Abstract
Autophagy is a highly conserved process that allows cells, tissues and organs to survive onslaughts such as nutrient deprivation, inflammation, hypoxia and other stresses. The core component proteins that regulate autophagy are well known, and the formation of a double-membrane structure that encompasses cytosolic cargo, including protein aggregates and organelles, has been intensively studied. However, less is known about the inputs that specifically alter recruitment of these components and how post-translational modifications can influence autophagy flux, or the rate at which autophagy substrates are turned over. We propose that three types of post-translational modifications – phosphorylation, ubiquitylation and acetylation – are crucial for autophagy induction, regulation and fine-tuning, and are influenced by a variety of stimuli. Understanding these novel mechanisms of autophagy regulation will give us deeper insights into this process and potentially open up therapeutic avenues.
Autophagy
Macroautophagy (henceforth, autophagy) is the catabolic process of delivering cytosolic cargo to the lysosome for degradation [1]. In this paper, we will refer primarily to ‘selective’ autophagy, which describes the removal of specific cargo, including protein aggregates, mitochondria, peroxisomes and intracellular pathogens. In general, selective autophagy acts as a quality-control mechanism for proteins and organelles. Non-selective autophagy is rapidly induced upon nutrient deprivation (amino acid removal, but not growth factor removal), and the contents of the induced autophagosomes include any protein or organelle that is in the vicinity of the expanding phagophore. This form of ‘garbage removal’ starts with the formation of an isolation membrane, the origin of which has been reported to be the endoplasmic reticulum [2], Golgi [3,4] and the mitochondria [5], all of which can independently produce autophagosomes. The double membrane structure then encapsulates, seals and eventually fuses with the lysosome, where the cargo is broken down into its constituent components and recycled to fuel the growth and proliferation of the cell [1] This process is highly conserved from yeast to mammals, and is essential for the proper development of the organism [1]. This is exemplified by the genetic knockout of either Atg5 or Atg7 in mice, which leads to neonatal lethality [6,7], and knockout of Beclin1 (mammalian Atg6) which is early embryonic lethal [8,9].
In yeast (Saccharomyces cerevisiae), the current total of autophagy (Atg) proteins is 34 (with more likely), all of which regulate aspects of autophagosome initiation, elongation, closure and fusion. However, the process in higher eukaryotes is even more complex, because of the presence of multiple isoforms of core autophagy components. In addition, the mammalian equivalents of several yeast Atg proteins have yet to be identified. For example, Atg1 in yeast has two mammalian isoforms (ULK1 and ULK2), and Atg8, a small ubiquitin-like protein that is conjugated to the autophagosomal membrane, has six mammalian isoforms [microtubule-associated proteins 1A/1B light chain 3A (MAP1LC3A), LC3B, LC3C, gamma-aminobutyric acid receptor-associated protein (GABARAP), GABARAP-L1, and GABARAP-L2], all of which appear to play crucial roles in the formation of autophagosomes [10,11]. In addition, the protease that systematically primes Atg8 proteins for conjugation, and removes Atg8 proteins from the outer autophagosomal membrane before fusion with the lysosome, has four isoforms in mammalian cells: Atg4A, B, C and D [12,13]. Therefore, the additional components present in the mammalian system add further layers of intricacy to a system that we are only just beginning to understand in yeast.
It has long been established that phosphorylation events are crucial for the initiation of autophagy, and that ubiquitylation of cargo proteins is required for their degradation. However, we believe that there is more finetuning involved throughout the autophagy process, including regulation by kinases, E3 ligases, acetylases and deacetylases that are not thought of as ‘classic’ autophagy regulators. Understanding this fine-tuning of autophagy might provide novel nodes for therapeutic intervention strategies for diseases that are refractory to conventional therapies, and deepen our insight into a fundamental cellular process.
Phosphorylation
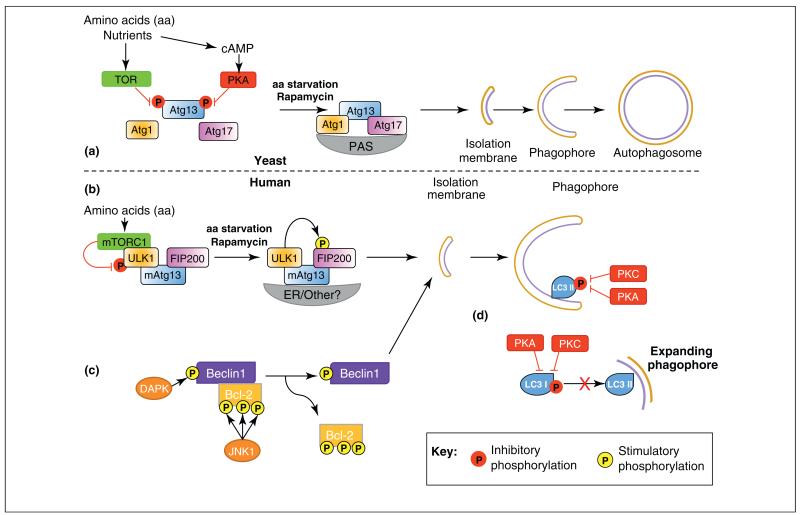
Autophagy has evolved in yeast as a mechanism to cope with various cellular stresses and it allows both yeast and mammalian cells to survive in conditions of low nutrient availability. The ability to sense low levels of nutrients and initiate autophagy is ultimately regulated by the primordial kinase target of rapamycin (Tor). Tor was identified in yeast and mammalian cells, where its inhibition is sufficient to induce cell cycle and growth arrest and autophagy, even in the presence of amino acids [14–16]. The ability of Tor to inhibit autophagy occurs via the phosphorylation of Atg13, which, in turn, reduces its affinity for Atg1 kinase and prevents formation of the Atg1–Atg13–Atg17 core complex [17] (Figure 1a).
Figure 1.
Regulation of autophagy by phosphorylation. (a) Under nutrient-rich conditions in yeast, autophagy is inhibited by phosphorylation of Atg13 by target of rapamycin (TOR), preventing Atg1–Atg13–Atg17 complex formation. Phosphorylation of Atg13 by PKA inhibits Atg1–Atg13–Atg17 association with the pre-autophagosomal structure (PAS). Upon amino acid starvation or rapamycin treatment these inhibitory phosphorylations are removed, Atg1–Atg13–Atg17 complex forms and autophagy can proceed with isolation membrane, phagophore and eventually autophagosome formation. (b) In mammals, phosphorylation by mammalian-TOR (mTOR) complex 1 (mTORC1) of ULK1 inhibits ULK1 and initiation of autophagy. Amino acid starvation or rapamycin treatment causes dissociation of mTORC1 and ULK1 can phosphorylate and activate FIP200 allowing autophagy to proceed. (b) Phosphorylation of either Beclin1 or Bcl-2 by DAPK and JNK1, respectively, cause the dissociation of the Bcl-2/Beclin complex and is sufficient for autophagy induction and isolation membrane formation. (c) Phosphorylation of LC3 by either PKA or PKC is sufficient to inhibit LC3 incorporation into autophagosomes by an undefined mechanism. Hypothetical mechanisms include inhibition of the interaction of LC3 with LC3 interacting proteins or with the conjugation/lipidation machinery.
Interestingly, Tor is not the only kinase that can regulate the Atg1 complex in S. cerevisiae. Recent evidence points to the cAMP-activated protein kinase A (PKA), as a novel regulator of autophagy [18]. Under nutrient-rich conditions, PKA phosphorylates Atg13 at phosphorylation sites distinct from those of Tor (Figure 1a). Importantly, PKA phosphorylation is sufficient to inhibit autophagy by preventing association of Atg13 with the pre-autophagosomal structure (PAS) [18]. Currently, there is no evidence of mammalian Atg13 (mAtg13) being similarly regulated by PKA phosphorylation.
Mammalian systems have mammalian TOR complex (mTORC) 1 and 2, which are distinct and characterized by the presence or absence of various subunits, the main components being raptor (in mTORC1) and rictor (in mTORC2). These complexes differ in their sensitivity to rapamycin; only mTORC1 is inhibited by the drug [19]. The mammalian equivalent of the Atg1–Atg13–Atg17 complex contains ULK1–mAtg13–FIP200, and mTORC1 is incorporated directly into the complex, where it mediates phosphorylation of ULK1 (unc51-like kinase) and mAtg13 (Figure 1b), thereby preventing the initiation of autophagy [20,21]. The phosphorylation of ULK1 by mTORC1 is inhibitory; and rapamycin treatment releases ULK1 to phosphorylate FIP200 (family-interacting protein of 200 kDa), and subsequently autophagy is induced [20] (Figure 1b). However, the precise mechanism by which ULK1 kinase activity is inhibited is not known. For more details of Atg1(ULK1) regulation, please consult the extensive reviews covering such topics [22,23].
As mentioned previously, there is no known role for PKA in the regulation of mAtg13 but other kinases have recently been shown to circumvent the need for mTOR and activate mammalian Atg6 (Beclin1). One mechanism of autophagy induction is via phosphorylation of the Beclin1 inhibitor protein Bcl-2 by the stress-activated c-Jun aminoterminal kinase 1 (JNK1) on three distinct sites [24] (Figure 1c). In addition to JNK1, death-associated protein kinase (DAPK) also causes dissociation of the Bcl-2–Beclin1 complex, in this case via direct phosphorylation of Beclin1 [25] (Figure 1c). It would be interesting to analyze the phosphorylation state of both Beclin1 and Bcl-2 at the identified sites, which might clarify whether organisms have developed a safety mechanism requiring both Beclin1 and Bcl-2 proteins to be phosphorylated to induce autophagy in this manner. Thus, key events in the genesis of autophagosomes are regulated by phosphorylation, highlighting the importance of this particular posttranslational modification in autophagy.
Another kinase involved in autophagy is I kappa B (IκB) kinase (IKK), which is best known for its role in nuclear factor kappa B (NF-κB) signaling pathways. Expression of a constitutively active form of IKK is sufficient to stimulate autophagy via phosphorylation of JNK1 and AMP kinase 1 (AMPK1) [26]. In addition, suppression of IKK expression via knockout or knockdown mechanisms is sufficient to inhibit autophagy under various stimuli [26].
One theme that has been cropping up recently with regard to the involvement of phosphorylation in autophagy is the modification of the small ubiquitin-like Atg8 proteins. This in itself is interesting, as there are no known post-translational modifications of ubiquitin other than conjugation to target proteins or incorporation into ubiquitin chains. The question is: why would Atg8 proteins require modification other than conjugation to phosphatidylethanolamine (PE)? Recent work has identified specific phosphorylation sites for members of the ABC kinase families, (PKA [27] and PKC [28]) in the N-terminal region of MAP1LC3 (LC3, a mammalian homologue of Atg8) (Figure 1c). In the case of PKC phosphorylation sites (Thr6 and Thr29), these appear to be specific for the LC3B isoform, and are not found in other members of the LC3 family [28].
It is interesting to note that the PKA site (Ser12 in MAP1LC3 proteins) is highly conserved in all LC3 (but not GABARAP, another mammalian homologue of Atg8) isoforms in human, mouse, rat and zebrafish, but is not present in yeast Atg8 or Drosophila melanogaster Atg8. Because PKA has not yet been shown to phosphorylate mAtg13, this regulation of LC3 proteins might be a way for PKA to ‘get in on the act’ of controlling autophagy in a mammalian system. Ser12 phosphorylation in MAP1LC3B (LC3B) inhibits its recruitment into autophagosomes [27]; however, the mechanism of this inhibition is unknown, and key questions remain. Does Ser12 phosphorylation inhibit interaction with conjugation/lipidation machinery at the autophagosomal membrane? Does this phosphorylation alter interaction with LC3-interacting proteins? The implications of phosphorylation of these sites by both PKA and PKC might include direct interference of the interaction of LC3 proteins with (LC3 interaction region) LIR-containing proteins, such as p62, and therefore exclusion of LC3 from autophagosomes. Two of the three phosphorylation sites (Thr6 and Ser12) lie directly in, or in the vicinity of, the N-terminal extension of LC3, which is essential for the binding of LC3-interacting proteins via their classic LIR motif [29]. These reports leave us with more questions than answers as to the role of both PKA- and PKC-mediated regulation of autophagy in mammalian systems. Further studies are required to fully understand this important modification and its role in autophagy.
It is interesting to speculate whether there are perhaps other points at which phosphorylation can specifically intervene in the autophagic process. For example, serine and threonine residues are often found in the proximity of the LIR motif of Atg8 interacting proteins, and a mechanism can be invisaged by which a kinase could phosphorylate these proteins and thereby regulate their interaction with Atg8/LC3/GABARAPs. This would be similar to phosphorylation-dependent regulation of small ubiquitin-like modifier (SUMO)-interacting motif (SIM) binding to SUMO [30], and again would offer an interesting node for regulating autophagic flux.
Ubiquitylation
As the name suggests, ubiquitin (Ub) is ubiquitous in both its expression and function. Ub is a small 8 kDa protein containing seven lysine residues that can be conjugated via the action of E1 (Ub-activating enzyme), E2 (Ub-conjugating enzyme) and E3 (Ub ligase, which dictates substrate specificity) enzymes into poly-lysine chains with various conformations and monoubiquitinate and multi-monoubiquitinate target proteins (reviewed in [31]). Ubiquitylation of target proteins results in their degradation, alteration of signaling properties and differential trafficking within the cell [31].
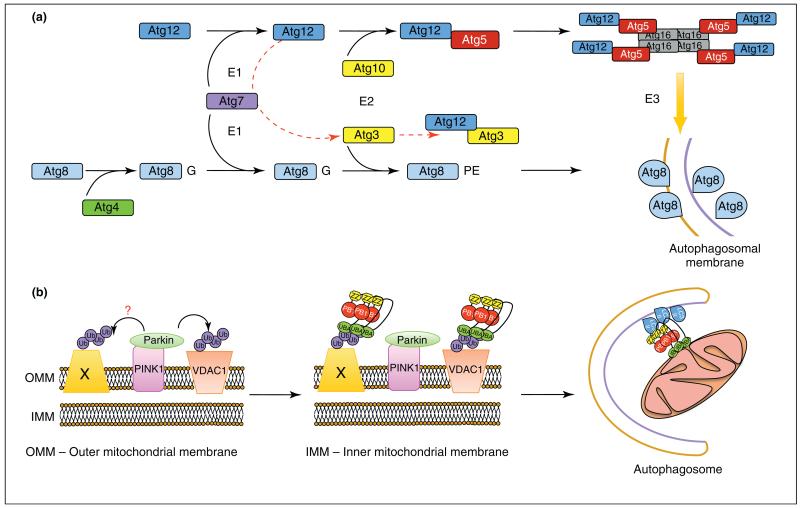
Autophagy has its own take on the same system. The autophagy proteins Atg8 (LC3, GABARAP in mammalian cells), Atg5 and Atg12 all undergo their own Ub-like conjugation reactions that are essential for expansion of the autophagosomal membrane. Both Atg8 and Atg12 proteins are activated by the E1-like enzyme Atg7, Atg12 is conjugated to Atg5 by the E2-like Atg10, and Atg8 is conjugated to PE by Atg3. Despite the presence of obvious E1- and E2-like proteins in this conjugation system, there are no obvious E3-like homologues. However, the Atg12–Atg5 conjugate forms a complex with Atg16 and this hierarchical assembly of proteins can drive the formation of Atg8–PE in an E3-like manner [32] (Figure 2a). Interestingly, a new complex of autophagy Ub-like proteins has been identified as a novel regulator of mitochondrial expansion and cell-death pathways, but has no role in the removal of depolarized mitochondria by autophagy (mitophagy) [33]. A single conjugation site within Atg3 is used by Atg7, and via the autocatalytic E2-like activity of Atg3, forms an Atg12–Atg3 complex [33]. The existence of this complex raises several questions, including whether other Atg12-based complexes exist in nature, whether there are other Atg-based complexes that have autophagy-independent roles and, if so, what exactly are these novel functions?
Figure 2.
Regulation of autophagy by ubiquitylation. (a) The Ub-like proteins Atg8 (LC3/GABARAP in higher eukaryotes), Atg5 and Atg12 all undergo a Ub-like conjugation via the action of E1-(Atg7) and E2 (Atg10, Atg3)-like enzymes. Atg8 is primed by C-terminal cleavage by the protease Atg4, which exposes a C-terminal glycine and is then conjugated to PE. Atg12 is conjugated directly to Atg5 and forms a complex with Atg16 that possesses E3-like activity, driving Atg8-PE incorporation into expanding autophagosomes. An alternative conjugation is reported between the E2-like enzyme Atg3 and Atg12, which requires Atg7 (red broken line). (b) The E3-ligase Parkin is recruited to damaged mitochondria via the serine/threonine kinase PINK1 where it can ubiquitylate the target substrate VDAC1 (voltage dependent anion channel 1) with K27- and K63-linked polyUb chains. These chains serve to recruit p62/SQSTM1 and incorporate damaged mitochondria into autophagosomes. However, recent evidence suggests that VDAC1 is dispensable for Parkin mediated-mitochondrial clearance and that there might be other ubiquitylated proteins (Protein X) that recruit p62 (PB1, Phox and Bem1p domain; Znf, Zinc Finger domain; UBA, Ubiquitin-associated domain).
The role of Ub in autophagy is best known for the removal of protein aggregates. The identification of p62/SQSTM1 and NBR1 (neighbour of BRCA1) as adaptor proteins that simultaneously bind LC3 (by means of an LIR) and ubiquitylated cargo (via their Ub binding domains) indicated that these could allow inclusion of ubiquitylated cargo into autophagosomes and their subsequent degradation by the lysosome [34–36]. In fact, despite reports of p62 and NBR1 binding to several Ub chains (K63 and K27) [35–37], there might be a case for monoubiquitin being sufficient for inclusion of ubiquitylated cargo into autophagosomes in a p62-dependent manner [38]. However, the in vivo specificity of p62/NBR1 toward Ub signals remains to be established under different physiological conditions. Furthermore, there are potentially many more Ub receptors implicated in regulation of autophagy processes. For example, NDP52 binds to ubiquitylated Salmonella upon their cytosolic infiltration [39]. Ubiquitylation might have a ubiquitous role in trafficking and signaling in autophagy.
The crosstalk between ubiquitylation and removal of damaged organelles is clearly depicted in the case of clearance of depolarized mitochondria mediated by Parkin, an E3 ligase [37,40]. Upon loss of mitochondrial membrane potential, Parkin is recruited and mediates the ubiquitylation of voltage-dependent anion channel 1 (VDAC1) in a K63- and K27-dependent manner. p62 is then recruited, and depolarized mitochondria are cleared by autophagy (mitophagy) [37]. However, a recent publication suggests that the picture is not as clear as once thought. Evidence has now surfaced that suggests p62 is responsible for mitochondrial clustering after the Parkin has ubiquitylated mitochondrial-anchored substrates [41]. Using VDAC1/3-deficient mouse embryonic fibroblasts, VDAC1 and VDAC3 were shown to be dispensable for Parkin-mediated p62 recruitment and subsequent mitochondrial clearance [41]. This is in stark contrast to data identifying ubiquitylated VDAC1 as an essential component for p62 recruitment and subsequent mitochondrial clearance [37]. The process of mitophagy is complex, and is both cell type- and context-specific. There are probably multiple targets for ubiquitylation on the outer mitochondrial membrane, and the presence of proteins that can specifically interact with the autophagy machinery can contribute to the clearance of damaged mitochondria. Indeed, Nix has been identified as a mitochondrial localized protein that directly interacts with the mammalian GABARAP and LC3 proteins, and is required for the clearance of mitochondria in reticulocytes in an autophagy- and LC3 interaction-dependent manner [42,43]. Nix is upstream of both Parkin- and p62-mediated mitochondrial clearance in cells treated with a chemical inducer of hypoxia, and is required for autophagy induction and subsequent clearance of mitochondria [44] (Figure 2b).
The initial recruitment of Parkin, which is distributed throughout the cytosol, to damaged mitochondria is dependent upon phosphatase and tensin homolog-induced putative kinase 1 (PINK1). Upon mitochondrial damage or depolarization, PINK1 recruits Parkin. The two proteins can be found in close proximity to each other on the mitochondria but PINK1 does not appear to phosphorylate Parkin and is not a substrate of Parkin [45]. Indeed, there is very little known about physiological substrates of PINK1, and it is interesting to speculate whether PINK1 might mediate phosphorylation and perhaps activation of autophagy. However, one recent study showed that PINK1 can bind to Beclin1 and enhance autophagy, but that this enhancement is not dependent upon PINK1 kinase activity, indicating that it might have more of an adaptor function [46].
From the evidence present in the literature, it appears that ubiquitylation serves as a signal for the degradation of protein aggregates, organelles, intracellular pathogens and a variety of other structures [47]. However, what is not clear is whether ubiquitylation of proteins directly involved in autophagy occurs, and whether this in some way alters autophagic flux. In addition, we have seen that monoubiquitylation, K63-linked and, more recently, K27-linked Ub chains all function in the removal of proteins and organelles via autophagy. This is strongly analogous to the endocytic pathway, where EGF-receptor trafficking is induced via monoubiquitylation, and signaling via K63-linked polyubiquitin chains is involved in NF-κB and DNA damage response pathways (reviewed in [48]). Could there be an E3 ligase that specifically targets autophagy components, and if so, what type of linkages might be used for this specialized pathway? Interestingly, at present it appears that Ub is a novel evolutionary signal in metazoan autophagy; in yeast there are currently no identified roles for ubiquitylation in autophagy [49].
Acetylation
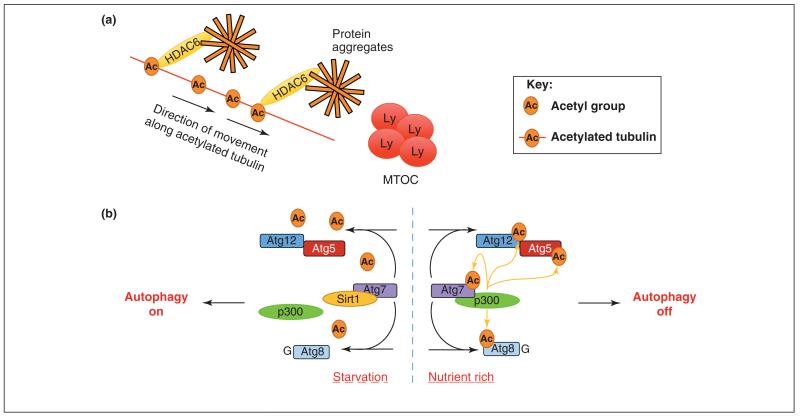
Acetylation (the addition of an acetyl group onto lysine or N-termini of target proteins) is an important post-translational modification for the alteration of protein function, gene transcription and receptor trafficking within the cell [50]. However, one area that has had limited exposure to the acetylome, until recently, is autophagy. Perhaps the best-known regulator of acetylation implicated in autophagy is histone de-acetylase 6 (HDAC6), which can regulate, in a selective autophagy-dependent manner, the retrograde transport of aggregate-containing inclusion bodies that are to be degraded [51] (Figure 3a). In addition, recent evidence indicates that HDAC6 is required for autophagosome maturation through induction of the filamentous actin (F-actin) network, and subsequent fusion of autophagosomes and lysosomes [52]. HDAC6 is also needed for Parkin/p62-mediated clearance of damaged mitochondria [53]. In D. melanogaster, impairment of the Ub proteasome system leads to an increase in the activity of autophagy degradation pathways, and the latter is dependent upon HDAC6 catalytic activity [54]. The evidence suggests that deacetylatase activity is essential for the degradation of protein aggregates via autophagy.
Figure 3.
Regulation of autophagy by acetylation. (a) Adaptor proteins such as HDAC6 can interact with acetylated tubulin and transport protein aggregates along microtubules towards the microtubule organizing centre (MTOC) and the lysosomes (LY). (b) Under nutrient-rich conditions, the acetyltransferase p300 interacts with Atg7, acetylates the key autophagy proteins Atg7, Atg8, Atg12 and Atg5, and inhibits autophagy. During starvation, p300 dissociates and Sirt1 deacetylase exerts its activity, removing acetyl groups from Atg7, Atg5, Atg12 and Atg8 proteins, and allowing autophagy to proceed.
In addition to HDAC6, there is now accumulating evidence for roles for other deacetylases and acetyltransferases in the regulation of core autophagy components. For example, the acetyltransferase p300 can acetylate Atg5, Atg7, Atg8 and Atg12 proteins when transfected into HeLa cells, and p300 can directly interact with Atg7 [55]. Acetylation by p300 inhibits autophagy, and silencing of p300 increases autophagy flux [55] (Figure 3b). In addition, acetylation of mutant Huntingtin protein at Lys9 and Lys444 promotes clearance of the mutant protein by autophagy, whereas a mutant version of Huntingtin that cannot be acetylated accumulates and leads to neurodegeneration [56]. However, the acetyltransferase that is involved in this process has not yet been identified.
Under starvation conditions, the microtubule network can recruit autophagy marker proteins, including PI3 kinase (phosphoinositide 3-kinase), WIPI (WD repeat domain phosphoinositide-interacting protein), and the Atg12–Atg5 conjugate, in a manner that is dependent upon tubulin acetylation, specifically on Lys40 [57]. This leads to the activation of kinesin and JNK1, which serves to release the Bcl-2–Beclin1 inhibitory complex and stimulate autophagy [57]. The acetylation of tubulin affects autophagosomal binding proteins that mediate the transport and eventual fusion of autophagosomes with the lysosomes. For example, HDAC6 controls the acetylation status of tubulin, and therefore the epidermal growth factor receptor [58,59], and autophagosome trafficking along microtubules [51] (Figure 3a).
In another link between autophagy and acetylation, the NAD-dependent deacetylase Sirt1 increases basal autophagy when transiently overexpressed, and can also form a complex with Atg5, Atg7 and Atg8 [59] (Figure 3b). In tissues from Sirt1 knockout mice, basal acetylation of autophagy component proteins is increased, and the phenotype of Sirt1 knockout mice partially resembles that of Atg5 knockout mice, indicating that Sirt1-dependent deacetylation is important for basal autophagy and neonatal survival [59]. Using high-resolution mass spectrometry (MS) to analyze global lysine acetylation in human acute myeloid leukemia cells, 3600 acetylated lysines on 1750 proteins were identified and quantified in response to deacetylase inhibitors. Interestingly, the small Ub-like modifier GABARAP-L2 and Atg7 were found in this screen (Supplemental table S1 in [60]), providing further evidence that acetylation of core proteins might play a key role in the regulation of autophagy. The emerging role of acetylation of key components of the autophagy pathway is an emerging field that we believe will play a crucial role in both the initiation and selective degradation of autophagy and its substrates.
Concluding remarks
To date, the study of autophagy has been dominated by the role of the primary kinase Tor/mTOR, considered the master regulator of autophagy both in yeast and in mammalian systems. However, a large and growing body of evidence points to other kinases, acetylases, deacetylases, and potentially even phosphatases having roles in regulating autophagy. We believe that these post-translational modifications, which can occur at multiple stages of autophagosome formation, lead to the induction, inhibition or finetuning of the autophagic response under a variety of conditions. For example, the phosphorylation of Beclin1 by either JNK1 or DAPK induces autophagy [24,25], PKA or PKC phosphorylation of LC3 proteins inhibits autophagy [27,28], and acetylation of Atg5, Atg7, Atg8 and Atg12 proteins by p300 also inhibits autophagy [55]. The future direction of this emerging area of autophagy research should be focused on developing new ways in which to identify modifications of autophagic proteins and monitoring them in real time, as well as studying their effect on autophagy flux. Use of high-resolution MS and the identification of interaction partners of modified autophagy proteins (and the consequences of such interactions) are crucial to increasing our understanding of this strongly dynamic process. Perhaps the biggest challenge will be integrating the growing number of modifications into a dynamic network that can be studied and, hopefully, exploited under both physiological and pathophysiological conditions, such as in Alzheimer’s disease, Parkinson’s disease and cancer.
Acknowledgment
We apologize to all scientists whose important contribution was not referenced in this review as a result of limitations in number of references. We are grateful to Philipp Wild, Christian Behrends, Ivana Novak and Zevi Elazar for comments and discussions. Research in the I.D. laboratory is supported by the Deutsche Forschungsgemeinschaft, the Cluster of Excellence ‘Macromolecular Complexes’ of the Goethe University Frankfurt (EXC115) and the European Research Council (ERC) grant agreement no. [250241-LineUb].
References
- 1.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayashi-Nishino M, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 3.Yen WL, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J. Cell Biol. 2010;188:101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Vaart A, et al. Exit from the golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2010;21:2270–2284. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 8.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue Z, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weidberg H, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betin VM, Lane JD. Atg4D at the interface between autophagy and apoptosis. Autophagy. 2009;5:1057–1059. doi: 10.4161/auto.5.7.9684. [DOI] [PubMed] [Google Scholar]
- 13.Tanida I, et al. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 14.Blommaart EF, et al. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 15.Brown EJ, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 16.Kunz J, et al. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 17.Kamada Y, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephan JS, et al. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wullschleger S, et al. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 20.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Y, et al. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zalckvar E, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Criollo A, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–631. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cherra SJ, 3rd, et al. Regulation of the autophagy protein LC3 by phosphorylation. J. Cell Biol. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang H, et al. Protein kinase C inhibits autophagy and phosphorylates LC3. Biochem. Biophys. Res. Commun. 2010;395:471–476. doi: 10.1016/j.bbrc.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shvets E, et al. The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J. Cell Sci. 2008;121:2685–2695. doi: 10.1242/jcs.026005. [DOI] [PubMed] [Google Scholar]
- 30.Stehmeier P, Muller S. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. Cell. 2009;33:400–409. doi: 10.1016/j.molcel.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanada T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 33.Radoshevich L, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 36.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 37.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 38.Kim PK, et al. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thurston TL, et al. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 40.Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narendra DP, et al. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1096. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweers RL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding WX, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J. Biol. Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michiorri S, et al. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 2010;17:962–974. doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- 47.Kirkin V, et al. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 48.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 49.Kraft C, et al. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 50.Deribe YL, et al. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 51.Iwata A, et al. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 52.Lee JY, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee JY, et al. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation and HDAC6-dependent mitophagy. J. Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pandey UB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 55.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J. Biol. Chem. 2009;284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeong H, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geeraert C, et al. Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J. Biol. Chem. 2010;285:24184–24194. doi: 10.1074/jbc.M109.091553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deribe YL, et al. Regulation of epidermal growth factor receptor trafficking by lysine deacetylase HDAC6. Sci. Signal. 2009;2:ra84. doi: 10.1126/scisignal.2000576. [DOI] [PubMed] [Google Scholar]
- 59.Lee IH, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]