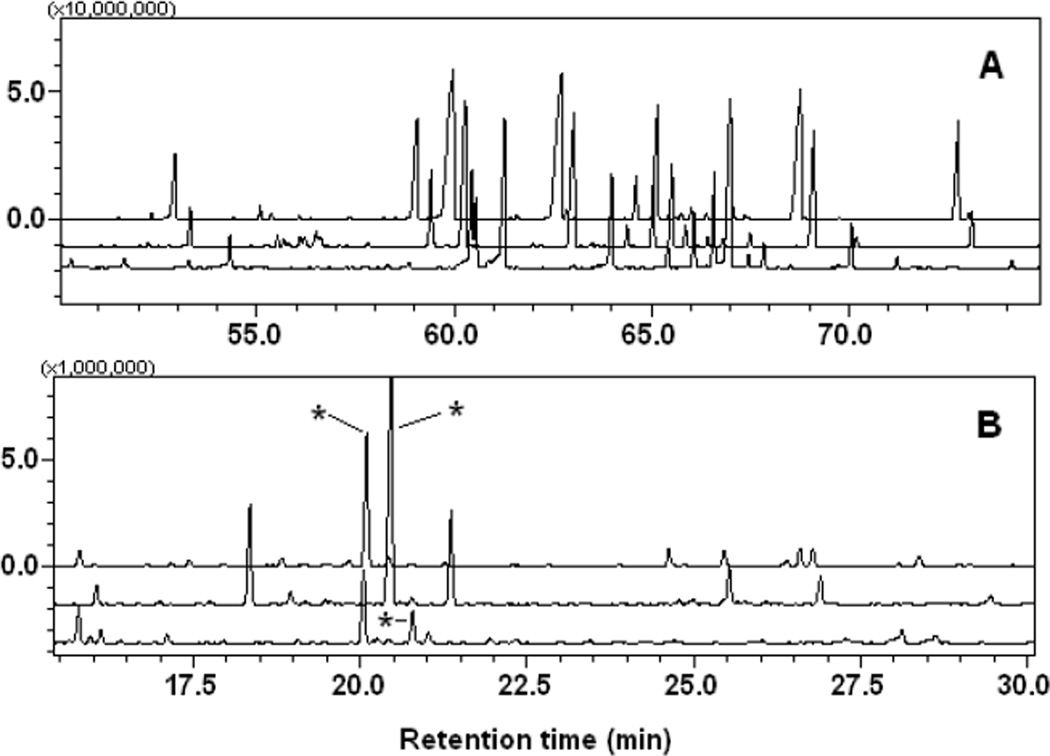
Major fatty acids in the strain C3 include 14:0, 16:0, 18:1, 19:0 cyclo, 14:0 3OH, and 16:0 3OH (Fig. 4A, Table S3). Other minor fatty acids including 15:0 and 17:0 were found in all treatments. Relative abundance of the major fatty acids differed largely and was substrate-dependent (Fig. 4A). For example, higher accumulation of 3-hydroxytetradecanoic acid and 3-hydroxyhexadecanoic acid (at 59.06 and 65.15 min, respectively, in Fig. 4A) was observed in carbaryl-supported cells, while the concentration of hexadecenoic acid (62.71 min) decreased as compared with that in NB- and glucose-supported cells.
Fig. 4.
Aligned GC-MS chromatograms of medium-chain to long-chain fatty acids (A) and 3-hydroxybutyrate from polyhydroxyalkanoates (B) from Burkholderia sp. C3, grown in nutrient broth (top), minimal salt medium with glucose (middle), and carbaryl (bottom) in A and B. Asterisk for the peaks for 3-hydroxybutyrate.
GC-MS analyses of purified PHAs indicated that the strain C3 can produce PHAs, mainly composed with 3-hydroxybutyric acid. The peak at 20.23 min gave a characteristic MS fragmentation (m/z 147) of hydroxyalkanoic acid TMS derivatives (Fig. 4B). PHA content was substrate-dependent. The strain C3 produced the highest amount of PHA when it was grown in MSM supplemented with glucose as a sole carbon source, followed by the NB- and carbaryl-supplemented media.
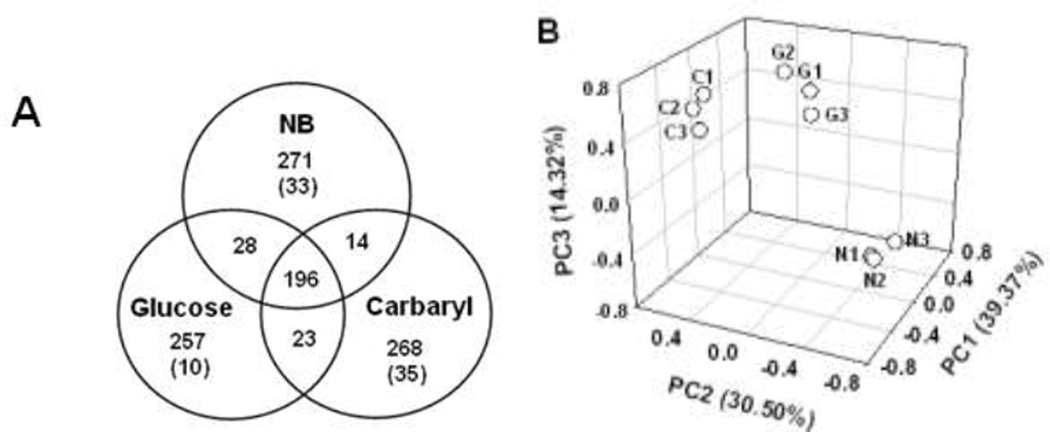
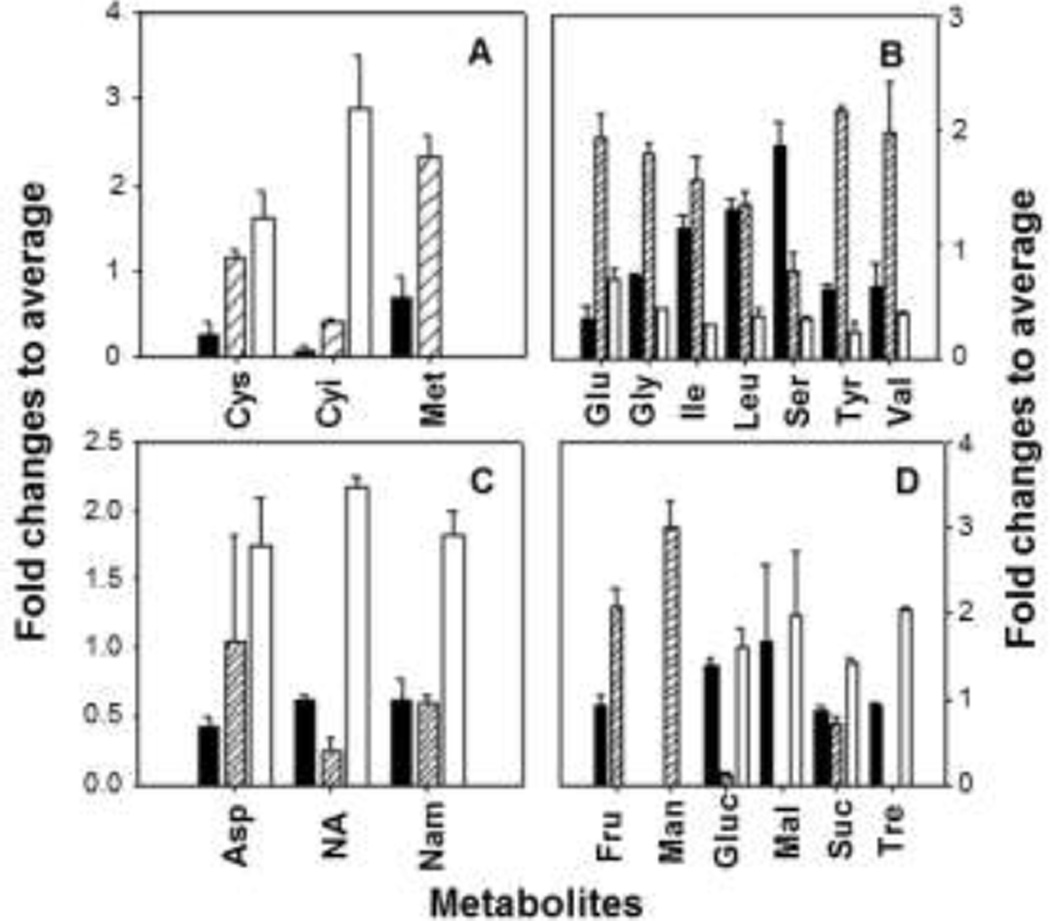
Approximately 339 peaks were observed in the GC-MS analyses of polar metabolites, where more than one half of them were tentatively identified by comparison of the mass spectra with those in the mass spectral library database and authentic standards and comparison of the retention times with those of the authentic standards (Fig. 5, Table S4). Approximately 196 metabolites were found in all three treatments, while 10–35 metabolites were unique in individual carbon sources (Fig. 6A). PCA showed that three groups of different carbon sources were clearly separated (Fig. 6B). Organic acids, alcohols, and related phosphates were the most frequently detected metabolites, followed by fatty acids, carbohydrates, amino acids, lipid precursors, general metabolic intermediates (Fig. S2). Numerous primary metabolites were differentially produced in response to carbon source variations. For example, the trehalose level was notably higher in carbaryl-supported cells than that in NB- or glucose-supported cells. Concentrations of most amino acids were lower in carbaryl-supported C3 cells than those in cells grown in the other carbon sources. Some exceptions included cysteine/cystine (cysteine dimmer) and aspartate of which the levels were significantly higher in carbaryl-supported cells than the other amino acids (Fig. 7). The concentrations of nicotineamide and nicotinic acid in carbaryl-grown cells were higher than those in NB- or glucose-supported cells.
Fig. 5.
GC-MS chromatograms of polar metabolites in primary metabolisms of Burkholderia sp. C3 grown in nutrient broth (A), minimal salt medium, supplemented with glucose (B), and carbaryl (C).
Fig. 6.
Venn-diagram of polar metabolites from Burkholderia sp. C3, grown in different carbon sources (A) and score plot of principal component analysis of polar metabolite profiles (B). Numbers of outskirt of cycles in Ben-diagram are the total number of peaks and unique metabolites, found only in the cells, grown with selected carbon sources. Data labels in score plot were as follows; C1–C3, carbaryl-supported, N1–N3, nutrient broth, G1–G3, glucose.
Fig. 7.
Concentrations of selected metabolites (A) sulfur-containing amino acids, (B) other amino acids, (C) aspartate, nicotinic acid and nicotinamide, and (D) mono- and disaccharides in Burkholderia sp. C3 grown in different carbon sources. Filled bar, glucose; blank with hash, nutrient broth; blank, carbaryl. Data were averages and standard errors of triplicate experiments (see Table S4 for statistical significance information). Abbreviations of metabolites: most amino acids were in the standard three letter codes, except cystine as Cyi; NA, nicotinic acid; Nam, nicotinamide; Fru, fructose; Man, mannose; Gluc, glucose; Mal, maltose; Suc, sucrose; Tre, trehalose.
Discussion
Catabolism of carbaryl and other N-methylcarbamates
A map of the putative breakdown of carbaryl and involvement of key enzymes is provided in Fig. 7. Various bacterial species can degrade N-methylcarbamates (Swetha and Phale 2005). Overall metabolic pathways of carbaryl in the strain C3 are very similar with those of the recently reported Pseudomonas (Swetha and Phale 2005). Analyses of these metabolites and the degradation products of 1-naphthol indicate that the strain C3 has enzymes for ester hydrolysis and phenol catabolism. The most common degradation enzymes of the N-methylcarbamates are metal-containing hydrolases, producing phenols and N-methylcarbamic acid, whereby the acid may be decomposed into methylamine and CO2 (Topp et al. 1993). In general, bacterial degradation of PAHs is initiated by the incorporation of one or two atoms of oxygen catalyzed by oxygenases (Krivobok et al. 2003). It is interesting that 1,2-dihydroxynaphthalene (IV), which is a dioxygenase-catalyzed product of phenanthrene in C3 (Tittabutr et al. 2011), was identified as a product of 1-naphthol (II) in the present study (Fig. 2). The genomic studies suggest that Burkholderia species have both dioxygenases and monooxygenases (Chain et al. 2006). It is not clear, however, whether a certain type of dioxygenase is involved in the transformation of 1-naphthol to naphthalene-1,2-diol. It is noteworthy that some bacteria have different regulatory systems for the degradation of 1-naphthol and naphthalene (Allen et al. 1997). Cytochrome P450s or other monooxygenases (e.g., FAD-containing monooxygenase) may also catalyze the transformation of 1-naphthol to naphthalene-1,2-diol or naphthoquinone. The detection of 4-hydroxycarbaryl and naphthoquinone suggests the involvement of the above enzymes (Table 1). Seo et al. (2007b) found the extradiol and intradiol cleavages of naphthalene-1,2-diol in the strain C3. Tittabutr et al. (2011) identified two dioxygenases in the strain C3 through gene cloning and protein analysis. The consecutive metabolic pathway of 1-naphthol in C3 is very similar with that in Pseudomonas spp. and Rhodococcus sp. NCIB 12038 in which salicylate is degraded into catechol and gentisate, respectively (Larkin and Day 1986; Swetha and Phale 2005). The strain C3 metabolizes salicylate into catechol (X) and gentisate (IX) (Table 1). Salicylate-1-hydroxylase can catalyze the transformation of salicylate or 1-hydroxy-2-naphthoate into their corresponding catechol (Balashova et al. 2001). Salicylate-5-hydroxylase, however, catalyzes the transformation of salicylate to gentisate (Zhou et al. 2002).
Primary metabolism-related proteins
N-Methylcarbamic acid and 1-naphthol from carbaryl are further metabolized into metabolites to enter primary metabolism pathways. The metabolites include carbon dioxide, formate, pyruvate and benozates. Ethanolamine utilization in Salmonella typhimurium was studied by Stojiljkovic et al. (1995). Mutations in EutE completely abolished the ability of mutants to utilize ethanolamine as a carbon source and reduced the ability to utilize ethanolamine as a nitrogen source. The genomic studies suggest that many Burkholderia and Ralstonia species have monoamine oxidases metabolizing methylamine into ammonia and carbon dioxide (Chain et al. 2006). In addition, amine dehydrogenases take part in the metabolism of alkylamine from which formaldehyde and ammonia are reaction products. It is not clear, however, whether the strain C3 has both enzymes or not. Over-expression of related proteins (e.g., EutE and FdhD) during carbaryl degradation (Table 2) indicates requirement of carbon source transformation through these enzymes and a concomitant change of primary metabolite profiles. EutE, for example, catalyzes biotransformation of phospholipids to acetate, which, in turn, is incorporated into the pyruvate metabolism and glycolysis. Up-production of EutE indicates possible changes of membrane properties when the role of phospholipids on membranes is considered (Stojiljkovic et al. 1995). EutE and polyphosphate-related proteins are strictly required for the survival of certain bacteria under acidic stresses (Price-Carter et al. 2005).
Formate dehydrogenase complex plays an important role in one-carbon metabolism. Formate and formaldehyde can be produced from different metabolic pathways, including the metabolism of methanol and methylamine. Burkholderia spp. are known to up-express specific proteins in response to formaldehyde and formate (Denef et al. 2004). Formate dehydrogenase, delta-subunit (FdhD) is involved in the metabolism of formate into CO2, formate-dependent nitrogen fixation in Rhizobium spp. (Manian et al. 1982), assimilation of formaldehyde in a photosynthetic bacterium (Barber and Donohue 1998) and C1 metabolism that is inter-connected with benzoate metabolism (Denef et al. 2004 and 2006). Rapid transformation of carbaryl to one carbon unit metabolites (e.g., methylamine, formaldehyde, and formate) may build up these metabolites at cytotoxic levels. Up-expression of Fdh may be an adaptive response of the strain C3 to formate, which the adaptive detoxification is indicated by up-regulation of stress related proteins (Table 2). Recent studies revealed over-expression of ribulose bisphosphate carboxylase (RuBisCo), presumably utilizing CO2 metabolized from formate in Burkholderia xenovorans LB400 and Ralstonia eutropha (Chain et al. 2006; Lee et al. 2006; Parnell et al. 2006). Because aldehyde-alcohol dehydrogenase (AdhE) involved in attenuation of oxidative stress and regulation of formate-pyruvate turn-over (Melchiorsen et al. 2000; Echave et al. 2002 and 2003), AdhE up-regulation again suggests the utilization of formate and oxidative stress during carbaryl catabolism (Table 2). Enzymes engaged in other steps of carbaryl catabolism were found to be not differentially expressed (Table S2), which suggests that they are constitutive.
Phasin and acetate kinase are down-expressed in the carbaryl-grown cells in comparison to glucose-grown cells. Both proteins are related to the removal and storage of excess carbon (Anderson and Dawes 1990; Grundy et al. 1993). PHAs are usually accumulated in high concentration carbohydrate-supplemented cultures. Phasins are localized at the surface of PHA granules, where they stabilize PHA granules and prevent non-specific binding of other proteins (Potter and Steinbüchel 2005). Reduced concentrations of PHAs and down-expression of phasin indicate limited carbon supply in carbaryl-supported cells. Among differentially expressed proteins, reduced production of aspartate oxidase is interesting since it is a key enzyme of de novo biosynthesis of NAD. Metabolomic analyses indicate complex regulations involved in NAD and aspartate metabolisms, which is discussed in the metabolomics sections below.
Chemosensor and regulator proteins
Among the up-expressed proteins in the adaptive sensing regulation systems are HTH-type transcriptional regulator (PcaQ) and methyl-accepting chemotaxis protein (TcP) during carbaryl catabolism (Table 2). Both are regulatory elements of bacterial phenol metabolism (Yamamoto and Imae 1993). Methyl-accepting chemotaxis proteins are known to regulate microbial motility from toxic phenols (e.g., 1-naphthol in the present study). Adaptive response protein (Ada) is to adapt alkylating agents and act as a DNA repair enzyme (Hakura et al. 1991). Although relationships between Ada and responses to carbaryl or its metabolites are not clear, the over-expression of Ada suggests that the strain C3 experience a high degree of DNA/RNA damages. Over-production of starvation sensing protein (e.g., RspA) is probably attributed to nutrient limitation in the strain C3 when carbaryl is a substrate. Bacteria under limited nutrients produce RspA to re-program the cell and adjust its physiology adaptive to the condition (Huisman and Kolter 1994). Regulatory proteins engaged in cell division are differentially expressed in C3 in response to carbaryl. Cell division inhibitor (SurA) is up-expressed, while chromosome partitioning protein (ParB) is down-expressed in carbaryl-supported cells. It is noteworthy that cell division is usually suppressed under highly stressed conditions.
Stress response and detoxification
Carbaryl catabolism produces 1-naphthol and formate (Table 1 and Fig. 7). Accumulation of those metabolites may damage physiologically significant macromolecules. Cellular responses to such a stress can be classified into (a) removal/repair of damaged parts, (b) attenuation of stress through the synthesis of protective chemicals, and (c) degradation of stressors. Various chaperones help correct folding of damaged proteins from chemical stress. Heat shock protein 70 (DnaK) and 60 kDa chaperonin 1 (GroL) are required for the survival of bacteria under stressed conditions (Kilstrup et al. 1997). Denatured proteins within the cytoplasm are subjected to the action of DnaK. ATPase (RavA) can act as a chaperone under acid-stressed conditions (Snider et al. 2006). Up-expression of RavA suggests toxic stresses to carbaryl-supported cells (Table 2). Over-production of such a chaperone is common phenomena under toxic conditions (Kim et al. 2004a; Lee et al. 2006).
Up-expression of polyphosphate metabolism proteins is also a very common response to phenols and PAHs in bacteria (Kim et al. 2004a; Santos et al. 2004). An increased expression of GTP pyrophosphokinase (RelA) was observed in carbaryl-supported cells (Table 2). The corresponding protein and polyphosphate kinase (Ppk) are key factors in stringent responses and related to various functions, including quorum sensing, biofilm formation, and virulence (Rashid et al. 2000). Strong expression of RelA, accompanied by up-expression of various chemosensors, regulators, and chaperones, indicates that the strain C3 is in a stringent condition during carbaryl metabolism.
Bacteria can utilize trehalose as the sole source of carbon and growth under both high-and low-osmolarity conditions (Rimmele and Boos 1994). Trehalose-6-phosphate hydrolase (TreC), trehalose synthase (TreS), and maltooligosyl trehalose synthase (TreY) participated in trehalose biosynthesis are up-expressed in carbaryl-supported cells (Table 2). In addition to the role as osmoprotectant, it is known that trehalose acts as an antioxidant in certain bacteria (Alvarez-Peral et al. 2002). Detection of up-expression of both intracellular trehalose and the associated enzymes (Table 2, Fig. 7) suggests that carbaryl is not an ideal substrate for C3 and causes high stresses. Regulation of trehalose influx/efflux is a possible mechanism to cope with toxicities. Bacterial cells adapt different strategies to meet this need, including the expression of specific transporters (e.g., ABC type transporters, porins) and changes of the membrane fluidity or hydrophobicity (Putman et al. 2000; Sokolovskä et al. 2003; Santos et al. 2004).
Up-expression of transporters is commonly found in bacteria under chemical stresses (Santos et al. 2004). Among the related proteins, iron and manganese transport proteins are up-expressed in carbaryl-supported cells (Table 2). Biological functions of the up-expression of these metal transporters require, however, further studies.
Oxidative stresses are well known phenomena when phenols and PAHs are substrates (Kim et al. 2004a; Santos et al. 2004). Glutathione plays diverse roles in cellular functions, especially under oxidative stresses (Ferguson and Booth 1998; Carmel-Harel and Storz 2000). Up-expression of GshB probably attenuates oxidative stresses in carbaryl-supported cells (Table 2). Superoxide dismutases (SOD), catalase (KatA), or bifunctional catalase-peroxidase (KatG) are the most commonly up-regulated proteins in response to oxidative stresses (Kim et al. 2004a; Santos et al. 2004). Over-expression of KatG indicates strong oxidative stressors in carbaryl-grown cells in comparison with glucose-supported cells.
Metabolomes
Comprehensive metabolomes have been studied in many fields (Weckwerth 2003; Robertson 2005; Keum et al. 2010). Few publications of bacterial metabolomes, however, have been found relevant to biodegradation of pesticides and organic pollut ants. Metabolomes are dynamic and responsive to nutrient and environment changes. Comparative analyses of metabolomes in response to different environments or substrates, aided by proteomics, can provide detailed insights into features of bacterial adaptation to, metabolic network, and metabolism of toxic chemicals such as N-methylcarbamates.
A total of 196 polar metabolites, 10 medium to long chain fatty acids, and one type of macromolecule (i.e., PHA) were detected in the strain C3 (Fig. 6A). Three distinct groups of polar metabolites depicted by PCA suggest that C3 can re-program its primary metabolism according to varying carbon sources (i.e., glucose, carbaryl and NB) (Fig. 6B). Metabolic patterns drawn over KEGG pathway database give large metabolic changes, depending on carbon sources (Fig. S1).
Among polar metabolites, accumulation of cysteine and cystine is one of the most noticeable metabolic responses to carbaryl in the strain C3 (Fig. 7A). Cysteine is an important amino acid to cope with various environmental stresses. Cysteine is a precursor of glutathione, the most important metabolite alleviating oxidative stresses. Over-expression of oxidative stress-related proteins and accumulation of cysteine and cystine indicate carbaryl-supported C3 cells are under strong oxidative stresses. Among several carbaryl metabolites, 1-naphthol and naphthoquinone probably cause such toxic stresses. It is known that PAH quinones can inhibit several reductases or consume reducing equivalents (e.g., NADH), and so that an excessive amount of quinones or its precursors can cause oxidative stress (Kim et al. 2004b).
Concentrations of the other amino acids, except aspartate, cysteine and cystine, decrease in carbaryl-supported C3 cells in comparison to glucose- and NB-supported cells (Fig. 7, Table S4). Production of the enzymes corresponding to the biosynthesis of these amino acids is depressed (Table 2). Bacterial cells under stringent conditions usually reduce protein synthesis, while the content of nucleic acid increases (Chatterji and Ojha 2001). The reduced amino acid content indicates that the strain C3 follows such a metabolomic response to carbaryl. Several carbohydrates also show differential responses to substrate variations in C3. In general, glucose-or NB-supported cells are rich in monosaccharide (e.g., fructose, glucose, and mannose), while carbaryl-supported cells produce an elevated amount of disaccharides (Fig. 7D). Trehalose accounts for a large portion of the polar metabolites (approximately 0.4% of the total peak area or 30% of disaccharides). Its concentration in carbaryl-supported cells is far higher than that in the others. Over-production of both the related proteomes and trehalose indicates high stresses to in carbaryl-grown C3 cells. Another remarkable change in C3 metabolomes is related to the over-production of aspartate, nicotinic acid and nicotinamide in carbaryl-grown cells in comparison to NB- and glucose-supported cells (Fig. 7C). Aspartate is an important precursor of nicotinamide adenine dinucleotide (NAD). The pyridine nucleotide precursors nicotinic acid and nicotinamide are also important intermediates for NAD biosynthesis. Rapid turnover of NAD and NADH is required for efficient bacterial PAH metabolism. Some enzymes being responsible for PAH catabolism (e.g., dioxygenases, hydroxylases) require the enzyme cofactor NADH, while others e.g., dehydrogenase) can regenerate NADH from NAD (Bugg 2003). Two separate pathways of NAD biosynthesis are known in bacteria, namely de novo biosynthesis from aspartate and the NAD salvage pathway (Lima et al. 2009). In addition to the role of cofactor, NAD can be incorporated to the TCA cycle through nicotinates (the acid and amide forms). It is difficult to confirm whether the high concentration of nicotinates in carbaryl-supported C3 cells is the consequence of a high rate of NAD and NADH biosynthesis through the salvage pathway or simply the rapid degradation of NAD to sustain other primary metabolisms. However, down-expression of aspartate oxidase (Table 2) suggests that de novo biosynthesis of NAD may be reduced during carbaryl metabolism. In addition, aspartate is an intermediate in numerous biochemical reactions (e.g., other amino acid biosynthesis, urea cycle, TCA cycle). An increased amount of aspartate may be an adaptive response to support these essential reactions, other than NAD biosynthesis. Although the possible involvement of the salvage pathways cannot be ruled out, a high concentration of nicotinates may be resulted from catabolic degradation of NAD to support other primary metabolisms.
PHAs are biopolymers commonly found in various bacterial species. PHAs are usually comprised with short chain 3-hydroxyalkanoic acids (e.g., 3-hydroxybutyric acid and 3-hydroxypentanoic acid). PHAs are generally considered as reservoirs of excess carbon although the exact biological role remains unclear (Anderson and Dawes 1990). Various rhizobial bacteria, including Sinorhizobium spp., produce PHAs (Lakshman and Shamala 2006; Keum et al. 2008). The monomeric composition of PHAs from the strain C3 is similar with that of the other Burkholderia spp. (Lakshman and Shamala 2006) and Sinorhizobium sp. C4 isolated from the same soil as the strain C3 (Keum et al. 2008). Reduced biosynthesis of PHAs in carbaryl-supported cells is in accordance with the results of the other polar metabolomes (Fig. 7), indicating limited carbon supply. Absence of phasin in proteome analyses also suggests that the carbon source supplement from carbaryl is insufficient for PHA accumulation. Concentrations of PHAs in NB-supported cells are lower than those in glucose-supported cells, indicating high dependence of PHA accumulation on carbon sources.
Profiles of cellular fatty acids in bacteria are dynamically responsive to exposure to toxic chemicals (Keum et al. 2008). Modulation of fatty acid composition results in changes of cell membrane fluidity, hydrophobicity, and permeability. The integrity of cellular membrane is essential for normal physiology. Differential productions of fatty acid profiles in C3 cells grown in the three substrates may reflect equilibrium networks among all primary metabolites to balance biological functions for cell survival and growth. Fatty acid profiles in NB-supported cells differ from those of the other carbon sources, indicating different metabolomes and metabolism reprogramming.
In summary, large differentiations of metabolomes and proteomes in the strain C3 have been observed among the cultures with carbaryl, glucose and NB. In general, the results showed the metabolic adaptation of Burkholderia sp. C3 to carbaryl in comparison with glucose and NB. The proteomic and metabolomic results indicate that carbaryl-supported C3 cells experience carbon supply shortage in comparison with glucose and NB. The metabolome changes are notably associated with biosynthesis and metabolisms of amino acids, sugars, PHAs, lipids, and cofactors. The proteome changes indicate utilization of one carbon metabolites produced from carbaryl for energy production and metabolic networks among primary metabolisms, defense and detoxification, and adaptation. Overall features of these changes are visualized via metabolomics and proteomics analyses. Detailed information of metabolic adaptation may provide better understanding of this environmentally relevant bacterium to enhance or modify for specific metabolic purposes. However, there remain several questions. For example, chemical compositions of natural matrices (e.g., soil particles, pore water, and organic matters in soil) differ from the media in this study, which will likely produce deviations from the metabolomic and proteomic responses observed in the present study. The complexity of microbial communities in the natural environment is also different from that of the closed single species systems. Those warrant further studies in a system that closely mimics the natural environment and by using an integrated genomic, proteomic and metabolomic approach.
Supplementary Material
Acknowledgments
This work was supported in part with US-EPA award no. 989512-01-1, US ONR HEET award N00014-09-1-0709, and grants from the National Institute on Minority Health and Health Disparities (8 G12 MD007601-26).
References
- Allen CCR, Boyd C, Larkin MJ, Reid KA, Sharma ND, Wilson K. Metabolism of naphthalene, 1-naphthol, indene, and indole by Rhodococcus sp. strain NCIMB 12038. Appl Environ Microbiol. 1997;63:151–155. doi: 10.1128/aem.63.1.151-155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Peral FJ, Zaragoza O, Pedreno Y, Argüelles J-C. Protective role of trehalose during severe oxidative stress caused by hydrogen peroxide and the adaptive oxidative stress response in Candida albicans. Microbiology. 2002;148:2599–2606. doi: 10.1099/00221287-148-8-2599. [DOI] [PubMed] [Google Scholar]
- Anderson AJ, Dawes EA. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashova NV, Stolz A, Knackmuss HJ, Kosheleva IA, Naumov AV, Boronin AM. Purification and characterization of a salicylate hydroxylase involved in 1-hydroxy-2-naphthoic acid hydroxylation from the naphthalene and phenanthrene-degrading bacterial strain Pseudomonas putida BS202-P1. Biodegradation. 2001;12:179–188. doi: 10.1023/a:1013126723719. [DOI] [PubMed] [Google Scholar]
- Barber RD, Donohue TJ. Function of a glutathione-dependent formaldehyde dehydrogenase in Rhodobacter sphaeroides formaldehyde oxidation and assimilation. Biochemistry. 1998;37:530–537. doi: 10.1021/bi971463t. [DOI] [PubMed] [Google Scholar]
- Bastiaens L, Springael D, Wattiau P, Harms H, deWachter L, Verachtert H, Diels L. Isolation of adherent polycyclic aromatic hydrocarbon (PAH)-degrading bacteria using PAH-sorbing carriers. Appl Environ Microbiol. 2000;66:1834–1843. doi: 10.1128/aem.66.5.1834-1843.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg TDH. Dioxygenase enzymes: catalytic mechanisms and chemical models. Tetrahedron. 2003;59:7075–7101. [Google Scholar]
- Carmel-Harel O, Storz G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol. 2000;54:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- Chain PSG, Denef VJ, Konstantinidis KT, Vergez LM, Agulló L, Reyes VL, Hauser L, Córdovag M, Gómez L, González MLM, Lao V, Larimer F, LiPuma JJ, Mahenthiralingam E, Malfatti SA, Marx CJ, Parnell JJ, Ramette A, Richardson P, Seeger M, Smith D, Spilker T, Sul WJ, Tsoi TV, Ulrich LE, Zhulin IB, Tiedje JM. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci USA. 2006;103:15280–15287. doi: 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji D, Ojha AK. Revisiting the stringent response, ppGpp and starvation signaling. Curr Op Microbiol. 2001;4:160–165. doi: 10.1016/s1369-5274(00)00182-x. [DOI] [PubMed] [Google Scholar]
- Denef VJ, Park J, Tsoi TV, Rouillard JM, Zhang H, Wibbenmeyer JA, Verstraete W, Gulari E, Hashsham SA, Tiedje JM. Biphenyl and benzoate metabolism in a genomic context: Outlining genome-wide metabolic networks in Burkholderia xenovorans LB400. Appl Environ Microbiol. 2004;70:4961–4970. doi: 10.1128/AEM.70.8.4961-4970.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef VJ, Patrauchan MA, Florizone C, Park J, Tsoi TV, Verstraete W, Tiedje JM, Eltis LD. Growth substrate and phase specific expression of biphenyl, benzoate and C1 metabolic pathways in Burkholderia xenovorans LB400. J Bacteriol. 2005;187:7996–8005. doi: 10.1128/JB.187.23.7996-8005.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef VJ, Klappenbach JA, Patrauchan MA, Florizone C, Rodrigues JLM, Tsoi TV, Verstraete W, Eltis LD, Tiedje JM. Genetic and genomic insights into the role of benzoate-catabolic pathway redundancy in Burkholderia xenovorans LB400. Appl Environ Microbiol. 2006;72:585–595. doi: 10.1128/AEM.72.1.585-595.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echave P, Esparza-Cerón MA, Cabiscol E, Tamarit J, Ros J, Membrillo-Hernández J, Lin ECC. DnaK dependence of mutant ethanol oxidoreductases evolved for aerobic function and protective role of the chaperone against protein oxidative damage in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:4626–4631. doi: 10.1073/pnas.072504199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echave P, Tamarit J, Cabiscol E, Ros J. Novel antioxidant role of alcohol dehydrogenase E from Escherichia coli. J Biol Chem. 2003;278:30193–30198. doi: 10.1074/jbc.M304351200. [DOI] [PubMed] [Google Scholar]
- Ferguson GP, Booth IR. Importance of glutathione for growth and survival of Escherichia coli cells: detoxification of methylglyoxal and maintenance of intracellular K+ J Bacteriol. 1998;180:4314–4318. doi: 10.1128/jb.180.16.4314-4318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Waters DA, Allen SH, Henkin TM. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J Bacteriol. 1993;175:7348–7355. doi: 10.1128/jb.175.22.7348-7355.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakura A, Morimoto K, Sofuni T, Nohmi T. Cloning and characterization of the Salmonella typhimurium ada gene, which encodes O6-methylguanine-DNA methyltransferase. J Bacteriol. 1991;173:3663–3672. doi: 10.1128/jb.173.12.3663-3672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Fukui M, Hayano K, Hayatsu M. Nucleotide sequence and genetic structure of a novel carbaryl hydrolase gene (cehA) from Rhizobium sp. strain AC100. Appl Environ Microbiol. 2002;68:1220–1227. doi: 10.1128/AEM.68.3.1220-1227.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman GW, Kolter R. Sensing starvation: a homoserine lactone-dependent signaling pathway in Escherichia coli. Science. 1994;265:537–539. doi: 10.1126/science.7545940. [DOI] [PubMed] [Google Scholar]
- Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, Hirasawa T, Naba M, Hirai K, Hoque A, Ho PE, Kakazu Y, Sugawara K, Igarashi S, Harada S, Masuda T, Sugiyama N, Togashi T, Hasegawa M, Takai Y, Yugi K, Arakawa K, Iwata N, Toya Y, Nakayama Y, Nishioka T, Shimizu K, Mori H, Tomita D. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- Keum Y-S, Seo J-S, Li QX. Synthesis of bacterial metabolites of polycyclic aromatic hydrocarbons: Benzochromenones, o-cartboxyvinylnaphthoates, o-substituted aryl-β-oxobutenoates. Synth Commun. 2005;35:2685–2693. [Google Scholar]
- Keum Y-S, Seo J-S, Hu Y, Li QX. Degradation pathways of phenanthrene by Sinorhizobium sp. C4. Appl Microbiol Biotechnol. 2006;71:935–941. doi: 10.1007/s00253-005-0219-z. [DOI] [PubMed] [Google Scholar]
- Keum Y-S, Seo J-S, Li QX, Kim JH. Comparative metabolomic analysis of Sinorhizobium sp. C4 during the degradation of phenanthrene. Appl Microbiol Biotechnol. 2008;80:863–872. doi: 10.1007/s00253-008-1581-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keum Y-S, Kim J-H, Li QX. Metabolomics in Pesticide Toxicology. In: Krieger R, editor. Hayes’ Handbook of Pesticide Toxicology. 3rd ed. New York: Academic Press; 2010. pp. 627–643. [Google Scholar]
- Kilstrup M, Jacobsen S, Hammer K, Vogensen FK. Induction of heat shock proteins DnaK, GroEL, and GroES by salt stress in Lactococcus latis. Appl Environ Microbiol. 1997;63:1826–1837. doi: 10.1128/aem.63.5.1826-1837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Jones RC, Cha C-J, Kweon O, Edmondson RD, Cerniglia CE. Identification of proteins induced by polycyclic aromatic hydrocarbon in Mycobacterium vanbaalenii PYR-1 using two-dimensional polyacrylamide gel electrophoresis and de novo sequencing methods. Proteomics. 2004a;4:3899–3908. doi: 10.1002/pmic.200400872. [DOI] [PubMed] [Google Scholar]
- Kim Y-H, Moody JD, Freeman JP, Brezna B, Engesser K-H, Cerniglia CE. Evidence for the existence of PAH-quinone reductase and catechol-O -methyltransferase in Mycobacterium vanbaalenii PYR-1. J Ind Microbiol Biotechnol. 2004b;31:507–516. doi: 10.1007/s10295-004-0178-x. [DOI] [PubMed] [Google Scholar]
- Krivobok S, Kuony S, Meyer C, Louwagie M, Willison JC, Jouanneau Y. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: Evidence for two ringhydroxylating dioxygenases. J Bacteriol. 2003;185:3828–3841. doi: 10.1128/JB.185.13.3828-3841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon O, Kim S-J, Jones RC, Freeman JP, Adjei MD, Edmondson RD, Cerniglia CE. A polyomic approach to elucidate the fluoranthene-degradative pathway in Mycobacterium vanbaalenii PYR-1. J Bacteriol. 2007;189:4635–4647. doi: 10.1128/JB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshman K, Shamala TR. Extraction of polyhydroxyalkanoate from Sinorhizobium meliloti cells using Microbispora sp culture and its enzymes. Enz Microbial Technol. 2006;39:1471–1475. [Google Scholar]
- Larkin MJ, Day MJ. The metabolism of carbaryl by three bacterial isolates, Pseudomonas spp. (NCIB 12042 & 12043) and Rhodococcus sp. (NCIB 12038) from garden soil. J Appl Bacteriol. 1986;60:233–242. doi: 10.1111/j.1365-2672.1986.tb01078.x. [DOI] [PubMed] [Google Scholar]
- Lee S-E, Li QX, Yu J. Proteomic examination of Ralstonia eutropha in cellular responses to formic acid. Proteomics. 2006;6:4259–4268. doi: 10.1002/pmic.200500824. [DOI] [PubMed] [Google Scholar]
- Lee S-E, Seo J-S, Keum Y-S, Lee KJ, Li QX. Fluoranthene metabolism and associated proteins in Mycobacterium sp. JS14. Proteomics. 2007;7:2059–2069. doi: 10.1002/pmic.200600489. [DOI] [PubMed] [Google Scholar]
- Lima WC, Varani AM, Menck CFM. NAD biosynthesis evolution in bacteria: lateral gene transfer of kynurenine pathway in Xanthomonadales and Flavobacteriales. Mol Biol Evol. 2009;26:399–406. doi: 10.1093/molbev/msn261. [DOI] [PubMed] [Google Scholar]
- Manian SS, Gumbleton R, O'Gara F. The role of formate metabolism in nitrogen fixation in Rhizobium spp. Arch Microbiol. 1982;133:312–317. [Google Scholar]
- Melchiorsen CR, Jokumsen KV, Villadsen J, Johnsen MG, Israelsen H, Arnau J. Synthesis and posttranslational regulation of pyruvate formate-lyase in Lactococcus lactis. J Bacteriol. 2000;182:4783–4788. doi: 10.1128/jb.182.17.4783-4788.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell JJ, Park JH, Denef V, Tsoi T, Hashsham S, Quensen J, III, Tiedje JM. Coping with polychlorinated biphenyl (PCB) toxicity: Physiological and genome-wide responses of Burkholderia xenovorans LB400 to PCB-mediated stress. Appl Environ Microbiol. 2006;72:6607–6614. doi: 10.1128/AEM.01129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter M, Steinbüchel A. Poly(3-hydroxybutyrate) granule-associated proteins: impacts on poly(3-hydroxybutyrate) synthesis and degradation. Biomacromolecules. 2005;6:552–560. doi: 10.1021/bm049401n. [DOI] [PubMed] [Google Scholar]
- Price-Carter M, Fazzio TG, Vallbona EI, Roth JR. Polyphosphate kinase protects Salmonella enterica from weak organic acid stress. J Bacteriol. 2005;187:3088–3099. doi: 10.1128/JB.187.9.3088-3099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putman M, van Veen HW, Konings WN. Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev. 2000;64:672–693. doi: 10.1128/mmbr.64.4.672-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S, Li QX. Proteomics in Pesticide Toxicology. In: Krieger R, editor. Hayes’ Handbook of Pesticide Toxicology. 3rd ed. New York: Academic Press; 2010. pp. 603–626. [Google Scholar]
- Qing Z, Yang L, Liu YH. Purification and characterization of a novel carbaryl hydrolase from Aspergillus niger PY168. FEMS Microbiol Lett. 2003;228:39–44. doi: 10.1016/S0378-1097(03)00718-3. [DOI] [PubMed] [Google Scholar]
- Rashid MH, Rumbaugh K, Passador L, Davies DG, Hamood AN, Iglewski BH, Kornberg A. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2000;97:9636–9641. doi: 10.1073/pnas.170283397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmele M, Boos W. Trehalose-6-phosphate hydrolase of Escherichia coli. J Bacteriol. 1994;176:5654–5664. doi: 10.1128/jb.176.18.5654-5664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DG. Metabonomics in toxicology: A review. Toxicol Sci. 2005;85:809–822. doi: 10.1093/toxsci/kfi102. [DOI] [PubMed] [Google Scholar]
- Santos PM, Benndorf D, Sa-Correia I. Insights into Pseudomonas putida KT2440 response to phenol-induced stress by quantitative proteomics. Proteomics. 2004;4:2640–2652. doi: 10.1002/pmic.200300793. [DOI] [PubMed] [Google Scholar]
- Seo J-S, Keum Y-S, Harada RM, Li QX. Isolation and characterization of bacteria capable of degrading polycyclic aromatic hydrocarbons (PAHs) and organophosphorus pesticides from PAH-contaminated soil in Hilo, Hawaii. J Agric Food Chem. 2007a;55:5383–5389. doi: 10.1021/jf0637630. [DOI] [PubMed] [Google Scholar]
- Seo J-S, Keum Y-S, Hu Y, Lee SE, Li QX. Degradation of phenanthrene by Burkholderia sp. C3: initial 1,2- and 3,4-dioxygenation and meta- and ortho-cleavage of naphthalene-1,2-diol. Biodegradation. 2007b;18:123–131. doi: 10.1007/s10532-006-9048-8. [DOI] [PubMed] [Google Scholar]
- Singh BK, Kuhad RC, Singh A, Lal R, Tripathi KK. Biochemical and molecular basis of pesticide degradation by microorganisms. Crit Rev Biotechnol. 1999;19:197–225. doi: 10.1080/0738-859991229242. [DOI] [PubMed] [Google Scholar]
- Snider J, Gutsche I, Lin M, Baby S, Cox B, Butland G, Greenblatt J, Emili A, Houry WA. Formation of a distinctive complex between the inducible bacterial lysine decarboxylase and a novel AAA+ ATPase. J Biol Chem. 2006;281:1532–1546. doi: 10.1074/jbc.M511172200. [DOI] [PubMed] [Google Scholar]
- Sokolovská I, Rozenberg R, Riez C, Rouxhet PG, Agathos SN, Wattiau P. Carbon source-induced modifications in the mycolic acid content and cell wall permeability of Rhodococcus erythropolis E1. Appl Environ Microbiol. 2003;69:7019–7027. doi: 10.1128/AEM.69.12.7019-7027.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I, Baumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: Nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swetha VP, Phale PS. Metabolism of carbaryl via 1,2-dihydroxynaphthalene by soil isolates Pseudomonas sp. strain C4, C5, and C6. Appl Environ Microbiol. 2005;71:5951–5956. doi: 10.1128/AEM.71.10.5951-5956.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittabutr P, Cho IK, Li QX. Phn and Nag-like dioxygenases metabolize polycyclic aromatic hydrocarbons in Burkholderia sp. C3. Biodegradation. 2011;22:1119–1133. doi: 10.1007/s10532-011-9468-y. [DOI] [PubMed] [Google Scholar]
- Topp E, Hanson RS, Ringelberg DB, White DC, Wheatcroft R. Isolation and characterization of an N-methylcarbamate insecticide-degrading methylotrophic bacterium. Appl Environ Microbiol. 1993;59:3339–3349. doi: 10.1128/aem.59.10.3339-3349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth W. Metabolomics in system biology. Ann Rev Plant Biol. 2003;54:669–689. doi: 10.1146/annurev.arplant.54.031902.135014. [DOI] [PubMed] [Google Scholar]
- Wilson B. Cholinesterases. In: Krieger R, editor. Hayes’ Handbook of Pesticide Toxicology. 3rd ed. New York: Academic Press; 2010. pp. 1457–1478. [Google Scholar]
- Yamamoto K, Imae Y. Cloning and characterization of the Salmonella typhimurium-specific chemoreceptor Tcp for taxis to citrate and from phenol. Proc Natl Acad Sci USA. 1993;90:217–221. doi: 10.1073/pnas.90.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou NY, Al-Dulayymi J, Baird MS, Williams PA. Salicylate 5-hydroxylase from Ralstonia sp. strain U2: a monooxygenase with close relationships to and shared electron transport proteins with naphthalene dioxygenase. J Bacteriol. 2002;184:1547–1555. doi: 10.1128/JB.184.6.1547-1555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.