Abstract
Regulated expression of glucose-6-phosphate dehydrogenase (G6PD) is due to changes in the rate of pre-mRNA splicing and not changes in its transcription. Starvation alters pre-mRNA splicing by decreasing the rate of intron removal, leading to intron retention and a decrease in the accumulation of mature mRNA. A regulatory element within exon 12 of G6PD pre-mRNA controls splicing efficiency. Starvation caused an increase in the expression of heterogeneous nuclear ribonucleoprotein (hnRNP) K protein and this increase coincided with the increase in the binding of hnRNP K to the regulatory element and a decrease in the expression of G6PD mRNA. HnRNP K bound to two C-rich motifs forming an ESS within exon 12. Overexpression of hnRNP K decreased the splicing and expression of G6PD mRNA, while siRNA-mediated depletion of hnRNP K caused an increase in the splicing and expression of G6PD mRNA. Binding of hnRNP K to the regulatory element was enhanced in vivo by starvation coinciding with a decrease in G6PD mRNA. HnRNP K binding to the C-rich motifs blocked binding of serine-arginine rich, splicing factor 3 (SRSF3), a splicing enhancer. Thus hnRNP K is a nutrient regulated splicing factor responsible for the inhibition of the splicing of G6PD during starvation.
Keywords: mRNA splicing, hnRNP K, SRSF3, nutrient regulation, liver
INTRODUCTION
Maintenance of energy homeostasis throughout the body is achieved by balancing the use of dietary energy to meet immediate needs and the storage of excess energy for use in the absence of dietary intake. A key pathway involved in this process is lipogenesis, by which excess energy in the form of carbohydrate is converted to triacylglycerol for storage in adipose tissue and as such this pathway is highly active and regulated in adipose tissue and liver. The enzymes involved in lipogenesis, ATP-citrate lyase, malic enzyme, fatty acid synthase, glucose-6-phosphate dehydrogenase (G6PD)2, and acetyl-CoA carboxylase increase in amount during consumption of high-carbohydrate diets in order to increase a cell’s capacity for lipogenesis; however, in response to starvation the amount of these proteins is decreased [1]. Dietary regulation of these proteins is controlled by both dietary nutrients and by changes in the amounts of key regulatory hormones such as insulin and glucagon. Regulation of most of the lipogenic enzymes is primarily through changes the transcriptional activity of their genes [1]. G6PD is an exception in this family. Nutrient and hormonal regulation of G6PD expression is exclusively by changes in the degree of splicing of its mRNA [2, 3].
RNA splicing is an essential step in gene expression that joins together the exons in the nascent transcript [4]. Splicing is a highly regulated process such that a single transcript can generate multiple mature mRNA via a process called alternative splicing. Evidence from global RNA sequencing indicates that alternative splicing is widespread and crucial for the proper function and fate of a cell [5–8]. In addition to alternative exon inclusion, the recognition and splicing of constitutive exons that are always present in the mature mRNA is regulated. Regulation of constitutive exon splicing can result in retained introns, which can trigger degradation of the mRNA presumably through nonsense-mediated decay [9]. This latter category is the type of alternative splicing that regulates expression of G6PD [3]. Alternative splicing and intron retention share regulatory mechanisms that involve the binding of splicing regulatory proteins to cis-acting elements within the mRNA. Within exons, these sequences are called exonic splicing enhancers (ESE) or exonic splicing silencers (ESS). Two protein families that include serine-arginine rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) are known to function as trans-acting factors, which regulate splicing through binding to the ESE and ESS elements, respectively [10–15]. As such, these proteins contain RNA recognition motifs by which they can bind to the pre-mRNA.
The mechanisms by which SR proteins and hnRNPs respond to environmental and cellular cues are only now emerging. Serine phosphorylation of SR proteins is a primary mechanism regulating their activity, and phosphorylation is associated with enhanced splicing of the SR protein targets [16]. In contrast, hnRNPs act both as enhancers or inhibitors of mRNA splicing as well as regulate other steps in gene expression [17–19]. Many hnRNPs can be phosphorylated, methylated on arginines and modified by sumoylation [20, 21]. Thus, hnRNPs could be candidates for nutritional regulation of splicing.
The observation that nutritional status can regulate RNA splicing has widespread implications for regulation of cellular function. G6PD provides a useful model to study splicing changes that occur in response to nutritional status because this is the sole mechanism regulating the accumulation of its mRNA. In mouse liver, accumulation of spliced G6PD mRNA occurs following consumption of a high carbohydrate, low-fat diet. Conversely, the amount of mature mRNA is decreased significantly by starvation or consumption of a diet high in polyunsaturated fatty acids [3, 22, 23]. The decrease in splicing of the nascent transcript is associated with a decrease in the removal of introns surrounding exon 12 of the G6PD mRNA and G6PD pre-mRNA containing these retained introns accumulates prior to its degradation [23]. Exon 12 contains the regulatory elements that are necessary for the nutrient regulation of splicing, which include both an ESS and an ESE [3, 24]. The SR protein, SR splicing factor 3 (SRSF3) binds to the ESE within exon 12, thus increasing splicing of G6PD pre-mRNA during refeeding [24]. The increase in splicing of G6PD pre-mRNA results in accumulation of mature mRNA and ultimately more G6PD enzyme activity. The question remains, is inefficient splicing of exon 12 the default state to which liver reverts during starvation? In addition to SRSF3, we identified 3 members of the hnRNP family of proteins, K, L and A2/B1 that bind to this element in vitro [25]. In the present report, we test the hypothesis that hnRNP K, L and/or A2/B1 inhibit splicing of the G6PD mRNA and are involved in the inhibition of G6PD expression during starvation. We found that expression of hnRNP K increased during starvation, which resulted in an increase in its binding to the ESS with exon 12. HnRNP K inhibited splicing of the G6PD nascent transcript as well as a splicing reporter that contains the exon 12 regulatory element. Conversely, siRNA-mediated depletion of hnRNP K resulted in increased splicing of the G6PD pre-mRNA. Finally, hnRNP K and SRSF3 bind to the same sequences within the regulatory element and do so in a mutually exclusive manner. We propose that hnRNP K is a nutrient-regulated silencer of RNA splicing.
2. MATERIAL AND METHODS
All animal experiments were conducted in conformity with the Public Health Service policy on Human Care and Use of Laboratory Animals, additionally the Institutional Animal Care and Use Committee of the Division of Laboratory Animal Resources at West Virginia University approved all experimental procedures.
2.1. RNA Electrophoretic Mobility Shift Assay (EMSA)
The RNA EMSA protocol is a modification of existing methods [26, 27]. Briefly, 100 fmol of a 5’-hexachlorofluorescein (HEX™) labelled-RNA probe (IDT) corresponding to nucleotides (nt) 50–84 of Exon 12 was mixed with 92 ng of purified recombinant FLAG-tagged hnRNP K (Origene) in 1× binding buffer (10 mM Tris pH 7.3, 1 mM MgCl2, 20 mM KCl, 1 mM DTT, 1 U of SuperRNasin (Ambion)) plus/minus unlabeled competitor oligonucleotides (0, 1 pmol, 2.5 pmol, 5 pmol, 7.5pmol, 10 pmol; IDT) in a total reaction volume of 20 µL. The reactions were incubated for 30 min at room temperature. Supershift reactions received 1 µg of anti-hnRNP K (3C2, Abcam) 20 min into the initial binding reaction and the reaction proceeded another 10 min. The samples were loaded onto a pre-running 5% native polyacrylamide gel. The gel was imaged directly on a Typhoon 9410 Imager and signals were quantified using ImageQuant TL software. RNA EMSAs involving competition between hnRNP K and SRSF3 were incubated in the conditions as previously described [24]. Purified recombinant FLAG-tagged hnRNP K (Origene) and purified recombinant glutathione S-transferase (GST) conjugated- SRSF3 (Abnova) were incubated in 1× binding buffer (10 mM Tris pH 7.5, 1 mM MgCl2, 100 mM KCl, 0.1 mM DTT, 5% glycerol; ref. 25) in a total reaction volume of 20 µL with 100 fmol of a 5’-hexachlorofluorescein (HEX™) labelled-RNA probe (IDT) corresponding to nt 50–84 of Exon 12. Reactions were incubated and analyzed as previously described [24].
2.2. Western analysis
Whole cell lysates [24] and nuclear extracts [25] were prepared as described. After gel electrophoresis, the proteins were transferred to PDVF membrane (Bio-Rad), and probed with the antibodies as indicated in the figure legends. HnRNP A2/B1, hnRNP L and hnRNP K specific antibodies (Santa Cruz Biotechnology) and β-tubulin antibody (Cell Signalling) were obtained from the indicated sources. Secondary antibodies were conjugated to horseradish peroxidase and the antibody interactions were detected using ECL plus (GE Healthcare) followed by visualization on film and a Typhoon 9410 Imager (GE Healthcare). Signals were quantified with ImageJ (NIH) and ImageQuant TL (Molecular Dynamics), respectively. To verify the accuracy of the protein quantitation in the nuclear extracts, the extracts were run on a polyacrylamide gel and silver stained. The overall intensity of the visualized bands was similar betweens starved and refed samples (data not shown).
2.3. RNA isolation and measurement
Total cellular RNA was isolated using TRI Reagent® (Molecular Research Center). The total RNA was then digested with DNase I (Turbo DNA-free, Invitrogen) according to the manufacture’s protocol. Specific RNA amounts were quantified by RT-qPCR (ICYCLER, Bio-Rad) using the QuantiTect SYBR Green kit (Qiagen) and primers listed in supplemental Table 1. All reactions were carried out in duplicate and annealing temperatures for each set of primers were determined by using melting curves. The amount of each mRNA was calculated using a relative standard curve. All RNA samples were also amplified in the absence of reverse transcriptase to test for DNA contamination. Samples with DNA contamination were digested with DNase I a second time.
2.4. HnRNP K overexpression and RNA reporters
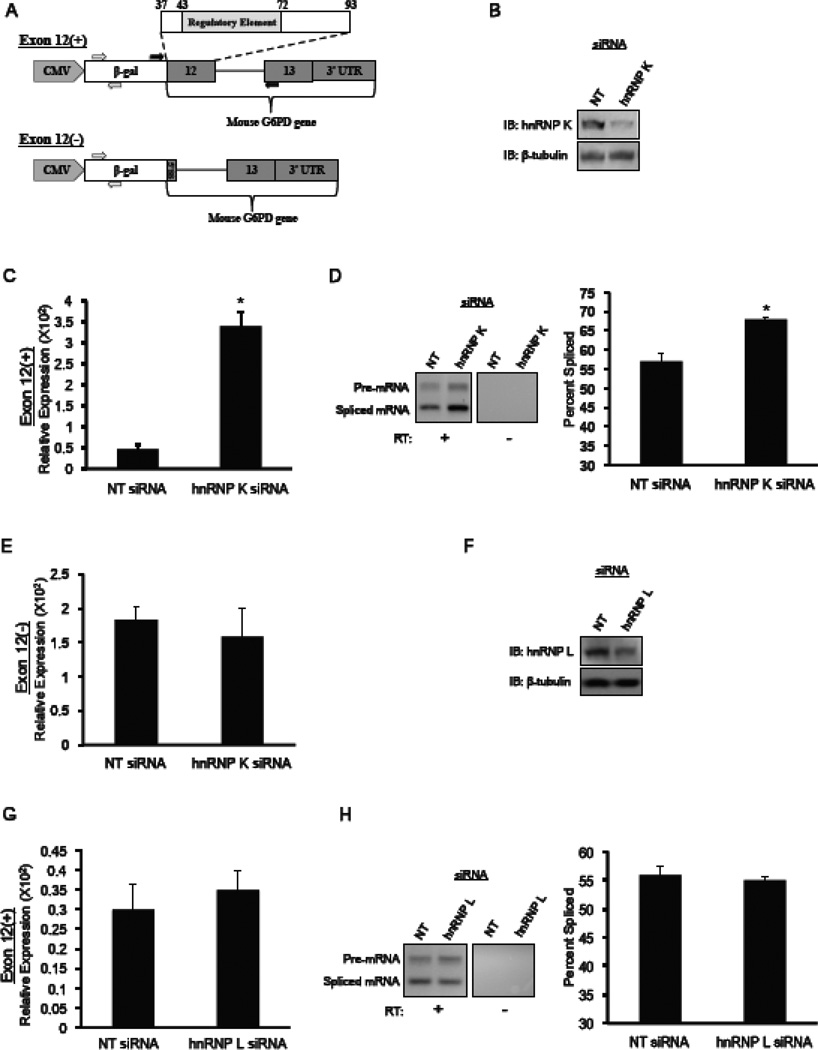
HepG2 (human hepatoma) cells were generated that stably express the RNA splicing reporters: Exon 12(+) or Exon 12(−) (Fig. 2) and were used for experiments testing the effect of overexpressing hnRNP K. The Exon 12(+) reporter contains mouse genomic DNA comprising nt 38–93 of exon 12 through the end of the G6PD gene as departed by a repetitive element, ligated to β-galactosidase and expression was driven by the CMV promoter. The cells were co-transfected with a plasmid expressing the neomycin resistance gene. Stable transformants were selected with G418 and then clonally isolated so that all cells reflect a single site of insertion of the RNA reporter. The Exon 12(−) reporter was constructed as the Exon 12(+) reporter but contains only the last 8 nt of exon 12 to preserve the 5’ splice site of the intron. One day after plating 2 × 105 cells in a 6 well plate, HepG2 cells were transfected with 3 µg of an expression vector for hnRNP K (Origene) or a control plasmid that expressed red fluorescent protein (RFP; pEGFP-N1, Clontech with RFP substituted for GFP; gift of Peter Stoilov) using the Express-in transfection reagent (Thermo Scientific) in DMEM containing 10% FBS. Total RNA and whole-cell protein extracts were collected after 48 h. Cell lysates were prepared using RIPA buffer plus phosphatase and protease inhibitors (Thermo Scientific). The quantitation of spliced versus unspliced reporter RNA used RT-qPCR and primers that produced 218 bp and 119 bp products, respectively.
Figure 2.
Depletion of hnRNP K by siRNA causes an increase in the splicing of G6PD mRNA. HepG2 cells stably expressing the G6PD reporter shown in (A) were transfected with 100nM siRNA against hnRNP K, hnRNP L, or a non-targeting (NT) control. After 48 hours, RNA and protein were isolated. The Exon 12(+) reporter contains mouse genomic DNA from exon 12 (nt 37–93), intron 12, and exon 13 including the entire 3’-untranslated region (3’-UTR) of the G6PD gene ligated to β-galactosidase (β-gal) and transcription is driven by the CMV promoter. The open arrows indicate the location of the primers used to detect overall reporter expression. The closed arrows represent primers used to detect the splicing of the reporter. The Exon 12(−) vector also contains mouse genomic DNA ligated to β-galactosidase but contains only the last 8 nt of exon 12 to preserve the 5’splice site (5’SS) of intron 12. (B) A representative immunoblot of hnRNP K depletion in HepG2 cells is shown. (C) Expression of mRNA produced by the Exon 12(+) reporter in HepG2 cells that were transfected with the indicated siRNA was measured with the primers to the β-galactosidase sequence (open arrows). The data are normalized to the amount of GAPDH mRNA. The data are the mean ± SE of n=6 independent siRNA knockdowns. (D) Following knockdowns with the indicated siRNA, splicing of the reporter RNA was with the primers indicated by black arrows. The products of a representative RT-PCR are shown. Samples were also amplified without prior treatment with reverse transcriptase (RT) to test for DNA contamination. The percentage of mRNA that was spliced is calculated as the intensity of the spliced band over the combined intensities of the bands for spliced and unspliced RNA. The data represent the mean ± SE of n=6 independent siRNA transfections. (E) Expression of the Exon 12(−) reporter in HepG2 cells that were transfected with the indicated siRNAs was measured by RT-qPCR and primers to the β-galactosidase portion of the reporter RNA. The amount of reporter mRNA is expressed relative to the amount of GAPDH mRNA. Each bar represents the mean ± SE n=3 independent siRNA transfections. (F–H) Knockdown of hnRNP L and measurement of Exon 12(+) reporter expression and splicing were carried out as described for hnRNP L. Each bar represents the mean ± SE n=4 independent siRNA transfections. The “*” symbol indicates a significant difference (p<0.05).
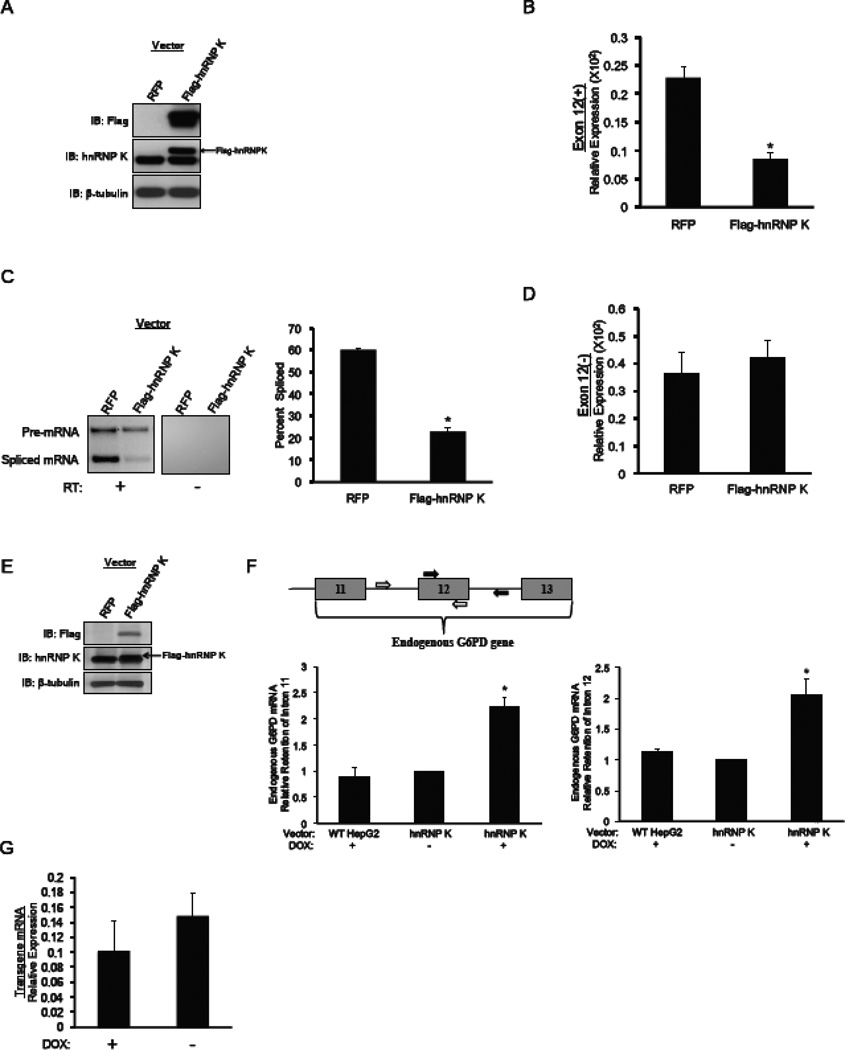
To test for effects of hnRNP K on mRNA stability apart from splicing, HepG2 cells stably expressing a doxycycline inducible hnRNP K (section 2.5) were treated with doxycycline for 72 h. During the last 24 h of doxycycline treatment, the cells were transiently transfected with 2 µg of reporter plasmid that contains the CMV promoter, the mouse G6PD cDNA, and the entire 3’-end of the G6PD gene [23]. Transfection was performed as described above and RNA and protein extracts were prepared 24 h after transfection. The accumulation of mouse G6PD mRNA produced by the reporter was measured with RT-qPCR and primers to exon 13 of the mouse RNA (supplemental Table 1) that differs significantly from the endogenous human G6PD mRNA.
2.5. Isolation of chromatin-associated RNA
The isolation of the chromatin fraction of nuclei and of the RNA in this fraction was by an existing protocol [28]. HepG2 cells were derived that stably express hnRNP K under a doxycycline inducible promoter. Cells (6.5 × 106) were plated in a 150mm plate with DMEM containing 10% FBS. Doxycycline (5 µg/ml) was added to the cells one day after plating. Wild type HepG2 cells were treated in the identical manner and were a control for non-specific effects of doxycycline. After 72 h with doxycycline, chromatin-associated RNA was collected. The quantitation of spliced versus unspliced RNA used RT-qPCR with primers to the endogenous G6PD mRNA (Fig. 3E). Primers detecting pre-mRNA retaining intron 11 produced a 156 bp product and primers that detected pre-mRNA retaining intron 12 produced a 168 bp product (supplemental Table 1).
Figure 3.
Ectopic expression of hnRNP K decreases the splicing of G6PD mRNA. HepG2 cells that stably express either the Exon 12(+) reporter or Exon 12(−) were transfected with a construct containing Myc-FLAG-tagged hnRNP K. After 48 hours, RNA and protein were isolated from the cells and the effects of ectopically expressing hnRNP K (Myc-FLAG-hnRNP K) or the control (RFP) on reporter splicing and expression were measured. (A) A representative immunoblot of the ectopic expression of Myc-FLAG-hnRNP K is shown. Ectopically expressed hnRNP K was detected with FLAG-specific antibody. An hnRNP K specific antibody was used to detect both the endogenous (lower band) and ectopically expressed (upper band) hnRNP K proteins. (B) Expression of the Exon 12(+) reporter in HepG2 cells that were transfected with the indicated vectors was measured by RT-qPCR and primers to the β-galactosidase portion of the RNA (see Fig. 2A). The amount of reporter mRNA is expressed relative to the amount of GAPDH mRNA, which is not regulated by these treatments. Each bar is the mean ± SE of n=3 independent transfections. (C) Splicing of the Exon 12(+) reporter was measured by RT-PCR and the primers indicated in Fig. 2A followed by separation of the products on a 2% agarose gel. A representative gel is shown. Samples were also amplified without prior treatment with reverse transcriptase (RT) to test for DNA contamination. The percentage of mRNA that was spliced is calculated as the intensity of the spliced band over the combined intensities of the bands for spliced and unspliced RNA. The data are the mean ± SE of n=3 independent hnRNP K transfections. (D) Expression of the Exon 12(−) reporter in HepG2 cells that were transfected with the indicated vectors was measured by RT-qPCR and primers to the β-galactosidase portion of the RNA. The amount of reporter mRNA is expressed relative to the amount of GAPDH mRNA. Each bar is the mean ± SE of n=3 independent hnRNP K transfections. (E) HepG2 cells that stably express doxycycline-inducible FLAG-hnRNP K were treated with doxycycline (Dox) to increase the amount of hnRNP K protein. The arrow indicates the FLAG-tagged hnRNP K detected with the hnRNP K antibody. (F) The primers used for RT-qPCR are indicated on the gene diagram. Chromatin-associated RNA that represents newly transcribed RNA was isolated and the amount of endogenous G6PD mRNA was measured by RT-qPCR. The data are expressed as the amount of mRNA containing intron 11 (graph on left) or intron 12 (graph on right) relative to the amount of cyclophilin mRNA. The data are the mean ± SE of n=3 independent experiments. The “*” symbol indicates a significant difference (p<0.05). The increase in hnRNP K expression by doxycycline is shown. (G) Following induction of hnRNP K expression with doxycycline (Dox), the HepG2 cells were transiently transfected with a reporter expressing the mouse G6PD cDNA as described in Materials and Methods. RNA was isolated 24 h later and mRNA produced by the reporter was measured by RT-pPCR and primers specific for the mouse mRNA. The data are expressed relative to the amount of GAPDH mRNA and are the mean ± SE of n=5 independent transfections.
2.6. SiRNA transfection
HepG2 cells stably expressing the RNA splicing reporter Exon 12(+) or Exon 12(−) were used for siRNA depletion of hnRNPs. One day after plating, the HepG2 cells (3.5 × 105) were transfected with a mixture of 2 siRNAs (Thermo Scientific) against hnRNP K or hnRNP L (50 nM each siRNA; supplemental Table 1). Transfections were performed with TransIT-siQUEST® transfection reagent (Mirus) in DMEM medium containing 10% FBS. SiGENOME SMART non-targeting siRNA pool #1 was used as a negative control (Thermo Scientific). Total RNA and whole-cell lysates were collected after 48h.
2.7. RNA Immunoprecipitation (RIP)
The RIP protocol is a modification of existing methods [29, 30]. For HepG2 cells, the formaldehyde crosslinking and cell lysis were performed as previously described [24]. Crosslinking was stopped by the addition of glycine to 125 mM. Following the cell lysis, lysate containing 500 µg of protein was suspended in buffer (1× PBS, 2× Halt™ Protease and Phosphatase Inhibitor Cocktail and 2U/uL SUPERaseIn™RNAse Inhibitor) and pre-cleared with protein G Dynabeads at 4°C for 2 h with rotation. A portion of the lysate was saved for the “input” sample. In a separate tube, protein G Dynabeads (Invitrogen) were washed twice with 1× PBS + 0.02% Tween 20 (PBS/T). Antibodies (10 µg, hnRNP K (3C2), Abcam and normal mouse IgG, Millipore) were added to the beads and incubated at room temperature for 2 h with rotation. The antibody-bound beads were washed three times with PBS/T and then mixed with the pre-cleared lysates. The bead/lysate mixtures were incubated overnight at 4°C with rotation and then washed six times with PBS/T. RNA bound to the immunoprecipitated proteins was released by treatment with proteinase K as previously described [24] and amplified by RT-qPCR using the primers in supplemental Table 1. Amplification without RT was used as a control for chromatin contamination.
For detection of hnRNP K binding to the regulatory element in whole liver, 8–10 week old male C57BL/6J (The Jackson Laboratory) were fasted for 18h or were fasted for 18h, and then refed a low-fat, high-carbohydrate diet (Basal Mix TD.00235, Harlan Laboratories; supplemented with 1% safflower oil) for 12 h. The livers were removed and processed as previously described [24]. All subsequent immunoprecipitations and amplifications of bound RNA were done as described for HepG2 cells.
2.8. Statistical Analysis
All statistical comparisons were made using the student’s t-test. Significance is defined as p<0.05.
3. RESULTS
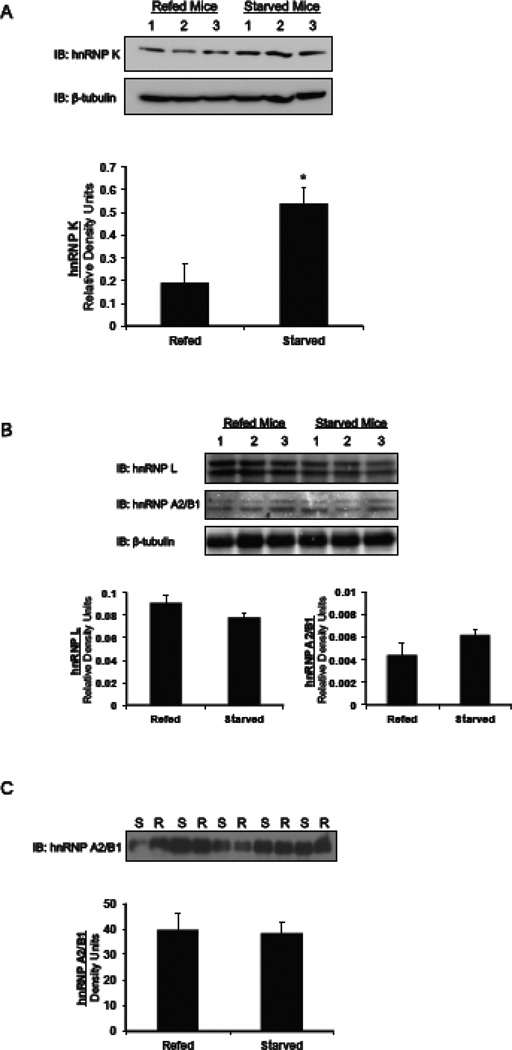
3.1. Starvation increases hnRNP K protein levels in vivo
To address the hypothesis that hnRNPs are starvation-induced splicing silencers, we measured the change in their expression in the livers of mice subjected to mild starvation. Starvation caused a 3-fold increase in the expression of hnRNP K protein as compared mice that were refed a high carbohydrate diet after a period of starvation (Fig. 1A). Antibodies to hnRNP L and hnRNP A2/B1 each detect 2 bands for the respective proteins. The reason for the doublet is not known. Regardless, expression of hnRNP L and A2/B1 did not change between starved and refed livers (Fig. 1B). Because hnRNP A2/B1 shuttles from the nucleus to the cytoplasm [31], we specifically measured its abundance in nuclear extracts to reflect the cellular pool in which the amount of this protein would be expected to change. Again, there was no change in the expression of hnRNP A2/B1 in the nucleus comparing the expression in 10 different mice (Fig. 1C). To control for protein quantitation, polyacrylamide gels of the nuclear extracts were silver-stained; overall protein abundance was similar between the dietary treatments. Thus abundance of hnRNP K is increased by a dietary paradigm that results in decreased splicing of the G6PD mRNA and decreased expression of the G6PD enzyme [3, 22, 23]. The fact that hnRNP A2/B1 does not change in abundance during starvation coupled with our previous data that hnRNP A2/B1 binds to multiple exons in the G6PD gene (Griffith and Salati, data not shown) argue that it does not play a major role in starvation induced silencing of G6PD expression. Thus, we have not pursued further experiments with this protein.
Figure 1.
Starvation causes an increase in the amount of hnRNP K protein in mice livers. Mice were either starved for 18 h or starved for 18 h and then refed a high carbohydrate diet for 12 h and protein extracts were made from the mouse livers as described in the materials and methods. (A) A western blot shows the amount of hnRNP K in the livers of mice. Each lane represents an individual mouse liver. Quantification of the changes in the amount of hnRNP K protein is shown. The amount of hnRNP K protein was normalized to the amount of β-tubulin. The data are the mean ± SE of n=3 mice per group. (B) A western blot shows the amounts of hnRNP L and A2/B1 in the livers of mice. Each lane represents an individual mouse liver. These antibodies detect 2 bands for each of these proteins. The reason for the doublet is not apparent. Quantification of the changes in the amount of hnRNP L and A2/B1 protein is shown. The amount of hnRNP L and A2/B1 protein was normalized to the amount of β-tubulin. The data are the mean ± SE of n=3 mice per group. (C) Nuclear extracts were prepared from mice that were starved or starved then refed and immunoblotted for hnRNP A2/B1. Each lane represents an individual mouse liver. Quantification of the changes in the amount of hnRNP A2/B1 protein was performed on the Typhoon imager and is shown. The data are the mean ± SE of n=4 mice per group. S, starved; R, refed. The “*” symbol indicated a significant difference (p<0.05).
3.2. SiRNA depletion of hnRNP K but not hnRNP L increases the splicing of G6PD
SiRNA-mediated depletions of hnRNP K and hnRNP L were used to test their functional significance on the expression of G6PD mRNA. Two HepG2 cell lines were developed that stably expressed splicing reporters either with (+) or without (−) exon 12 ligated to the heterologous gene, β-galactosidase. The Exon 12(+) reporter permits the evaluation of changes in the splicing and the overall expression of the reporter mRNA driven solely by the splicing regulatory element within exon 12 and not affected by regulatory elements in other exons of the G6PD mRNA (Fig. 2A). While it only contains one of the introns surrounding exon 12, the splicing and expression of the Exon 12(+) reporter was regulated by insulin and fatty acids in an identical fashion to the endogenous gene following transfection into primary rat hepatocytes [3, 23]. Knockdown of hnRNP K with siRNA resulted in a 69 ± 1% depletion of hnRNP K protein (Fig. 2B). The depletion of hnRNP K was accompanied by a 7-fold or more increase in the amount of RNA produced by the Exon 12(+) reporter (Fig. 2C). This measurement does not discriminate between spliced and unspliced RNA and, thus this increase reflects primarily RNA that has been spliced, as the unspliced RNA is ultimately degraded. The overall increase in the accumulation of the reporter mRNA correlated with a significant increase in the amount of splicing of mRNA from the Exon 12(+) reporter, indicating that RNA splicing was no longer being silenced (Fig 2D). The increase in reporter RNA accumulation over and above what can be attributed to a change in splicing ratio may reflect that the less efficiently spliced RNA was being degraded in the cell without hnRNP K knockdown. HepG2 cells that stably express the Exon 12(−) reporter were used as a negative control to show that the regulatory element in exon 12 is required for this regulation. As expected, the depletion of hnRNP K did not alter the accumulation of mRNA expressed from the Exon 12(−) reporter (Fig 2E). SiRNA against hnRNP L caused a 55 ± 6% depletion of hnRNP L but no change in the expression or splicing of the reporter RNA (Fig. 2 F–H). Depletion of hnRNP L to amounts greater than this compromised cellular viability. The absence of a change in hnRNP L amount by nutritional status and the lack of an effect of hnRNP L depletion on G6PD splicing suggest that hnRNP L per se is not a required splicing regulator albeit it may play a regulatory role in combination of that provided by hnRNP K. These data support a hypothesis whereby hnRNP K inhibits splicing of G6PD mRNA and that the exon 12 regulatory element is involved in this response.
3.3. Overexpression of hnRNP K decreases the splicing of G6PD mRNA
A gain-of–function approach was used to test if hnRNP K inhibits splicing of G6PD mRNA in cells. Overexpression of hnRNP K was performed in the HepG2 cells that stably express the G6PD reporters, Exon 12(+) and Exon 12(−) (Fig. 2A). Transfection of HepG2 cells with a vector that results in expression of FLAG-hnRNP K caused a 53 ± 4% increase in hnRNP K protein over endogenous levels (Fig. 3A). The increase in hnRNP K protein levels caused a 60% decrease in the expression of mRNA from the Exon 12(+) reporter as compared to control cells that were transfected with a vector expressing an unrelated protein, RFP (Fig. 3B). To test for changes in splicing that could explain the decrease in expression of mRNA from the Exon 12(+) reporter, primers were designed that would amplify exon 12, intron 12, and exon 13 (Fig. 2A, black arrows). Amplification of the mRNA by RT-PCR with these primers results in 2 products representing pre-mRNA and spliced mRNA. After ectopic expression of hnRNP K, the extent of splicing of mRNA from the Exon 12(+) reporter decreased 80% when compared to control cells transfected with the vector expressing RFP (Fig. 3C). Overexpression of hnRNP K in HepG2 cells that express the Exon 12(−) reporter did not alter the expression of Exon 12(−) reporter mRNA (Fig. 3D). Thus, even small increases in hnRNP K can inhibit splicing and the regulatory element within exon 12 is involved in the inhibition of splicing by hnRNP K.
We next asked if overexpression of hnRNP K could decrease splicing of the endogenous G6PD gene. A clonal derivative of HepG2 cells was developed that ectopically expresses hnRNP K under the control of a doxycycline inducible promoter. Incubation of the HepG2 cells with doxycycline caused a 40 ± 4% increase in the expression of hnRNP K (Fig. 3E). Splicing of the endogenous mRNA could not be measured in total RNA isolates. Thus, chromatin associated RNA was isolated to enrich the samples in newly transcribed RNA. RT-PCR and primers that resulted in amplification across the exon-intron boundaries surrounding exon 12 was used to measure the amount of endogenous G6PD pre-mRNA. Following the induction of hnRNP K expression, G6PD mRNA retaining intron 11 and intron 12 increased by 2-fold or more compared to the clonal HepG2 cells without doxycycline treatment or wild type HepG2 cells treated with doxycycline (Fig. 3F). The presence of these retained introns indicates a decrease in splicing efficiency.
A final possibility is that hnRNP K action involves stabilization of the mRNA and this could confound the interpretation of splicing data. To test for mRNA stabilization apart from splicing, the HepG2 cells were induced to express hnRNP K by doxycycline treatment. The cells were transfected with an RNA reporter that contained the G6PD cDNA as well as the G6PD 3’-UTR through the end of the gene. As stability elements can be within 3’-UTR, this plasmid would contain any such potential elements; however, the plasmid does not have introns from the G6PD gene and thus would not undergo exon 12-directed splicing. Following doxycycline treatment, FLAG-hnRNP K expression was increased 136 ± 4 - fold (n=5; blot not shown), but expression of mRNA from the cDNA reporter was not changed (Fig. 3G). Thus, hnRNP K does not regulate stability of mRNA containing exon 12. Together, these data demonstrate that hnRNP K is involved in the inhibition of splicing of G6PD mRNA.
3.4. HnRNP K binds to the exon 12 regulatory element in cells
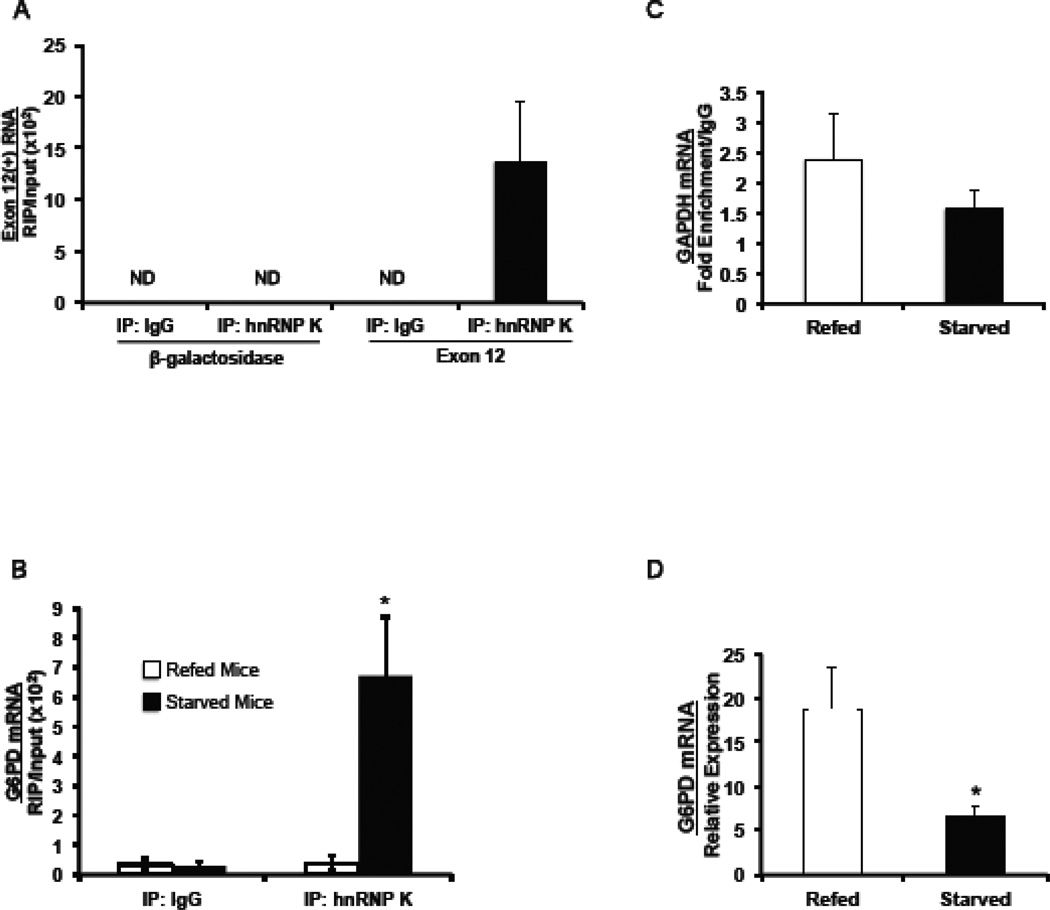
Our previously reported RNA affinity assays demonstrated that hnRNP K can bind to the splicing regulatory element in vitro [25]. To determine if hnRNP K binds to the regulatory element in intact cells, we subjected the cells to chemical crosslinking and immunoprecipitated hnRNP K. RNA crosslinked to hnRNP K was detected by RT-qPCR using primers that amplify exon 12 RNA produced by the Exon 12(+) reporter. Because the reporter contains mouse sequences, reporter RNA can be amplified without interference of the endogenous gene. In HepG2 cells expressing the Exon 12(+) reporter, hnRNP K binding to the regulatory element was enriched 14-fold or more as compared to the IgG control (Fig. 4A). This result was not due to incompletely fragmented RNA, as primers to the β-galactosidase portion of the mRNA did not amplify RNA in the immunoprecipitate. Thus hnRNP K is binding to the exon 12 regulatory element in intact cells.
Figure 4.
HnRNP K binds to exon 12 of the G6PD mRNA and in mouse liver; the binding is enhanced by starvation. (A) HepG2 cells expressing the Exon 12(+) reporter were crosslinked with formaldehyde and hnRNP K was immunoprecipitated from the lysates. G6PD mRNA bound to hnRNP K was measured by RT-qPCR and primers, which amplify the region of the reporter RNA from the end of β-galactosidase to the end of exon 12. Immunoprecipitation with IgG was used as a control for non-specific binding. Immunoprecipitation with hnRNP K or IgG followed by RT-qPCR of an upstream region of β-galactosidase (open arrows Fig 2A) was used as a control for non-specific pull-down of reporter RNA. The amount of RNA detected is expressed relative to input RNA in each sample. The abbreviation “ND” represents not detectable. (B) Mice were either starved for 18 h or starved for 18 h and then refed a high carbohydrate diet for 12 h. Following the formaldehyde crosslinking of the livers, RNA bound to hnRNP K was precipitated using an antibody against hnRNP K, or IgG as a control. Equal amounts of protein from the crosslinked lysates were added to each immunoprecipitation. G6PD mRNA immunoprecipitated with hnRNP K or IgG was detected with RT-qPCR and primers nucleotides 18–89 of exon 12, which contains the regulatory element (nucleotides 43 to 72). The amount of detected RNA is expressed relative to the input RNA in each sample. The “*” symbol indicates a significant difference (p<0.05) between the IgG control and the hnRNP K RIP. The data are the mean ± SE of n=3 mice per group. (C) GAPDH mRNA immunoprecipitated by the hnRNP K antibody was measured by RT-qPCR using primers to GAPDH (nt 2281–2348) in order to assess specificity of hnRNP K binding. Data are expressed as fold increase in enrichment relative to the amount of IgG enrichment. (D) Total RNA was isolated from a portion of liver used in the RIP analysis. G6PD mRNA was measured using RT-qPCR as previously described. The data are the mean ± SE of n=3 mice per group. The “*” symbol indicates a significant difference (p<0.05).
3.5. Starvation enhances the binding of hnRNP K to the splicing regulatory element in vivo
In intact animals, starvation inhibits G6PD splicing and causes retention of introns surrounding exon 12 [23]. To test if hnRNP K interaction with the regulatory element is regulated by nutritional status in intact liver, we performed RIP using mice that had been starved or starved and then refed a high-carbohydrate diet. The livers were subjected to chemical crosslinking and the amount of hnRNP K bound to the exon 12 regulatory element was measured by RT-qPCR. Exon 12 RNA was enriched in the hnRNP K immunoprecipitates by 23-fold or more compared to the IgG control (Fig. 4B). Furthermore, starvation caused a 12.5-fold increase in hnRNP K bound to regulatory element compared to refed mice. Binding of hnRNP K in the livers of refed mice was barely detectable and was similar to the amounts detected following immunoprecipitation with IgG (Fig. 4B). To test if the starvation-induced increase in hnRNP K binding was specific to G6PD, RNA corresponding to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also measured. Expression of GAPDH is not regulated by starvation and refeeding [22, 32]. Starvation did not significantly alter the presence of GAPDH mRNA in hnRNP K immunoprecipitates and enrichment relatively to IgG was low in both dietary conditions (Fig. 4C). Our previous data has clearly demonstrated that decreased splicing of exon 12 is the primary mechanism by which starvation decreases G6PD mRNA abundance [3, 22, 23]. In this experiment, the increase in hnRNP K binding correlated with a 60% decrease in the abundance of G6PD mRNA in the same livers (Fig. 4D). In summary, hnRNP K inhibits the splicing of G6PD mRNA, it binds to the regulatory element in exon 12 of this RNA, and this binding is enhanced by starvation. Together these data are consistent with the hypothesis that hnRNP K is a starvation-induced splicing silencer.
3.6. HnRNP K binds to C-rich motifs within exon 12 of G6PD mRNA
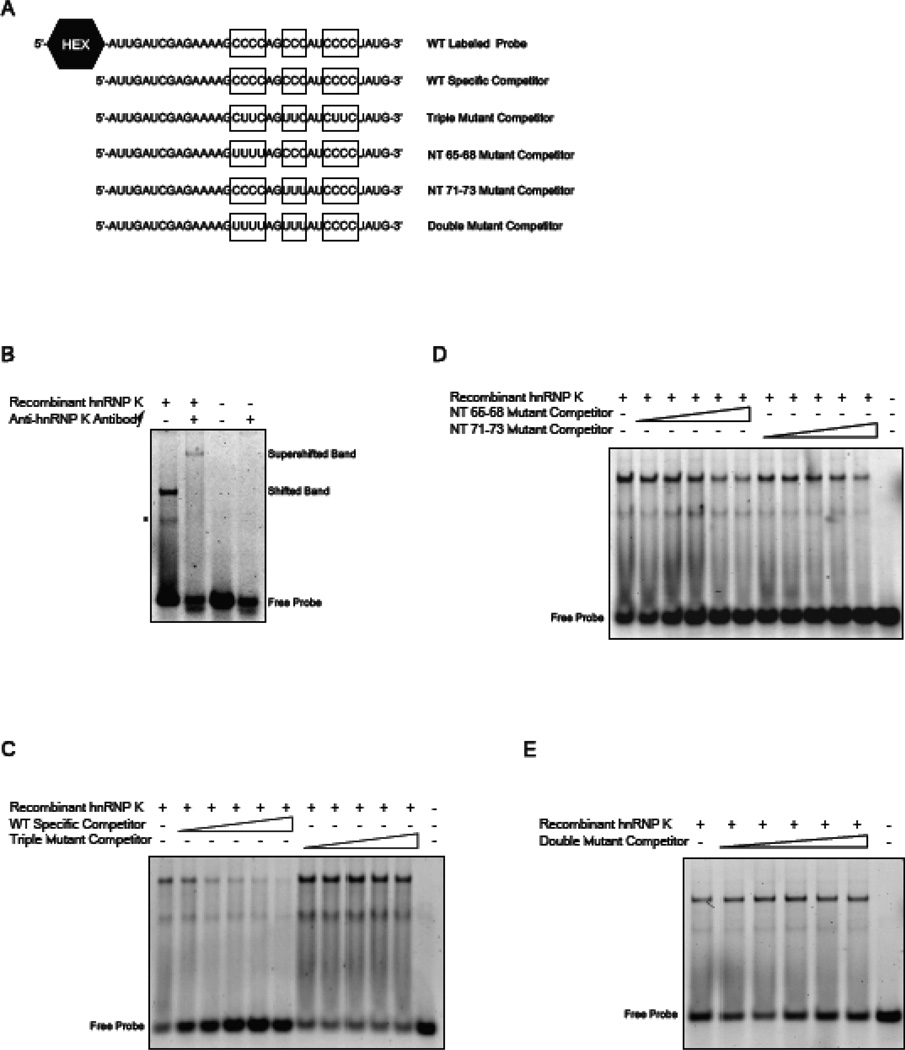
We have previously demonstrated the SRSF3 also binds to exon 12 of the G6PD mRNA and in contrast to hnRNP K, its binding is decreased by starvation and increased by refeeding [24]. HnRNP K is known to form complexes with other proteins and therefore we asked if hnRNP K per se could bind to G6PD RNA and if its binding site co-localized with the sequences required for SRSF3 binding. The splicing regulatory element in exon 12 of the G6PD mRNA encompasses nt 43–72 of the 93 nt exon and contains two C-rich sequences, which are known to be hnRNP K binding motifs [19, 33]. A third motif is located downstream of the splicing regulatory element; thus the entire region was used in RNA EMSAs to evaluate binding of hnRNP K to exon 12 (Fig. 5A). Incubation of a HEX-labelled probe representing nt 50 to 83 and recombinant hnRNP K protein resulted in two shifted bands (Fig. 5B, lane 1). The reason for 2 bands was not clear, thus a supershift was performed to determine which band represents the hnRNP K and RNA complex. The addition of hnRNP K specific antibody resulted in a supershift of both bands suggesting that they are protein:RNA complexes although this does not rigorously eliminate probe secondary structure or a peptide from hnRNP K degradation as a cause of the lower band (Fig. 5B, lane 2). Increasing concentrations of the WT specific competitor (Fig. 5A) were able to compete for binding of hnRNP K (Fig. 5C). In contrast, mutation of all three C-rich motifs within this RNA oligonucleotide abrogated binding to hnRNP K (92% decrease in the shifted band with the WT competitor versus a 12% decrease for the triple mutant at 100×; Fig. 5C). To further localize the sequences involved in binding, competitions were performed with oligonucleotides containing mutations of the C-rich motif at nt 65–68 or the C-rich motif at nt 71–73. Mutants missing only a single C-rich motif were able to reduce binding of hnRNP K to the HEX-labelled probe by 52% and 40% at 100×, respectively (Fig. 5D); however, when both of these C-rich motifs were mutated, the oligonucleotide failed to compete for hnRNP K binding (Fig. 5E). Thus the C-rich motif downstream of the regulatory element was unable to compensate for the loss of the C-rich motifs at nt 65–68 and 71–73, which is consistent with our functional data that defined the silencing element to the nt 43–72 region of exon 12 [3]. These results indicate that hnRNP K binds directly to the RNA and further the region between nt 65–73 co-localizes with the sequence to which SRSF3 binds.
Figure 5.
HnRNP K binds to C-rich sequences with in the regulatory element of G6PD mRNA. RNA EMSA assays were carried out using a fluorescently labelled probe, encompassing nt 50 to 83 of exon 12 and purified recombinant hnRNP K. (A) The sequence of the wild type (WT) labelled probe and the various competitor probes are shown. The boxes highlight the C-rich regions within exon 12 that have been mutated in the competitor oligonucleotides. (B) Purified hnRNP K protein was incubated with the WT labelled probe or the WT labelled probe plus an antibody specific for hnRNP K. A representative of two RNA EMSAs is shown. The asterisk represents a band of unknown origin that disappears upon supershift with hnRNP K antibody. (C) Competition for hnRNP K binding by the WT specific competitor, or the triple mutant competitor with the same results is shown. All competitor probes were added at 10×, 25×, 50×, 75×, and 100× molar excess. A representative of two RNA EMSAs is shown. (D) Competitors that contain mutations to a single C-rich motif (NT 65–68 or NT 71–73) were incubated with WT labelled probe and purified hnRNP K protein. (E) Competition for hnRNP K binding between the WT labelled probe and the double mutant competitor is shown.
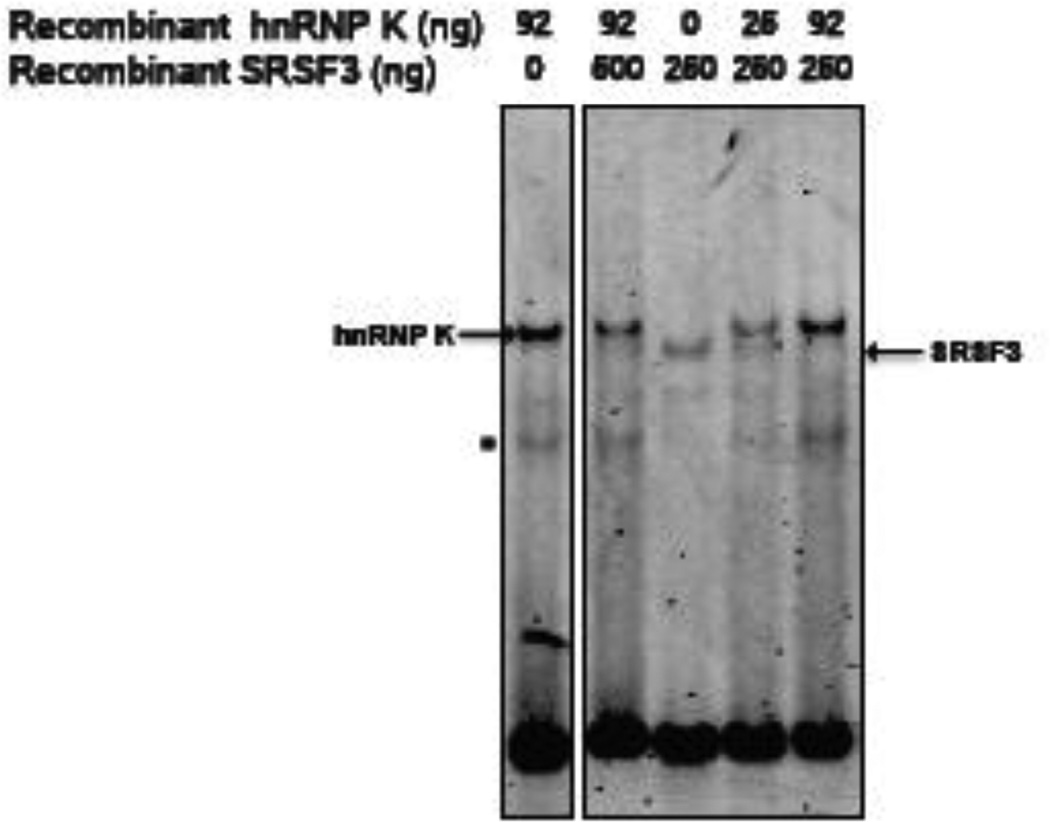
3.7. HnRNP K and SRSF3 compete for binding to the exon 12 regulatory element
SRSF3 binds to nt 62–72 of exon 12, which contains the first C-rich motif that was necessary for maximal hnRNP K binding [24]. RNA EMSAs were used to test for competition between hnRNP K and SRSF3 for binding to the regulatory element. Addition of SRSF3 to the binding reaction failed to decrease the binding of K to the HEX-labelled probe (Fig. 6, lanes 1 and 2). In contrast, even small amounts of hnRNP K effectively competed with SRSF3 for binding (Fig. 6, lanes 3,4 and 5). This is despite the fact that SRSF3 was present in 28 and 7-fold molar excess over hnRNP K. Thus, hnRNP K and SRSF3 binding appears to be mutually exclusive and the ability of hnRNP K to block SRSF3 binding suggests a dominant effect of hnRNP K on regulating splicing of G6PD pre-mRNA.
Figure 6.
Binding of hnRNP K and SRSF3 to the regulatory element is mutually exclusive. RNA EMSA assays were carried out using the fluorescently labelled probe, encompassing nt 50 to 83 of exon 12 (Fig. 5A). Purified recombinant hnRNP K protein and purified recombinant SRSF3 were incubated with the WT labelled probe with the amounts of protein indicated on the figure. A representative of two RNA EMSAs both with identical results is shown. The band corresponding to the protein:RNA complexes are indicated with arrows. The asterisk represents a band of unknown origin that disappears upon supershift with hnRNP K antibody.
4. DISCUSSION
The starved and fed states are regulatory challenges faced by all vertebrates. The need to switch between fuel storage and use occurs on a daily basis as the organism responds to limited periods of food consumption such as between meals or prolonged starvation during illness or lack of food availability. We have developed a body of evidence that demonstrates that the RNA splicing machinery is an intracellular target for regulation during starvation. The G6PD gene has provided a useful tool for these studies because starvation and refeeding result in large changes in the accumulation of G6PD mRNA and do so solely by regulating the rate of splicing of the G6PD transcript [34]. During starvation, decreased splicing of exon 12 results in the retention of the surrounding introns and degradation of the transcript. A cis-acting element in exon 12 is required for regulated splicing of G6PD pre-mRNA [3]. In this report, we demonstrate that hnRNP K is a starvation-induced splicing factor that binds to this cis-acting element and inhibits splicing of the G6PD pre-mRNA.
HnRNP K binds to the regulatory element via 2 C-rich motifs (Fig. 5). These C-rich binding sites are similar to other known binding sites that have been identified for hnRNP K by SELEX. Of note, a third C-rich motif just downstream of the nt 42–73 regulatory element did not bind hnRNP K effectively. Thus, hnRNP K binding does exhibit some selectivity. In contrast to hnRNP K, hnRNPs L and A2/B1 also bound to the regulatory element in RNA affinity assays [25] but failed to show regulation of G6PD expression or selectivity in further analysis. An interaction between these proteins and hnRNP K has been demonstrated and thus their appearance in the RNA affinity assay may only occur in vitro [35, 36]. Alternatively, they may play a role enhancing the action of hnRNP K or in combination with other splicing regulators. Nonetheless, a sharp change in splicing regulation was not seen with hnRNP L deletion alone to the extent observed with hnRNP K. Highlighting the role for hnRNP K in G6PD expression, an interaction between hnRNP K and the RNA was observed in both intact cells and liver tissue (Fig. 4). The binding of hnRNP K to the regulatory element in exon 12 is enhanced following starvation, and this increase in binding coincides with decreased accumulation of G6PD mRNA. In addition to the direct interaction between hnRNP K and the RNA, loss of function and gain of function experiments further highlight the role of hnRNP K in silencing splicing of G6PD pre-mRNA. While, hnRNP K was relatively abundant in the refed livers, our data with overexpression of hnRNP K in HepG2 cells demonstrate that even small increases in the amount of this protein in the cells, can cause changes in the splicing of G6PD mRNA (Fig. 3). Our results demonstrate that hnRNP K plays an important role in regulating splicing in response to nutritional stimuli.
The fact that expression of hnRNP K protein is increased following periods of starvation provides additional evidence for a regulatory role linking splicing changes to dietary cues. To our knowledge, this is the first evidence that hnRNP K expression is regulated by dietary changes in the liver. Nutritional status may regulate hnRNP K via the hormones, insulin and glucagon, which change in abundance between the starved and refed states. In this regard, diabetes is associated with an increase in hnRNP K expression in proximal renal tubular cells and insulin reverses this effect [37]. The regulation of hnRNP K in liver may involve a similar effect as starvation and diabetes both involve decreased insulin action. Because hnRNP K has other cellular targets such as inhibition of the production of the pro-apoptotic Bcl-xs splice isoform [38], as well as, enhancing the transcriptional activity of p53 [18, 39, 40], regulation of its activity by starvation may have multiple additional targets.
The changes observed in the expression of hnRNP K protein may only be partially responsible for the increase in hnRNP K binding to the G6PD regulatory element and the decrease in accumulation G6PD mRNA. HnRNP K is subject to various posttranslational modifications, including phosphorylation, sumoylation and methylation, which could play a role in regulating it actions [17–19, 21, 40–42]. Preliminary studies in primary rat hepatocytes did not show a change in either serine or tyrosine phosphorylation during insulin treatment (Cyphert and Salati, unpublished). We also tested for palmitoylation of hnRNP K, but none was observed (Cyphert and Salati, unpublished). An additional possibility is the recruitment of other splicing silencers. HnRNP L also bound to the regulatory element. While its absence from the cells did not enhance G6PD expression, its role may be complimented by other splicing regulatory factors, such as the hnRNP L-like protein [27]. Establishing these mechanisms are active areas for future research.
Strong evidence exists for a model whereby regulated splicing involves a competition between hnRNP K and SRSF3 for binding to the exon 12 regulatory element. Both proteins bind between nt 63 and 73 and the binding is regulated by starvation and refeeding (Fig. 5 and ref. [24]. In contrast with hnRNP K, SRSF3 binding in vivo is enhanced by refeeding and inhibited by starvation [24]. The binding of SRSF3 to the regulatory element in intact liver coincides with an increase in the splicing of the G6PD mRNA and the regulating splicing is dependent on the presence of exon 12 [3, 23]. These results support the concept of mutually exclusive binding of these regulatory factors as a mechanism to control splicing of the exon. This hypothesis is also consistent with the observed competition between SRSF3 and splicing silencers in the regulation of the alternative splicing of the insulin receptor [43]. With respect to G6PD, hnRNP K appears to play a dominant role inhibiting splicing. HnRNP K competes against even molar excesses of SRSF3 for binding to the regulatory element (Fig. 6). Effective competition by SRSF3 may require phosphorylation in response to stimulatory signals. G6PD expression in liver is very low until stimulated to increase by insulin or a high carbohydrate diet [22, 44]. Competition between hnRNP K and SRSF3 may regulate G6PD expression beyond the requirements for energy homeostasis. In this regard, G6PD expression is also elevated in liver tumors as compared to normal liver tissue [45]. HnRNP K amount is decreased in tumors as compared to normal liver [45]; in contrast, SRSF3 is increased in human tumors and rat hepatoma cells and it abundance is very low in normal liver and in rat hepatocytes (Jia, 2010 and Cyphert and Salati, unpublished).
In conclusion, hnRNP K joins SRSF3 as a nutrient regulated splicing factor. In addition, it establishes a role for hnRNP K for regulation of splicing via intron retention. Nutrient regulated splicing is likely occurring with other metabolic genes and the understanding of how these inhibitory and enhancing factors interact is essential for understanding how nutrients regulate metabolic processes.
Supplementary Material
Highlights.
The expression of hnRNP K is regulated by nutritional status.
HnRNP K inhibits the splicing and expression of G6PD mRNA during starvation.
Starvation increases binding of hnRNP K to a cytosine-rich element in G6PD mRNA.
HnRNP K competes with SRSF3 for binding to the cytosine-rich element in exon 12.
ACKNOWLEDGEMENTS
We thank Dr. Peter Stoilov for helpful discussions and critically reading the manuscript. We thank Joseph Greschner for help developing the protocols for RNA EMSA. We than Douglas Poe for help developing the immunoprecipitation protocol for hnRNP K. This work was supported by the National Institutes of Health, National Institute for Diabetes and Digestive and Kidney Diseases [DK46897 to L.M.S.] and an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences [2P20RR016477-13]. Funding for open access charge: National Institutes of Health [DK46897}.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CMV, cytomegalovirus; EMSA, electrophoretic mobility shift assay; ESE, exonic splicing enhancer; ESS, exonic splicing silencer; G6PD, glucose-6-phosphate dehydrogenase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, glutathione S-transferase; HEX™, hexachlorofluorescein; hnRNP, heterogeneous nuclear ribonucleoprotein; RFP, red fluorescent protein; RIP, RNA immunoprecipitation; SRSF3, serine-arginine rich, splicing factor 3
REFERENCES
- 1.Hillgartner FB, Salati LM, Goodridge AG. Physiological and molecular mechanisms involved in nutritional regulation of fatty acid synthesis. Physiol Rev. 1995;75:47–76. doi: 10.1152/physrev.1995.75.1.47. [DOI] [PubMed] [Google Scholar]
- 2.Salati LM, Amir-Ahmady B. Dietary regulation of expression of glucose-6-phosphate dehydrogenase. Annu Rev Nutr. 2001;21:121–140. doi: 10.1146/annurev.nutr.21.1.121. [DOI] [PubMed] [Google Scholar]
- 3.Szeszel-Fedorowicz W, Talukdar I, Griffith BN, Walsh CM, Salati LM. An exonic splicing silencer is involved in the regulated splicing of glucose 6-phosphate dehydrogenase mRNA. J Biol Chem. 2006;281:34146–34158. doi: 10.1074/jbc.M603825200. [DOI] [PubMed] [Google Scholar]
- 4.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 5.Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore MJ, Silver PA. Global analysis of mRNA splicing. RNA. 2008;14:197–203. doi: 10.1261/rna.868008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blencowe BJ, Ahmad S, Lee LJ. Current-generation high-throughput sequencing: deepening insights into mammalian transcriptomes. Genes Dev. 2009;23:1379–1386. doi: 10.1101/gad.1788009. [DOI] [PubMed] [Google Scholar]
- 8.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 9.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 10.Zahler AM, Damgaard CK, Kjems J, Caputi M. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J Biol Chem. 2004;279:10077–10084. doi: 10.1074/jbc.M312743200. [DOI] [PubMed] [Google Scholar]
- 11.Rothrock CR, House AE, Lynch KW. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashima T, Rao N, David CJ, Manley JL. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet. 2007;16:3149–3159. doi: 10.1093/hmg/ddm276. [DOI] [PubMed] [Google Scholar]
- 13.Chen CD, Kobayashi R, Helfman DM. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo S, Yamamoto N, Murakami T, Okumura M, Mayeda A, Imaizumi K. Tra2 beta, SF2/ASF and SRp30c modulate the function of an exonic splicing enhancer in exon 10 of tau pre-mRNA. Genes Cells. 2004;9:121–130. doi: 10.1111/j.1356-9597.2004.00709.x. [DOI] [PubMed] [Google Scholar]
- 15.Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 16.Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostareck-Lederer A, Ostareck DH, Cans C, Neubauer G, Bomsztyk K, Superti-Furga G, Hentze MW. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol Cell Biol. 2002;22:4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelisch F, Pozzi B, Risso G, Munoz MJ, Srebrow A. DNA damage-induced heterogeneous nuclear ribonucleoprotein K sumoylation regulates p53 transcriptional activation. J Biol Chem. 2012;287:30789–30799. doi: 10.1074/jbc.M112.390120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 21.Li T, Evdokimov E, Shen RF, Chao CC, Tekle E, Wang T, Stadtman ER, Yang DC, Chock PB. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc Natl Acad Sci U S A. 2004;101:8551–8556. doi: 10.1073/pnas.0402889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stabile LP, Hodge DL, Klautky SA, Salati LM. Posttranscriptional regulation of glucose-6-phosphate dehydrogenase by dietary polyunsaturated fat. Arch Biochem Biophys. 1996;332:269–279. doi: 10.1006/abbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- 23.Tao H, Szeszel-Fedorowicz W, Amir-Ahmady B, Gibson MA, Stabile LP, Salati LM. Inhibition of the splicing of glucose-6-phosphate dehydrogenase precursor mRNA by polyunsaturated fatty acids. J Biol Chem. 2002;277:31270–31278. doi: 10.1074/jbc.M203196200. [DOI] [PubMed] [Google Scholar]
- 24.Walsh CM, Suchanek AL, Cyphert TJ, Kohan AB, Szeszel-Fedorowicz W, Salati LM. Serine Arginine Splicing Factor 3 (SRSF3) is involved in enhanced splicing of glucose-6-phosphate dehydrogenase (G6PD) RNA in response to nutrients and hormones in liver. J Biol Chem. 2013;288:2816–2828. doi: 10.1074/jbc.M112.410803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffith BN, Walsh CM, Szeszel-Fedorowicz W, Timperman AT, Salati LM. Identification of hnRNPs K, L and A2/B1 as candidate proteins involved in the nutritional regulation of mRNA splicing. Biochim Biophys Acta. 2006;1759:552–561. doi: 10.1016/j.bbaexp.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motta-Mena LB, Smith SA, Mallory MJ, Jackson J, Wang J, Lynch KW. A disease-associated polymorphism alters splicing of the human CD45 phosphatase gene by disrupting combinatorial repression by heterogeneous nuclear ribonucleoproteins (hnRNPs) J Biol Chem. 2011;286:20043–20053. doi: 10.1074/jbc.M111.218727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selth LA, Gilbert C, Svejstrup JQ. RNA immunoprecipitation to determine RNA-protein associations in vivo. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5234. pdb prot5234. [DOI] [PubMed] [Google Scholar]
- 30.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 31.Cui H, Wu F, Sun Y, Fan G, Wang Q. Up-regulation and subcellular localization of hnRNP A2/B1 in the development of hepatocellular carcinoma. BMC Cancer. 2010;10:356. doi: 10.1186/1471-2407-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodge DL, Salati LM. Nutritional regulation of the glucose-6-phosphate dehydrogenase gene is mediated by a nuclear posttranscriptional mechanism. Arch Biochem Biophys. 1997;348:303–312. doi: 10.1006/abbi.1997.0373. [DOI] [PubMed] [Google Scholar]
- 33.Thisted T, Lyakhov DL, Liebhaber SA. Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and alphaCP-2KL, suggest Distinct modes of RNA recognition. J Biol Chem. 2001;276:17484–17496. doi: 10.1074/jbc.M010594200. [DOI] [PubMed] [Google Scholar]
- 34.Amir-Ahmady B, Salati LM. Regulation of the processing of glucose-6-phosphate dehydrogenase mRNA by nutritional status. J Biol Chem. 2001;276:10514–10523. doi: 10.1074/jbc.M010535200. [DOI] [PubMed] [Google Scholar]
- 35.Mikula M, Dzwonek A, Karczmarski J, Rubel T, Dadlez M, Wyrwicz LS, Bomsztyk K, Ostrowski J. Landscape of the hnRNP K protein-protein interactome. Proteomics. 2006;6:2395–2406. doi: 10.1002/pmic.200500632. [DOI] [PubMed] [Google Scholar]
- 36.Shnyreva M, Schullery DS, Suzuki H, Higaki Y, Bomsztyk K. Interaction of two multifunctional proteins. Heterogeneous nuclear ribonucleoprotein K and Y-box-binding protein. J Biol Chem. 2000;275:15498–15503. doi: 10.1074/jbc.275.20.15498. [DOI] [PubMed] [Google Scholar]
- 37.Wei CC, Zhang SL, Chen YW, Guo DF, Ingelfinger JR, Bomsztyk K, Chan JS. Heterogeneous nuclear ribonucleoprotein K modulates angiotensinogen gene expression in kidney cells. J Biol Chem. 2006;281:25344–25355. doi: 10.1074/jbc.M601945200. [DOI] [PubMed] [Google Scholar]
- 38.Revil T, Pelletier J, Toutant J, Cloutier A, Chabot B. Heterogeneous nuclear ribonucleoprotein K represses the production of pro-apoptotic Bcl-xS splice isoform. J Biol Chem. 2009;284:21458–21467. doi: 10.1074/jbc.M109.019711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moumen A, Masterson P, O'Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y, Zhou X, Liu N, Wang C, Zhang L, Mo W, Hu G. Arginine methylation of hnRNP K enhances p53 transcriptional activity. FEBS Lett. 2008;582:1761–1765. doi: 10.1016/j.febslet.2008.04.051. [DOI] [PubMed] [Google Scholar]
- 41.Habelhah H, Shah K, Huang L, Ostareck-Lederer A, Burlingame AL, Shokat KM, Hentze MW, Ronai Z. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat Cell Biol. 2001;3:325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 42.Ostareck-Lederer A, Ostareck DH, Rucknagel KP, Schierhorn A, Moritz B, Huttelmaier S, Flach N, Handoko L, Wahle E. Asymmetric arginine dimethylation of heterogeneous nuclear ribonucleoprotein K by protein-arginine methyltransferase 1 inhibits its interaction with c-Src. J Biol Chem. 2006;281:11115–11125. doi: 10.1074/jbc.M513053200. [DOI] [PubMed] [Google Scholar]
- 43.Sen S, Talukdar I, Webster NJ. SRp20 and CUG-BP1 modulate insulin receptor exon 11 alternative splicing. Mol Cell Biol. 2009;29:871–880. doi: 10.1128/MCB.01709-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stabile LP, Klautky SA, Minor SM, Salati LM. Polyunsaturated fatty acids inhibit the expression of the glucose-6-phosphate dehydrogenase gene in primary rat hepatocytes by a nuclear posttranscriptional mechanism. J Lipid Res. 1998;39:1951–1963. [PubMed] [Google Scholar]
- 45.Jia R, Li C, McCoy JP, Deng CX, Zheng ZM. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci. 2010;6:806–826. doi: 10.7150/ijbs.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.