Summary
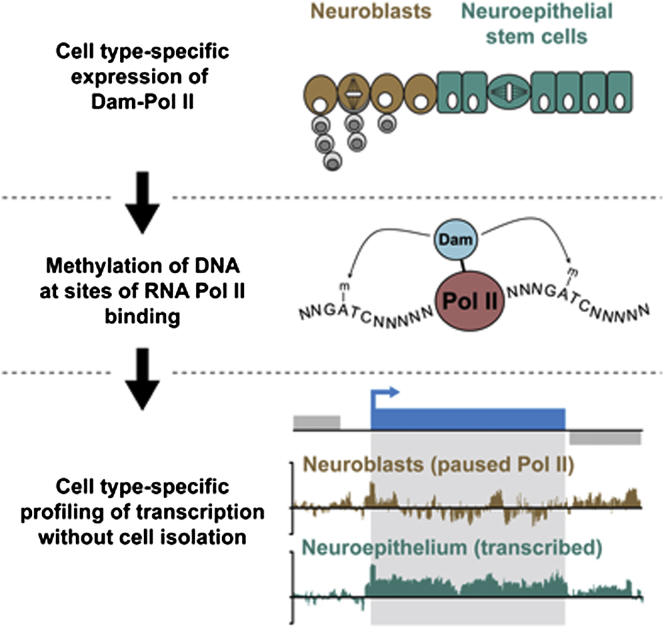
Cell-type-specific transcriptional profiling often requires the isolation of specific cell types from complex tissues. We have developed “TaDa,” a technique that enables cell-specific profiling without cell isolation. TaDa permits genome-wide profiling of DNA- or chromatin-binding proteins without cell sorting, fixation, or affinity purification. The method is simple, sensitive, highly reproducible, and transferable to any model system. We show that TaDa can be used to identify transcribed genes in a cell-type-specific manner with considerable temporal precision, enabling the identification of differential gene expression between neuroblasts and the neuroepithelial cells from which they derive. We profile the genome-wide binding of RNA polymerase II in these adjacent, clonally related stem cells within intact Drosophila brains. Our data reveal expression of specific metabolic genes in neuroepithelial cells, but not in neuroblasts, and highlight gene regulatory networks that may pattern neural stem cell fates.
Graphical Abstract
Highlights
-
•
TaDa is a method for cell-type-specific profiling of chromatin binding proteins
-
•
TaDa does not require cell sorting, fixation, or affinity purification
-
•
This is a highly sensitive and robust technique for transcriptional profiling
-
•
We report differential RNA Pol II binding in clonally related stem cells
Southall et al. developed “TaDa,” a technique that enables cell-specific genome-wide profiling of DNA- or chromatin-binding proteins without cell isolation, fixation, or affinity purification. TaDa genome-wide profiles of RNA polymerase II binding reveal spatially and temporally precise distinctions between adjacent, clonally related neural stem cells in intact Drosophila brains.
Introduction
During the development of multicellular organisms, each cell acquires its specific fate through a precisely controlled pattern of gene expression. To examine the transcriptional profile and/or chromatin state of specific cells and tissues, many techniques require some form of cell isolation, such as fluorescent activated cell sorting (FACS) (Bryant et al., 1999) or laser capture microdissection (LCM) (Neira and Azen, 2002). These methods can be technically challenging, can yield a mixed population of cells, and may also disturb the transcriptional state of the cells or tissues being isolated. Other methods for assaying transcription are based on RNA pull-down, relying on targeted expression of a tagged RNA-binding (Roy et al., 2002) or a ribosomal protein (Thomas et al., 2012), or an RNA modifying enzyme (Miller et al., 2009). These kinds of approaches cannot assess genome-wide binding of transcription factors or chromatin-binding proteins. Techniques that can assay both chromatin binding and transcriptional profiling (including INTACT [Deal and Henikoff, 2010; Henry et al., 2012; Steiner et al., 2012] and BiTS-ChIP [Bonn et al., 2012a, 2012b]) involve affinity purification of tagged nuclei, requiring fixation and FACS or magnetic-activated cell sorting (MACS), as well as large amounts of starting material (e.g., 4–6 million nuclei) (Bonn et al., 2012b).
We have developed “TaDa” to assess genome-wide protein binding in vivo in a cell type-specific way without cell purification. It is simple and requires no cell isolation, fixation, cell sorting, or immunoprecipitation. TaDa is based on DNA adenine methyltransferase identification (DamID) (van Steensel and Henikoff, 2000; van Steensel et al., 2001), an in vivo chromatin profiling technique (Choksi et al., 2006; Germann et al., 2006; Guelen et al., 2008; Schuster et al., 2010; Woolcock et al., 2011) in which an Escherichia coli DNA adenine methyltransferase is fused to a DNA- or chromatin-binding protein of interest. When the fusion protein is expressed in vivo, its binding site is tagged by adenine methylation. Expression of Dam methylase at high levels is toxic, however, and can lead to nonspecific methylation (van Steensel and Henikoff, 2000). As a result, DamID requires the Dam methylase-fusion protein to be expressed at extremely low levels. This has been achieved by expression from basal promoters (Bianchi-Frias et al., 2004; Choksi et al., 2006; Vogel et al., 2007) with the drawback that the Dam-fusion protein is then expressed constitutively in all cell types. Previous attempts to control the spatial specificity of Dam fusions using targeted expression systems, such as GAL4 (Brand and Perrimon, 1993), have resulted in excessively high levels of the methylase and toxicity.
We have devised a means of reducing the level of translation of the Dam-fusion protein, thereby enabling us to express the fusion protein at very low levels in a cell- or tissue-specific fashion with both spatial and temporal control (TaDa, Targeted DamID). TaDa is robust, reproducible and sensitive, requires no crosslinking, immunoprecipitation or cell sorting, and avoids the difficulties associated with isolating small quantities of RNA and protein. TaDa can be completed in 3 days from start to finish and requires fewer than 10,000 cells, possibly far fewer, in contrast to the 4–6 million required by other methods. It can be adapted for use in any model system, as all of the elements of the method are transferable and endogenous adenine methylation is rarely found in eukaryotes (van Steensel and Henikoff, 2000). Conditional expression of the Dam-fusion protein can be achieved using the GAL4 system (Brand and Perrimon, 1993), which has been adapted successfully for use in other model systems, or with recombination systems such as Cre/Lox or Flp/FRT. TaDa can be used to map genome-wide binding of any DNA- or chromatin-binding protein and also to assay gene expression by profiling RNA polymerase II occupancy. We use TaDa to assess differential transcription in neighboring, clonally related stem cells in the optic lobes of intact Drosophila brains. We identified genes in all of the signaling pathways known to be active in optic lobe neuroepithelial cells as well as noncanonical metabolic pathways and genes in the retinal determination network, which had not previously shown to be active. By assaying transcription in neuroepithelial cells over time, we reveal a switch in the activity of signaling pathways that control the fate of these symmetrically dividing neural stem cells.
Results
Attenuation of Dam Methylase Translation
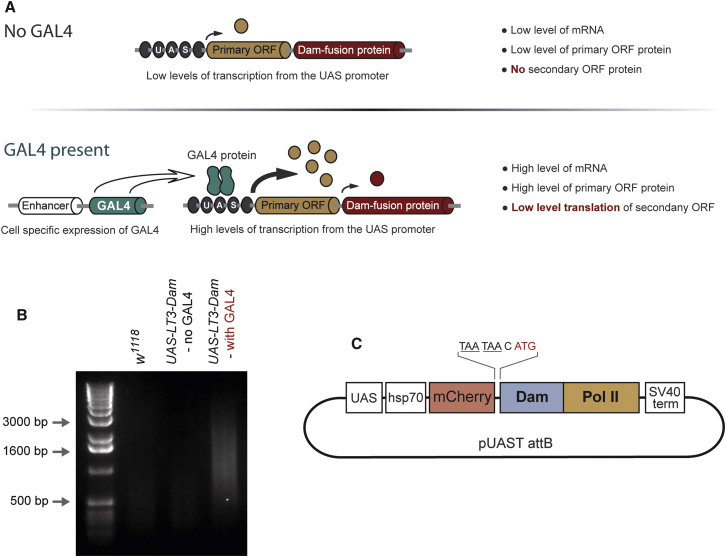
In order to benefit from targeted expression with the GAL4 system, we aimed to reduce the level of translation of the Dam fusion protein, thereby avoiding the toxicity associated with high levels of the methylase. We took advantage of the fact that, at a low frequency, eukaryotic ribosomes are able to reinitiate translation on bicistronic messages lacking an obvious IRES (Luukkonen et al., 1995; Child et al., 1999; Van Blokland et al., 2011). The degree of reinitiation is dependent upon the size of the primary ORF (Kozak, 2001), therefore we designed GAL4-inducible constructs containing primary ORFs of different lengths followed by a secondary ORF encoding the Dam methylase. Translation of the secondary ORF must be essentially null in uninduced cells and sufficiently low upon induction to avoid toxicity (Figure 1A). At levels where the Dam methylase is functional and nontoxic, the protein is undetectable by western blotting or immunofluorescence (Vogel et al., 2007).
Figure 1.
TaDa Allows Cell-Type-Specific Profiling of Protein-DNA Interactions
(A) Schematic representation of the TaDa method. Translation of the Dam-fusion protein is greatly reduced by the addition of an upstream ORF and by ribosome re-initiation. This prevents expression of the Dam-fusion in uninduced cells and nonspecific methylation in induced cells.
(B) Adenine methylated DNA can only be amplified from cells when UAS-LT3-Dam expression is driven by GAL4. DamID samples (from 50 larval brains) were prepared from wild-type (w1118), UAS-LT3-Dam with no GAL4 (w1118x UAS-LT3-DAM), and UAS-LT3-Dam driven in neuroblasts (insc-GAL4 x UAS-LT3-Dam). A small amount of DNA is detectable in the wild-type sample (53 ng/μl) and a similar amount in the w1118x UAS-LT3-DAM sample (49 ng/ul). This DNA is the remaining template genomic DNA plus any nonspecifically amplified DNA (unrelated to adenine methylation). In the presence of GAL4, methylated DNA is observed (301 ng/μl).
(C) Design of the TaDa construct for profiling RNA Pol II occupancy in the genome.
See also Figure S1.
We generated a series of transgenes in which a primary ORF (ORF1) is followed by two stop codons and a secondary ORF (ORF2) encoding the Dam methylase (Figure 1A). In UAS-LT1-Dam (low level translation version 1) ORF1 encodes six amino acids (MGGSAG), in UAS-LT2-Dam ORF1 encodes the first 80 amino acids of mGFP6 (Schuldt et al., 1998) and in UAS-LT3-Dam ORF1 encodes full-length mCherry (246 amino acids) (Shaner et al., 2004). Transgenic lines were generated for each construct.
As a first step, we crossed each line to ase-GAL4 (for neural stem cell expression) and assayed toxicity. Expression of UAS-Dam (Choksi et al., 2006), in which the Dam methylase is encoded by ORF1, leads to 100% embryonic lethality. This is also the case for UAS-LT1-Dam, suggesting that ORF2 expression is too high. ase-GAL4 driving UAS-LT2-Dam causes 100% lethality at pupal stages. Expression of UAS-LT3-Dam is not toxic at any stage of development and has the added advantage of marking the expressing cells with mCherry.
We next tested whether UAS-LT3-Dam is expressed in uninduced cells. UAS-LT3-Dam was crossed to insc-GAL4 (GAL4MZ1407, expressed in neural stem cells from the embryo onward) and to w1118 (control). Fifty brains were collected from third instar larvae and genomic DNA extracted. DNA was digested with DpnI, which only recognizes methylated GATC sites, followed by amplification of the methylated DNA by PCR (see Experimental Procedures). We were able to amplify DNA from insc-GAL4 x UAS-LT3-Dam brains (Figure 1B), suggesting that the Dam methylase has been expressed and is active. In contrast, DNA from w1118 x UAS-LT3-Dam brains was equivalent to that from control brains (w1118) and barely detectable (Figure 1B). Therefore, by controlling the level of translation of the Dam methylase, Dam-fusion proteins can be targeted temporally and spatially using the GAL4 system: GAL4-driven expression of LT3-Dam is sufficient to methylate genomic DNA and there is no detectable DNA methylation in the absence of GAL4.
Dam-Polymerase II Accurately Reflects Endogenous Polymerase II Occupancy
One of the goals of modern stem cell biology is to manipulate cells down particular developmental pathways, for example by inducing pluripotency and driving cells to differentiate into a specific cell type or by trans-differentiating cells from one cell type to another. In order to do this, we need to understand as much as possible about the transcriptional states of different cells and cell populations and how they change over time.
To assay genome-wide transcription in a cell-specific fashion, we used TaDa to profile RNA polymerase II occupancy in vivo. A GFP-tagged version of the core subunit of Pol II (RpII215) has been shown to be functional in vertebrate cells (Sugaya et al., 2000). We fused Dam-methylase to the core subunit of Pol II to create Dam-Pol II (pUAST-LT3-NDam-Pol II; Figure 1C). To confirm that Dam-Pol II is being expressed at very low levels in our system, we drove expression of UAS-LT3-Dam-Pol II in all cells throughout larval development using tubulin-GAL4 (Lee and Luo, 1999). Western blot analysis reveals that Dam-Pol II levels are undetectable relative to endogenous Pol II (Figure S1 available online).
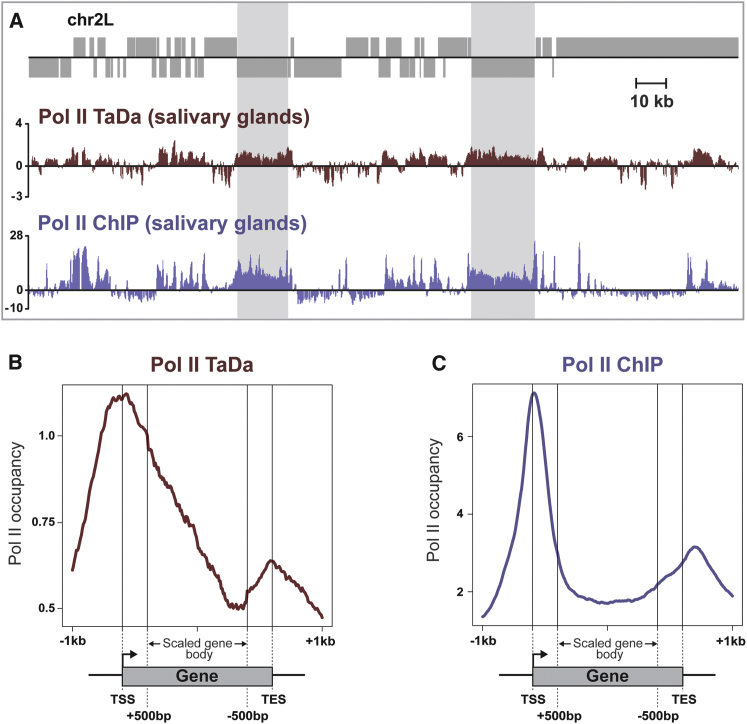
To assess whether Dam-Pol II recapitulates endogenous Pol II binding, we performed Pol II TaDa on third instar larval salivary glands and compared our results with the previously published Pol II ChIP-seq data (Conrad et al., 2012). We used hs-GAL4 (Brand et al., 1994) to express UAS-LT3-Dam-Pol II in salivary glands (Costantino et al., 2008). Genomic DNA was extracted, digested with Dpn I, and PCR-amplified. Amplified DNA from Dam-only (reference) and Dam-Pol II (experimental) tissue was labeled and cohybridized to Nimblegen whole genome tiling arrays. The binding profiles are extremely similar (Figure 2A), with a highly significant correlation between the two data sets (r = 0.586, p < 1 × 10−7, signal across genes). A meta-analysis of Dam-Pol II occupancy reveals similar binding profiles for TaDa and ChIP-seq (Figures 2B and 2C). Therefore, Dam-Pol II accurately reflects endogenous Pol II occupancy.
Figure 2.
Dam-Polymerase II Accurately Reflects Endogenous Polymerase II Occupancy
(A) Comparison of RNA Pol II occupancy in salivary glands as determined by TaDa and ChIP-seq.
(B) Metagene profile for Dam-Pol II across transcribed genes in salivary glands, from 1 kb upstream of the transcription start site (TSS) to 1 kb downstream of the transcription end site (TES). Y axis represents the log2 ratio of Dam-Pol II/Dam.
(C) Metagene profile for Pol II ChIP-seq across transcribed genes in salivary glands.
Mapping RNA Polymerase II Occupancy in Neural Stem Cells
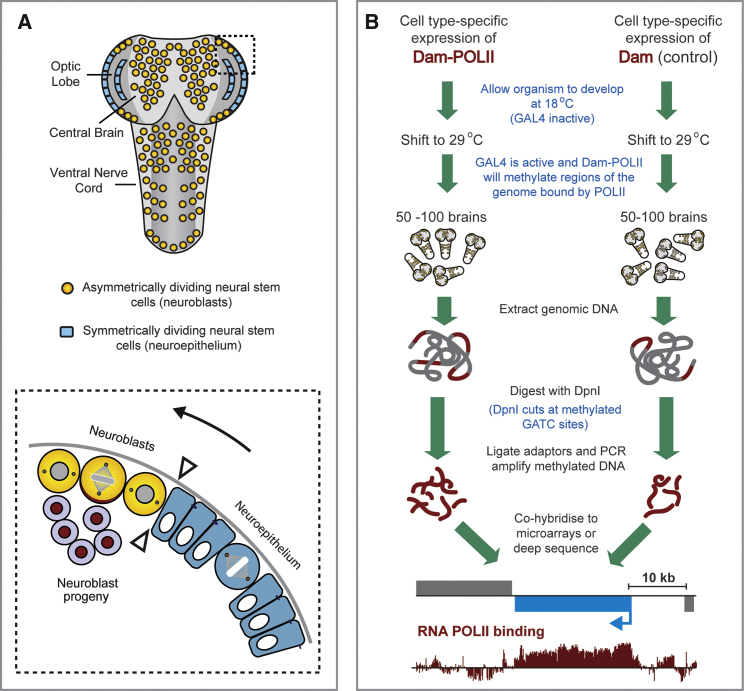
The developing Drosophila optic lobe contains two clonally related populations of stem cells: symmetrically dividing neuroepithelial cells that transform over time into asymmetrically dividing neural stem cells (neuroblasts) (Egger et al., 2007; Yasugi et al., 2008) (Figure 3A). This cell fate transition is remarkably similar to that seen in the developing mammalian cerebral cortex, where neuroepithelial cells divide symmetrically to expand the stem cell pool before transforming into asymmetrically dividing radial glial cells (Farkas and Huttner, 2008). Understanding the common molecular mechanisms that promote neurogenesis is crucial in efforts to regenerate or repair the nervous system after injury or disease. Comparing the transcriptomes of neuroepithelial cells and neuroblasts is a first step in revealing the transcriptional changes leading from stem cell self-renewal to differentiation.
Figure 3.
Experimental Design for Assessing RNA Pol II Binding in Clonally Related Stem Cell Populations in the Drosophila Brain
(A) The Drosophila developing optic lobe contains symmetrically dividing neural stem cells (neuroepithelium) that undergo a transition (see arrowheads) to asymmetrically dividing stem cells (neuroblasts).
(B) Experimental design: a cell-specific GAL4 driver line is used to drive expression of Dam and Dam-Pol II. A temperature sensitive GAL80 is ubiquitously expressed to allow temporal control of expression.
We drove expression of Dam-Pol II in the neuroepithelium or in neuroblasts, with GAL4c855a and GAL4MZ1407, respectively (Egger et al., 2007, 2010). Expression was temporally regulated with GAL80ts, a temperature-sensitive negative regulator of GAL4 (Matsumoto et al., 1978; McGuire et al., 2003). Dam-Pol II was expressed for 24 hr, from mid third instar to wandering third instar larval stages. Genomic DNA was extracted from ∼100 brains, digested with Dpn I and amplified (Figure 3B).
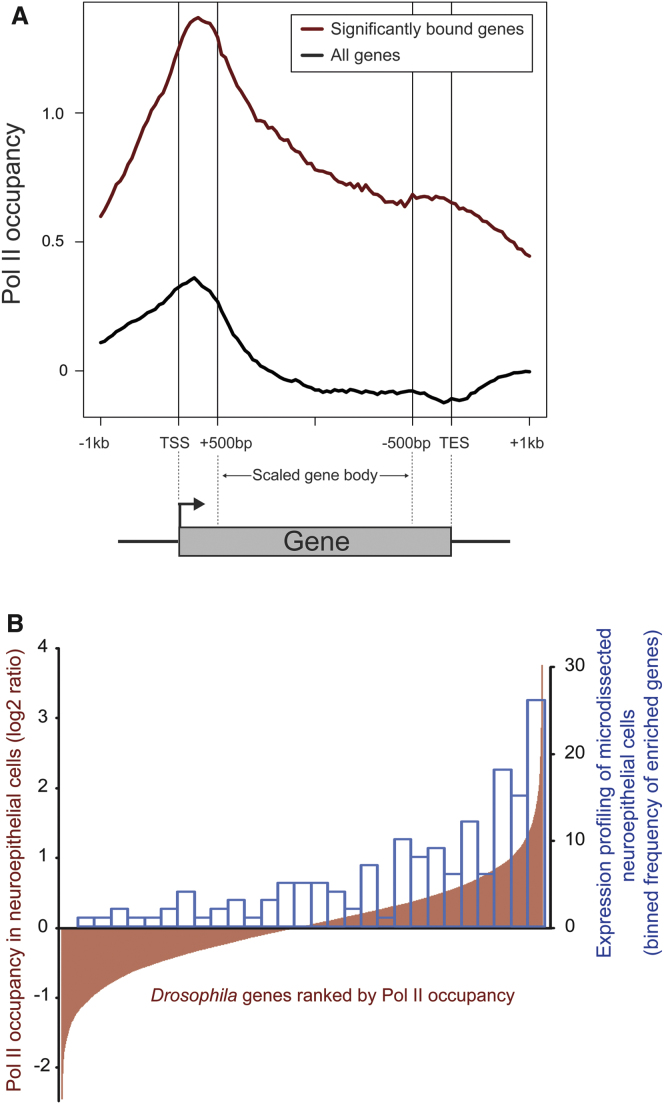
We performed a meta-analysis of Dam-Pol II occupancy in neuroepithelial cells (Figure 4A). The profile of Dam-Pol II binding across all annotated genes is consistent with previous ChIP studies (Gilchrist et al., 2010; Nechaev et al., 2010; Lam et al., 2012). We find a peak immediately downstream of the transcriptional start site (TSS) and a dip at the transcriptional stop (Lam et al., 2012), demonstrating that Dam-Pol II binding accurately reflects Pol II occupancy in vivo. The mean ratios for binding across annotated transcripts were determined (Dam-Pol II/Dam-only) and a false discovery rate (FDR) assigned (see Experimental Procedures). A mean ratio change >1.3-fold and an FDR <5% was considered significant. We found a high degree of reproducibility between biological replicates, with an average correlation coefficient of 0.88 when comparing three neuroepithelial cell replicates.
Figure 4.
Metagene Profiles of Dam-Pol II Occupancy and Comparison with mRNA Expression Profiling
(A) Metagene profiles are shown for all transcribed neuroepithelial genes, and for all genes, from 1 kb upstream of the transcription start site (TSS) to 1 kb downstream of the transcription end site (TES). Y axis represents the log2 ratio of Dam-Pol II/Dam.
(B) Comparison of Dam-Pol II occupancy with mRNA expression profiling. All Drosophila genes are represented on the x axis ranked, from left to right, based on Pol II occupancy in the neuroepithelium (log2 ratio of Dam-Pol II/Dam shown on right y axis). In order to compare Pol II data with the 157 genes identified by mRNA expression profiling of neuroepithelial cells (Egger et al., 2010), the frequency of these genes falling into bins of 500 genes on the x axis were calculated (left y axis).
In total, 2,711 genes with significant Pol II occupancy were identified in the neuroepithelium and 4,142 in neuroblasts. A literature search identified 42 genes as expressed in the neuroepithelium. We found Pol II binding to 88% of these genes in the neuroepithelium (Table S1). A total of 157 genes were previously identified as expressed in microdissected neuroepithelial cells (Egger et al., 2010) (Figure 4B). Of these, at least two (trol and dsx) are false positives (Voigt et al., 2002; Rideout et al., 2010). With TaDa, we observed Dam-Pol II binding at 80% of genes identified in microdissected neuroepithelial cells and trol and dsx were correctly assigned (not expressed in the neuroepithelium).
patj, mer, aPKC, dome, and Stat92E are reported to be expressed in the neuroepithelium but exhibited a Dam-Pol II/Dam log2 ratio that was below that deemed to be significant (Table S1). It is possible that these genes produce stable mRNAs and/or proteins and only require a low level of active transcription. We were concerned whether perdurance of GAL4 and its target gene, in this case the Dam-fusion, might generate false positive signals. However, we saw no signal for the neuroblast genes eyeless and insc in neuroepithelial cells (Figures 6E and 6E′ and Egger et al., 2007, respectively). Dam-Pol II was bound to ase, wor, brat, and mira, in the neuroepithelium as well as in neuroblasts (Table S2). These genes may be regulated posttranscriptionally in the neuroepithelium (e.g., by one of the microRNAs that we found to be differentially expressed between neuroblasts and neuroepithelium). If we discover a signal that is indeed due to perdurance, then it can be eliminated by expression of GAL80 in neuroblasts.
Figure 6.
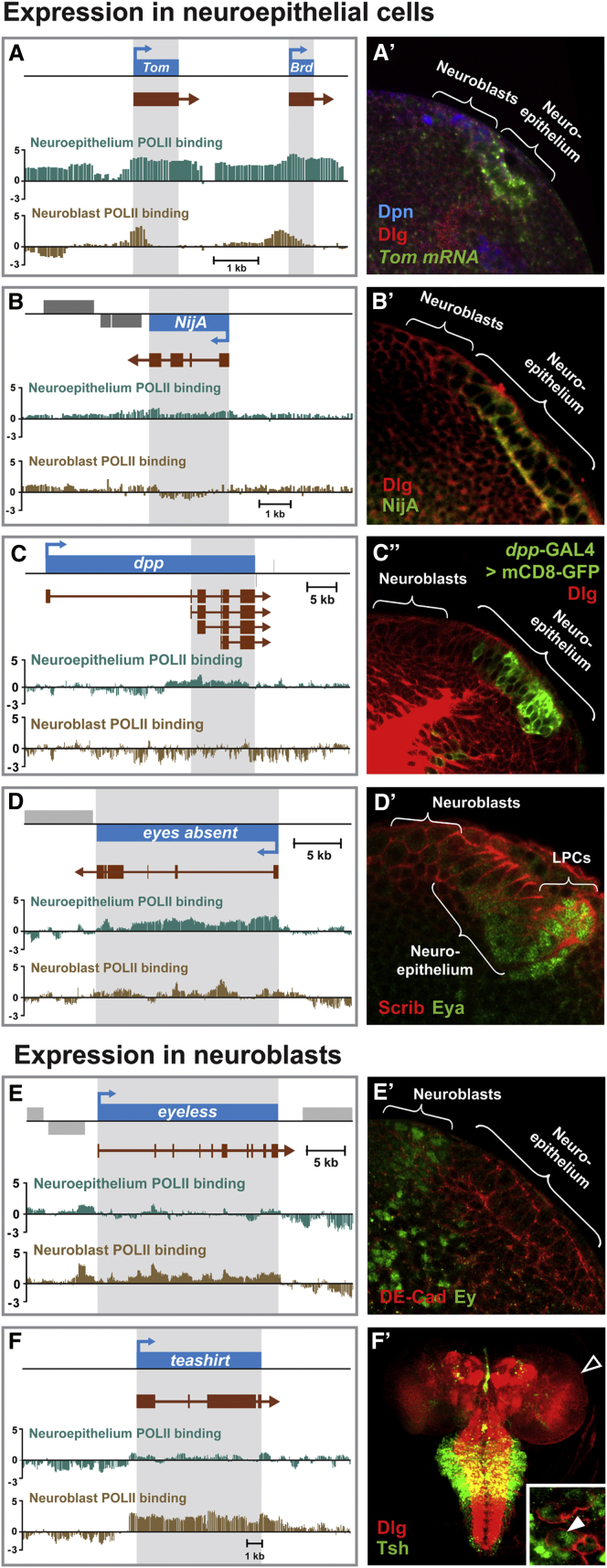
Cell-Type-Specific Differences in RNA Pol II Occupancy Reflect Differences in Gene Expression
(A–F) Differential Pol II occupancy in neuroepithelial cells and neuroblasts. Scale bars represent log2 ratio change between Dam-Pol II and Dam (reference) samples. (A′–F′) Expression patterns of the respective genes in the larval brain and ventral nerve cord.
To identify genes that are specifically expressed or highly enriched in either the neuroepithelium or in neuroblasts, we compared Pol II occupancy between these two stem cell subtypes. Of the 2,711 genes bound by Pol II in the neuroepithelium, 352 genes show a significantly enriched signal relative to neuroblasts (Table S2). This is nearly twice as many genes as identified by transcriptome analysis of microdissected neuroepithelial cells. Of the 4,142 neuroblast genes, 1,802 are preferentially bound by Pol II in neuroblasts as compared to neuroepithelial cells. Among these are the known neuroblast genes ase, wor, mira, brat, insc, and pros (Table S2). None of these genes were identified in microdissected optic lobe neuroblasts, in which only 36 genes were found to be expressed (Egger et al., 2010).
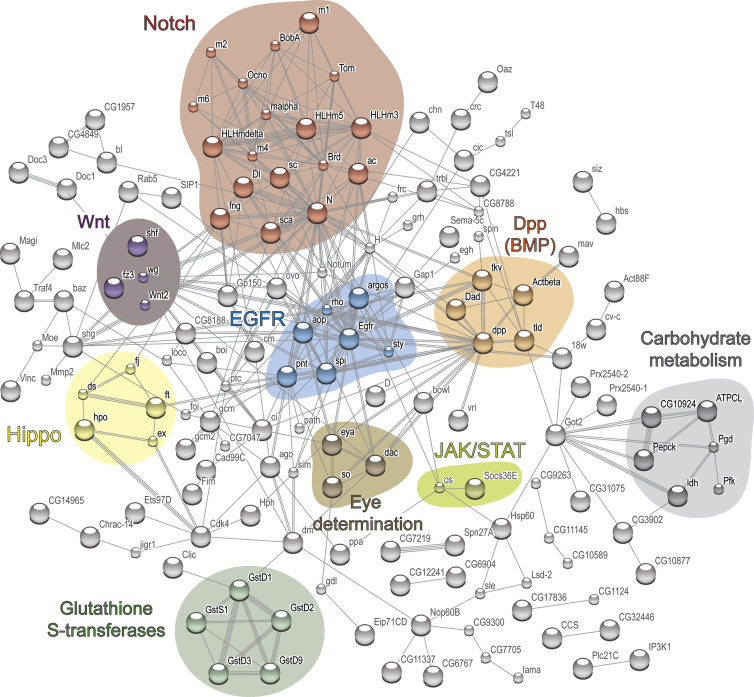
To obtain an integrated view of the 352 genes enriched in the neuroepithelium, we used STRING (Szklarczyk et al., 2011) to generate a network of interactions based on experimental data as well as predicted interactions from coexpression and literature searches. The STRING analysis highlighted signaling pathways known to be active in the neuroepithelium (Figure 5), such as Notch (Egger et al., 2010; Ngo et al., 2010; Reddy et al., 2010; Yasugi et al., 2010), EGFR (Yasugi et al., 2010), Hippo (Reddy et al., 2010), JAK-STAT (Yasugi et al., 2008; Ngo et al., 2010; Wang et al., 2011), Wnt (Kaphingst and Kunes, 1994), and Dpp (Kaphingst and Kunes, 1994).
Figure 5.
Known and Predicted Interactions between the Products of the 352 Genes Bound Preferentially by Pol II in Neuroepithelial Cells
Analysis was performed using STRING with a medium confidence score of 0.4 and all interaction methods available. Clusters representing select signaling pathways, the eye determination pathway, glutathione S-transferases and carbohydrate metabolism genes are highlighted. Single nodes are not displayed.
Notch maintains neuroepithelial cell fate and prevents premature differentiation into neuroblasts and neurons (Egger et al., 2010; Ngo et al., 2010; Reddy et al., 2010; Wang et al., 2011; Yasugi et al., 2010). In the neuroepithelium, Pol II binds to the Tom and Brd genes (Figure 6A). Tom and Brd are Bearded family proteins that block Delta endocytosis in cells where Notch is active (Bardin and Schweisguth, 2006; De Renzis et al., 2006). We confirmed transcription of Tom in the neuroepithelium by RNA in situ hybridization (Egger et al., 2010) (Figure 6A′). As predicted by Dam-Pol II occupancy, neither of these genes is transcribed in neuroblasts (Figure 6A′ and data not shown). Intriguingly, however, we observe Dam-Pol II paused at the 5′ ends of both Tom and Brd in neuroblasts (Figure 6A), suggesting that transcription of these genes may resume at a later stage.
The JAK/STAT ligand Unpaired (Upd) inhibits the transition of neuroepithelial cells into neuroblasts (Yasugi et al., 2008). Dam-Pol II bound upd in neuroepithelial cells but not neuroblasts (Figure 7B). In support of this result, we found that Upd-GAL4 is expressed in a subset of neuroepithelial cells but not in neuroblasts (Figure 7B″). Similar results were found for Egfr (Figures 7C and 7C″) and dpp (Figures 6C and 6C′), whereas hairy was expressed in neuroepithelial cells in the inner proliferation center (Figures S3B and S3B′). We found that even low level binding of Pol II is predictive of gene expression, as seen at the NijA locus (Figures 6B and 6B′). We also found genes that are expressed in neuroblasts but not neuroepithelial cells, e.g., eyeless (Figures 6E and 6E′) and tsh (Figures 6F and 6F′). Of the ten genes identified by TaDa and shown in Figure 6 only two, Tom and NijA, were identified by mRNA expression profiling (Egger et al., 2010).
Figure 7.
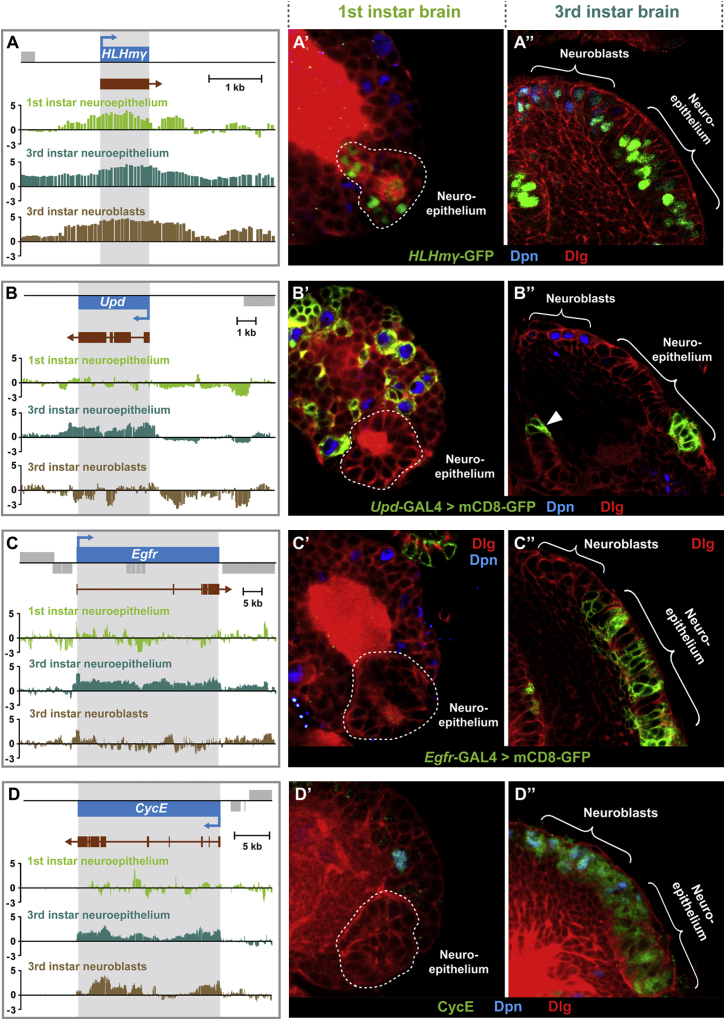
Temporal and Cell-Type-Specific Differences in RNA Pol II Occupancy Reflect Differences in Gene Expression
(A–D) Pol II occupancy in first instar larval neuroepithelial cells, third instar larval neuroepithelial cells, and neuroblasts. Scale bars represent log2 ratio change between Dam-Pol II and Dam (reference) samples. (A′–D′) Expression patterns of the respective genes in the first instar larval brain. (A″–D″) Expression patterns of the respective genes in the third instar larval brain.
Importantly, TaDa revealed the expression of numerous genes not previously shown to be active in the optic lobe. These include genes in the retinal determination network (Figure S2), such as twin of eyeless, Optix, eyes absent (Figures 6D and 6D′), and dachshund in both the neuroepithelium and in neuroblasts, wg, dpp, and sine oculis in neuroepithelium alone, and eyeless in neuroblasts alone (Figures 6E and 6E′). We also found a set of glutathione S-transferase genes (GSTs) and genes involved in carbohydrate metabolism expressed in the neuroepithelium (Figure 5).
Temporal Changes in Neuroepithelial Cell Transcription
The neuroepithelium does not begin to generate neuroblasts until the second instar larval stage (Hofbauer and Campos-Ortega, 1990; Nassif et al., 2003). To investigate the transcriptional changes in neuroepithelial cells before and after neuroblast production, we compared Pol II occupancy in the neuroepithelium at first and third instar larval stages. Several of the newly identified neuroepithelial genes are transcribed both in early and late neuroepithelial cells, including glutathione S-transferase GstD1 and the retinal determination genes eya, Optix, dac, and toy, as are the Notch target genes HLHmgamma (Figures 7A and 7A′), HLHmbeta, and E(spl). Strikingly, however, the majority of genes involved in the signaling pathways that are active in the third instar neuroepithelium are not transcribed in the first instar (Figures 7 and S2; Table S2); these include genes in the Notch, EGFR, Fat/Hippo, JAK/STAT, Dpp, and Wnt pathways. For example, the Notch targets HLHm5, HLHmdelta, and HLHmalpha are expressed at third, but not first, instar. Transcription of the gene encoding the JAK/STAT ligand Upd, which controls the timing of the neuroepithelial to neuroblast transition, is also absent from the early neuroepithelium (Figures 7B and 7B′), as is Egfr (Figures 7C and 7C′), dpp (Figures S3C–S3C″), members of the Hippo pathway (ds, hpo, and ft), and the Wnt pathway (wg, fz3, and Wnt2). This suggests that there is switch during neuroepithelial development when these signaling pathways are activated. It is likely that this is coordinated with the beginning of optic lobe neuroblast production at the second instar larval stage and may be induced by an ecdysone pulse toward the end of the first instar.
We also found that the cell cycle genes CycE (Figure 7) and E2f are not expressed in the early neuroepithelium, which is consistent with little or no cell division at this stage.
Discussion
TaDa is designed to express Dam-fusion proteins at very low levels in a spatially and temporally restricted manner, allowing the identification of protein-DNA interactions in a cell-specific fashion in vivo without the need for cell purification. We show here that TaDa enables transcriptome analysis and chromatin profiling in any cell type or lineage that can be genetically labeled. With TaDa, we identified actively transcribed genes in neuroepithelial cells and neuroblasts and genes at which RNA Pol II was paused. TaDa can be applied to the study of chromatin-associated factors (van Steensel et al., 2001; Tolhuis et al., 2006; Vogel et al., 2006; Filion et al., 2010), including nuclear envelope proteins (Guelen et al., 2008) and components of the RNAi pathway (Woolcock et al., 2011). We have also used TaDa to assess genome-wide binding of transcription factors (E. Contreras Sepulveda, A.R.C. Thompson, T.D.S., B.E., O.J.M., and A.H.B., unpublished data) and are currently assessing the binding of chromatin modifiers (T.D.S., O.J.M., and A.H.B., unpublished data).
TaDa is highly sensitive; we could detect Pol II binding in ∼250 neuroblasts (Miller et al., 2009) of ∼15,000 cells in the larval brain with only small amounts of starting material (50 larval brains). We recently succeeded in profiling ∼100 neurons of the ∼150,000 cells in intact adult heads (T.D.S. and A.H.B., unpublished data). TaDa generates a robust and reproducible signal exclusively in cells where GAL4 is active. The thousands of available GAL4 lines ensures that most cell types can be profiled.
TaDa can be adapted for use in any model organism amenable to transformation, as well as at any stage of development from embryo to adult. DamID works effectively in Drosophila (Bianchi-Frias et al., 2004; Choksi et al., 2006; Pym et al., 2006; Southall and Brand, 2009), yeast (Woolcock et al., 2011), plants (Germann et al., 2006), Caenorhabditis elegans (Schuster et al., 2010), and human tissue culture cells (Vogel et al., 2006; Guelen et al., 2008). Ribosome reinitiation, which makes TaDa possible, has been observed in mammalian cell lines (Luukkonen et al., 1995; Van Blokland et al., 2011). We are currently adapting TaDa for use in mammalian systems.
Our data confirm the expression of signaling pathways known to be active and required for the development of the proliferating neuroepithelium, such as Notch, JAK/STAT, EGFR, Wnt, and Dpp. We also identified many more genes, such as the transcription factors of the retinal determination network, which may pattern the neuroepithelium and cell adhesion molecules, such as NinjurinA, which may regulate tissue integrity and polarity in this rapidly dividing tissue.
The discovery of the spatially restricted expression of retinal determination genes in the optic lobe is intriguing. The retinal determination network is a highly dynamic and cross-regulatory gene network, with multiple feedback loops acting to regulate gene expression in the developing eye disc (Kumar, 2009). It is unclear whether these same regulatory relationships are recapitulated in the optic lobe, particularly because many of the transcription factors are expressed in distinct populations of neural precursors. It will be interesting to determine whether retinal determination gene function contributes to the patterning of distinct regions and cell types within the optic lobe.
Our STRING analysis also identified two unexpected clusters of genes as being expressed in the neuroepithelium, those encoding glutathione S-transferases (GSTs) and genes involved in carbohydrate metabolism. GSTs have roles in protecting cells against chemical toxicity and oxidative stress, in modulating JNK signaling and in cancer progression (for review, see Hayes et al., 2005; Laborde, 2010). Of the carbohydrate metabolism genes, Pepck and CG10924 are involved in gluconeogenesis and glyceroneogenesis, the generation of glucose and glycerol from noncarbohydrate carbon substrates. This pathway is usually associated with nutritional deprivation (for review, see Jitrapakdee, 2012). Another gene in the cluster is Pgd, which is part of the pentose phosphate pathway (PPP), an alternative to glycolysis for metabolizing glucose. PPP generates NADPH, an important player in protecting cells from oxidative stress. Isocitrate dehydrogenase (Idh) is part of the citric acid cycle and is also important for generating cytosolic NADPH and protecting cells against oxidative damage (Kim et al., 2012). These results suggest that noncanonical metabolic pathways are active in the neuroepithelium and that there may be mechanisms in place to protect against oxidative stress (GSTs and the carbohydrate metabolism genes Idh and Pgd).
TaDa has revealed dramatic differences in the activity of signaling pathways in the neuroepithelium during brain development. The Notch, EGFR, Fat-Hippo, and JAK/STAT signaling pathways work in coordination to control the transition of neuroepithelial cells to neuroblasts (for review, see Egger et al., 2011). The Dpp and Wnt pathways play a role in patterning the developing optic lobe (Kaphingst and Kunes, 1994). Strikingly, the majority of the genes in these pathways are not expressed in the first instar neuroepithelium. This suggests that there is a switch during neuroepithelial development, when these pathways are activated, likely in response to a developmentally regulated pulse of ecdysone (Truman et al., 1994). Many of the retinal determination genes (e.g., eya, Optix, dac, and toy) are expressed in the early neuroepithelium suggesting that they are responsible for patterning the neuroepithelium at this stage, or that they act to maintain neuroepithelial cell fate.
TaDa has permitted us to assay, simply and quickly, the transcriptomes of closely related neural stem cell pools in the developing brain, revealing the expression of both known and unknown pathways in these cells. These data provide the basis for future studies to uncover the function of retinal determination factors and noncanonical metabolism in neural stem cell regulation. Furthermore, we anticipate that the ease and adaptability of TaDa will open new horizons for researchers exploring the chromatin landscapes and transcriptomes of specific cell types during development and in the adult organism.
Experimental Procedures
Expression Constructs
Details and sequences of all primers used for generating constructs are shown in Supplemental Experimental Procedures. pUAST-LT1-NDam was generated by PCR amplifying NDamMyc from pUAST-Ndam (Choksi et al., 2006), using a 5′ primer that incorporates the coding sequence for MGGSAG, followed by two stop codons before the NdamMyc ORF. The resulting PCR product was cloned into pUAST (Brand and Perrimon, 1993) with EcoRI and BglII sites. pUAST-LT2-NDam was generated by amplifying the first 240 bp of mGFP6 (Schuldt et al., 1998) and PCR fusing it to a NdamMyc PCR fragment using a second PCR reaction. ΔmGFP6 is followed by two stop codons and a single nucleotide before the NDamMyc start codon. The fusion PCR product was cloned into pUAST using EcoRI and BglII. pUASTattB-LT3-NDam (see Figure 1C) was generated in a similar manner to pUAST-LT2-NDam, however, the full mCherry CDS was used as the primary ORF and the pUASTattB vector (Bischof et al., 2007) was used to allow site specific integration of the construct into the Drosophila genome. To generate pUAST-mGFP6-RpII215, pUAST-NmGFP6 was first made by PCR amplifying mGFP6 and cloning it into pUAST with EcoRI and BglII. RpII215 was PCR amplified from an embryonic cDNA library and cloned into pUAST-NmGFP6 using NotI and XbaI sites. RpII215 was cut out from pUAST-mGFP6-RpII215 with NotI and XbaI to generate pUASTattB-LT3-NDam-RpII215.
DamID
Salivary glands: UAS-LT3-NDam and UAS-LT3-NDam-RpII215 were crossed to hs-GAL4 (Brand et al., 1994; Costantino et al., 2008) and reared at 25°C. Thirty-five third instar larvae were dissected for each sample. Third instar larval brains: UAS-LT3-NDam and UAS-LT3-NDam-RpII215 were crossed to the appropriate GAL4 line containing GAL80ts. Embryos were collected over a 4 hr period at 25°C and then shifted to 18°C (restrictive temperature) for 2 days. Hatched larvae were then transferred to a food plate containing standard fly food and left at 18°C for a further 5 days. The food plate was then shifted to 29°C (permissive temperature) for 24 hr before dissection (third instar larvae). One hundred brains were dissected for each sample. First instar larval brains: same as for third instar except that hatched larvae were immediately shifted to 29°C for 24 hr, and 300 brains were dissected. Genomic DNA was extracted and methylated DNA processed and amplified as previously described (Sun et al., 2003; Choksi et al., 2006) apart from the following modifications: After precipitation of the gDNA, the DpnI digest was set up in 50 μl (rather than 10 μl). It is especially important not to shear the gDNA, therefore when the 50 μl of DpnI mix was added, the pellet was not disturbed by vortexing or mixing. Following overnight digestion, an extra 0.5 μl of DpnI enzyme was added and the sample gently mixed by pipetting. After denaturation of DpnI, the DNA was purified (QIAGEN PCR purification) into 30 μl of H2O, of which 15 μl was used for the ligation step.
Amplified DNA from experimental and Dam-only controls were labeled with either Cy3 or Cy5 and cohybridized to Nimblegen HD2 Drosophila whole genome tiling arrays (performed at FlyChip, Cambridge, UK). Arrays were scanned using a Genepix 4000B dual laser scanner (Axon) and data probe intensities extracted using NimbleScan software (Nimblegen). Three biological replicates were performed for profiling of the neuroepithelium (one dye swap) and two replicates for neuroblasts and salivary glands (with dye-swap).
DamID Analysis
Log2 ratio files were generated (Dam-Pol II over Dam-only) and median normalized (NimbleScan software). Each replicate data set was analyzed separately: Using Drosophila genome annotation release 5.47, the mean ratio change across each annotated transcript was calculated. To assign a FDR value, the frequency of transcripts with a mean ratio over specific values (from 0.1 to 0.75 log2 increase) were calculated within a randomized data set (for each chromosome arm) using 10 iterations and 1,000 sampling events. This was repeated for a range of gene sizes (250 to 2,500 bp). These data were used to model FDR values relative to the Dam-Pol II enrichment across a transcript and gene sizes, therefore enabling extrapolation of FDR values for larger ratio changes and larger transcripts. After being performed for each replicate, each transcript was assigned a mean ratio between the biological replicates and the highest associated FDR. Perl script available on request. To compare data sets, log2 ratios were subtracted and the data median normalized. These data were then analyzed as described above to identify genes with significantly different Pol II occupancy. Due to the presence of negative log2 ratios in DamID experiments, these genes were filtered to check that any significantly enriched genes were also bound by Pol II in the experiment of interest (numerator data set). A gene list was generated from the transcript data using the values from the associated transcript with the most significant FDR. Metagene profiles were created from nonoverlapping genes with a size between 1,500 bp and 20 kb. Where multiple splice variants existed, metagene profiles of transcribed genes were created using only the transcript with the lowest FDR from each gene.
Statistics
The correlation between Dam-Pol II and Pol II-ChIP log2 ratios across gene structures was analyzed by first determining the average ratio across each gene structure (using the Perl script described above) and then calculating the correlation coefficient using Excel. Significantly bound genes were determined using the FDR program described above.
Fly Lines and Germline Transformation
asense-GAL4 (Zhu et al., 2006) was used for testing toxicity of UAS-Dam lines. For POLII profiling, we used the GAL4 driver lines hs-GAL4 (Brand et al., 1994; Costantino et al., 2008), GAL4c855a (Manseau et al., 1997; Egger et al., 2007), and insc-GAL4 (GAL4MZ1407) (Luo et al., 1994). tubulin-GAL4 (Lee and Luo, 1999) was used for analysis of Dam-Pol II protein levels. w-; insc-GAL4; tubulin-GAL80ts and w-; tubulin-GAL80ts; GAL4c855a were used for temporally restricted expression of UAS-LT3-NDam and UAS-LT3-NDam-RpII215 in neuroblasts and the neuroepithelium, respectively. GAL4 reporter lines used upd-GAL4 (Halder et al., 1995), h-GAL4 (Brand and Perrimon, 1993), yw; rA87-GAL4/CyO (gcm-GAL4) (Chotard et al., 2005), EGFR-GAL4 (Bloomington stock 45123; Jenett et al., 2012), and dpp-GAL4 (Takaesu et al., 2002). HLHmgamma-GFP (Almeida and Bray, 2005) was used to assess the expression pattern of HLHmgamma. UAS-LT1-NDam, UAS-LT2-NDam, and UAS-mGFP6-RpII215 were generated by injection of w1118 embryos with the respective constructs and Δ2-3 helper plasmid as a transposase source. UAS-LT3-NDam and UAS-LT3-NDam-RpII215 were generated by injection of w-; +; attP2 (Markstein et al., 2008) embryos with the respective constructs and phiC31 integrase helper plasmid pBS130 as an integrase source.
Immunohistochemistry and In Situ Hybridization
Immunohistochemistry of Drosophila larval brains was carried out as previously described (Van Vactor et al., 1991), using the following antisera: mouse anti-Dlg (1/100), (Developmental Studies Hybridoma Bank), chicken anti-GFP (1/2,000) (ab13970, Abcam), guinea pig anti-Dpn (1/500) (gift from J.B. Skeath), guinea pig anti-NijA (1/500) (Zhang et al., 2006), rabbit anti-Tsh (1/300) (Wu and Cohen, 2000), rabbit anti-Eyeless (1/300) (gift from Uwe Walldorf), mouse anti-Eyes absent (1/75) (DSHB), rat anti-DE-Cadherin (DCAD2) (1/20) (DSHB), anti-CycE (1/30) (Richardson et al., 1995), and rabbit anti-Scribble (1/2,000). Secondary antibodies coupled to Alexa-488, Alexa-546, or Alexa-633 (Invitrogen) were used at a dilution of 1/200. Fluorescent in situ hybridization was performed as previously described (Egger et al., 2007). Images were acquired with a Leica SP2 or a Zeiss 510 confocal microscope. Figures and illustrations were assembled using Adobe Photoshop and Adobe Illustrator.
Acknowledgments
We thank Andrea Page McCaw, Helen Skaer, Uwe Walldorf, Chris Doe, and DSHB for antibodies, Steve Russell and Bettina Fischer at FlyChip, Barret Pfeiffer for advice about GAL4 lines, and Iris Salecker and the Bloomington Stock Center for Drosophila stocks. This work was funded by Wellcome Trust Programme grants 068055 and 092545 (to A.H.B.) and by a Wellcome Trust PhD Studentship (to K.S.G), Swiss National Fund Fellowship (to B.E.), EMBO Long Term Fellowship (to O.J.M.), and Herchel Smith Postdoctoral Research Fellowships (to T.D.S. and E.E.C.). A.H.B acknowledges core funding to the Gurdon Institute from the Wellcome Trust (092096) and CRUK (C6946/A14492).
Published: June 20, 2013
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes Supplemental Experimental Procedures, three figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2013.05.020.
Accession Numbers
The DamId-chip data have been deposited under NCBI GEO accession number GSE46803.
Supplemental Information
References
- Almeida M.S., Bray S.J. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech. Dev. 2005;122:1282–1293. doi: 10.1016/j.mod.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Bardin A.J., Schweisguth F. Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev. Cell. 2006;10:245–255. doi: 10.1016/j.devcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Bianchi-Frias D., Orian A., Delrow J.J., Vazquez J., Rosales-Nieves A.E., Parkhurst S.M. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS Biol. 2004;2:E178. doi: 10.1371/journal.pbio.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S., Zinzen R.P., Girardot C., Gustafson E.H., Perez-Gonzalez A., Delhomme N., Ghavi-Helm Y., Wilczyński B., Riddell A., Furlong E.E. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Bonn S., Zinzen R.P., Perez-Gonzalez A., Riddell A., Gavin A.C., Furlong E.E. Cell type-specific chromatin immunoprecipitation from multicellular complex samples using BiTS-ChIP. Nat. Protoc. 2012;7:978–994. doi: 10.1038/nprot.2012.049. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Manoukian A.S., Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- Bryant Z., Subrahmanyan L., Tworoger M., LaTray L., Liu C.R., Li M.J., van den Engh G., Ruohola-Baker H. Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis. Proc. Natl. Acad. Sci. USA. 1999;96:5559–5564. doi: 10.1073/pnas.96.10.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child S.J., Miller M.K., Geballe A.P. Translational control by an upstream open reading frame in the HER-2/neu transcript. J. Biol. Chem. 1999;274:24335–24341. doi: 10.1074/jbc.274.34.24335. [DOI] [PubMed] [Google Scholar]
- Choksi S.P., Southall T.D., Bossing T., Edoff K., de Wit E., Fischer B.E., van Steensel B., Micklem G., Brand A.H. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Chotard C., Leung W., Salecker I. glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron. 2005;48:237–251. doi: 10.1016/j.neuron.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Conrad T., Cavalli F.M., Vaquerizas J.M., Luscombe N.M., Akhtar A. Drosophila dosage compensation involves enhanced Pol II recruitment to male X-linked promoters. Science. 2012;337:742–746. doi: 10.1126/science.1221428. [DOI] [PubMed] [Google Scholar]
- Costantino B.F., Bricker D.K., Alexandre K., Shen K., Merriam J.R., Antoniewski C., Callender J.L., Henrich V.C., Presente A., Andres A.J. A novel ecdysone receptor mediates steroid-regulated developmental events during the mid-third instar of Drosophila. PLoS Genet. 2008;4:e1000102. doi: 10.1371/journal.pgen.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Renzis S., Yu J., Zinzen R., Wieschaus E. Dorsal-ventral pattern of Delta trafficking is established by a Snail-Tom-Neuralized pathway. Dev. Cell. 2006;10:257–264. doi: 10.1016/j.devcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Deal R.B., Henikoff S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B., Boone J.Q., Stevens N.R., Brand A.H., Doe C.Q. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B., Gold K.S., Brand A.H. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development. 2010;137:2981–2987. doi: 10.1242/dev.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B., Gold K.S., Brand A.H. Regulating the balance between symmetric and asymmetric stem cell division in the developing brain. Fly (Austin) 2011;5:237–241. doi: 10.4161/fly.5.3.15640. [DOI] [PubMed] [Google Scholar]
- Farkas L.M., Huttner W.B. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr. Opin. Cell Biol. 2008;20:707–715. doi: 10.1016/j.ceb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Filion G.J., van Bemmel J.G., Braunschweig U., Talhout W., Kind J., Ward L.D., Brugman W., de Castro I.J., Kerkhoven R.M., Bussemaker H.J., van Steensel B. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germann S., Juul-Jensen T., Letarnec B., Gaudin V. DamID, a new tool for studying plant chromatin profiling in vivo, and its use to identify putative LHP1 target loci. Plant J. 2006;48:153–163. doi: 10.1111/j.1365-313X.2006.02859.x. [DOI] [PubMed] [Google Scholar]
- Gilchrist D.A., Dos Santos G., Fargo D.C., Xie B., Gao Y., Li L., Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W., van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Halder G., Callaerts P., Gehring W.J. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hayes J.D., Flanagan J.U., Jowsey I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- Henry G.L., Davis F.P., Picard S., Eddy S.R. Cell type-specific genomics of Drosophila neurons. Nucleic Acids Res. 2012;40:9691–9704. doi: 10.1093/nar/gks671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer A., Campos-Ortega J.A. Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Rouxs Arch. Dev. Biol. 1990;198:264–274. doi: 10.1007/BF00377393. [DOI] [PubMed] [Google Scholar]
- Jenett A., Rubin G.M., Ngo T.T., Shepherd D., Murphy C., Dionne H., Pfeiffer B.D., Cavallaro A., Hall D., Jeter J. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitrapakdee S. Transcription factors and coactivators controlling nutrient and hormonal regulation of hepatic gluconeogenesis. Int. J. Biochem. Cell Biol. 2012;44:33–45. doi: 10.1016/j.biocel.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Kaphingst K., Kunes S. Pattern formation in the visual centers of the Drosophila brain: wingless acts via decapentaplegic to specify the dorsoventral axis. Cell. 1994;78:437–448. doi: 10.1016/0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Shin J.Y., Kim M., Hann S.K., Oh S.H. Expression of cytosolic NADP(+)-dependent isocitrate dehydrogenase in melanocytes and its role as an antioxidant. J. Dermatol. Sci. 2012;65:118–125. doi: 10.1016/j.jdermsci.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J.P. The molecular circuitry governing retinal determination. Biochim. Biophys. Acta. 2009;1789:306–314. doi: 10.1016/j.bbagrm.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laborde E. Glutathione transferases as mediators of signaling pathways involved in cell proliferation and cell death. Cell Death Differ. 2010;17:1373–1380. doi: 10.1038/cdd.2010.80. [DOI] [PubMed] [Google Scholar]
- Lam K.C., Mühlpfordt F., Vaquerizas J.M., Raja S.J., Holz H., Luscombe N.M., Manke T., Akhtar A. The NSL complex regulates housekeeping genes in Drosophila. PLoS Genet. 2012;8:e1002736. doi: 10.1371/journal.pgen.1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Luo L., Liao Y.J., Jan L.Y., Jan Y.N. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Luukkonen B.G., Tan W., Schwartz S. Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J. Virol. 1995;69:4086–4094. doi: 10.1128/jvi.69.7.4086-4094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manseau L., Baradaran A., Brower D., Budhu A., Elefant F., Phan H., Philp A.V., Yang M., Glover D., Kaiser K. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev. Dyn. 1997;209:310–322. doi: 10.1002/(SICI)1097-0177(199707)209:3<310::AID-AJA6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Markstein M., Pitsouli C., Villalta C., Celniker S.E., Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Toh-e A., Oshima Y. Genetic control of galactokinase synthesis in Saccharomyces cerevisiae: evidence for constitutive expression of the positive regulatory gene gal4. J. Bacteriol. 1978;134:446–457. doi: 10.1128/jb.134.2.446-457.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S.E., Le P.T., Osborn A.J., Matsumoto K., Davis R.L. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Miller M.R., Robinson K.J., Cleary M.D., Doe C.Q. TU-tagging: cell type-specific RNA isolation from intact complex tissues. Nat. Methods. 2009;6:439–441. doi: 10.1038/nmeth.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif C., Noveen A., Hartenstein V. Early development of the Drosophila brain: III. The pattern of neuropile founder tracts during the larval period. J. Comp. Neurol. 2003;455:417–434. doi: 10.1002/cne.10482. [DOI] [PubMed] [Google Scholar]
- Nechaev S., Fargo D.C., dos Santos G., Liu L., Gao Y., Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neira M., Azen E. Gene discovery with laser capture microscopy. Methods Enzymol. 2002;356:282–289. doi: 10.1016/s0076-6879(02)56941-x. [DOI] [PubMed] [Google Scholar]
- Ngo K.T., Wang J., Junker M., Kriz S., Vo G., Asem B., Olson J.M., Banerjee U., Hartenstein V. Concomitant requirement for Notch and Jak/Stat signaling during neuro-epithelial differentiation in the Drosophila optic lobe. Dev. Biol. 2010;346:284–295. doi: 10.1016/j.ydbio.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pym E.C., Southall T.D., Mee C.J., Brand A.H., Baines R.A. The homeobox transcription factor Even-skipped regulates acquisition of electrical properties in Drosophila neurons. Neural Dev. 2006;1:3. doi: 10.1186/1749-8104-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy B.V., Rauskolb C., Irvine K.D. Influence of fat-hippo and notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development. 2010;137:2397–2408. doi: 10.1242/dev.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson H., O’Keefe L.V., Marty T., Saint R. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development. 1995;121:3371–3379. doi: 10.1242/dev.121.10.3371. [DOI] [PubMed] [Google Scholar]
- Rideout E.J., Dornan A.J., Neville M.C., Eadie S., Goodwin S.F. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P.J., Stuart J.M., Lund J., Kim S.K. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- Schuldt A.J., Adams J.H., Davidson C.M., Micklem D.R., Haseloff J., St Johnston D., Brand A.H. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 1998;12:1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster E., McElwee J.J., Tullet J.M., Doonan R., Matthijssens F., Reece-Hoyes J.S., Hope I.A., Vanfleteren J.R., Thornton J.M., Gems D. DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol. Syst. Biol. 2010;6:399. doi: 10.1038/msb.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Southall T.D., Brand A.H. Neural stem cell transcriptional networks highlight genes essential for nervous system development. EMBO J. 2009;28:3799–3807. doi: 10.1038/emboj.2009.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner F.A., Talbert P.B., Kasinathan S., Deal R.B., Henikoff S. Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res. 2012;22:766–777. doi: 10.1101/gr.131748.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugaya K., Vigneron M., Cook P.R. Mammalian cell lines expressing functional RNA polymerase II tagged with the green fluorescent protein. J. Cell Sci. 2000;113:2679–2683. doi: 10.1242/jcs.113.15.2679. [DOI] [PubMed] [Google Scholar]
- Sun L.V., Chen L., Greil F., Negre N., Li T.R., Cavalli G., Zhao H., Van Steensel B., White K.P. Protein-DNA interaction mapping using genomic tiling path microarrays in Drosophila. Proc. Natl. Acad. Sci. USA. 2003;100:9428–9433. doi: 10.1073/pnas.1533393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaesu N.T., Johnson A.N., Newfeld S.J. Posterior spiracle specific GAL4 lines: new reagents for developmental biology and respiratory physiology. Genesis. 2002;34:16–18. doi: 10.1002/gene.10109. [DOI] [PubMed] [Google Scholar]
- Thomas A., Lee P.J., Dalton J.E., Nomie K.J., Stoica L., Costa-Mattioli M., Chang P., Nuzhdin S., Arbeitman M.N., Dierick H.A. A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PLoS ONE. 2012;7:e40276. doi: 10.1371/journal.pone.0040276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B., de Wit E., Muijrers I., Teunissen H., Talhout W., van Steensel B., van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Truman J.W., Talbot W.S., Fahrbach S.E., Hogness D.S. Ecdysone receptor expression in the CNS correlates with stage-specific responses to ecdysteroids during Drosophila and Manduca development. Development. 1994;120:219–234. doi: 10.1242/dev.120.1.219. [DOI] [PubMed] [Google Scholar]
- Van Blokland H.J., Hoeksema F., Siep M., Otte A.P., Verhees J.A. Methods to create a stringent selection system for mammalian cell lines. Cytotechnology. 2011;63:371–384. doi: 10.1007/s10616-011-9354-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B., Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Delrow J., Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- Van Vactor D.L., Jr., Cagan R.L., Krämer H., Zipursky S.L. Induction in the developing compound eye of Drosophila: multiple mechanisms restrict R7 induction to a single retinal precursor cell. Cell. 1991;67:1145–1155. doi: 10.1016/0092-8674(91)90291-6. [DOI] [PubMed] [Google Scholar]
- Vogel M.J., Guelen L., de Wit E., Peric-Hupkes D., Lodén M., Talhout W., Feenstra M., Abbas B., Classen A.K., van Steensel B. Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res. 2006;16:1493–1504. doi: 10.1101/gr.5391806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel M.J., Peric-Hupkes D., van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- Voigt A., Pflanz R., Schäfer U., Jäckle H. Perlecan participates in proliferation activation of quiescent Drosophila neuroblasts. Dev. Dyn. 2002;224:403–412. doi: 10.1002/dvdy.10120. [DOI] [PubMed] [Google Scholar]
- Wang W., Li Y., Zhou L., Yue H., Luo H. Role of JAK/STAT signaling in neuroepithelial stem cell maintenance and proliferation in the Drosophila optic lobe. Biochem. Biophys. Res. Commun. 2011;410:714–720. doi: 10.1016/j.bbrc.2011.05.119. [DOI] [PubMed] [Google Scholar]
- Woolcock K.J., Gaidatzis D., Punga T., Bühler M. Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 2011;18:94–99. doi: 10.1038/nsmb.1935. [DOI] [PubMed] [Google Scholar]
- Wu J., Cohen S.M. Proximal distal axis formation in the Drosophila leg: distinct functions of teashirt and homothorax in the proximal leg. Mech. Dev. 2000;94:47–56. doi: 10.1016/s0925-4773(00)00311-7. [DOI] [PubMed] [Google Scholar]
- Yasugi T., Umetsu D., Murakami S., Sato M., Tabata T. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135:1471–1480. doi: 10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]
- Yasugi T., Sugie A., Umetsu D., Tabata T. Coordinated sequential action of EGFR and Notch signaling pathways regulates proneural wave progression in the Drosophila optic lobe. Development. 2010;137:3193–3203. doi: 10.1242/dev.048058. [DOI] [PubMed] [Google Scholar]
- Zhang S., Dailey G.M., Kwan E., Glasheen B.M., Sroga G.E., Page-McCaw A. An MMP liberates the Ninjurin A ectodomain to signal a loss of cell adhesion. Genes Dev. 2006;20:1899–1910. doi: 10.1101/gad.1426906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Lin S., Kao C.F., Awasaki T., Chiang A.S., Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.