Abstract
OBJECTIVES:
The aim of this cross-sectional study was to evaluate whether interleukin 10 (IL10) and transforming growth factor β1 (TGFβ1) gene polymorphisms were associated with persistent IgE-mediated cow's milk allergy in 50 Brazilian children. The diagnostic criteria were anaphylaxis triggered by cow's milk or a positive double-blind, placebo-controlled food challenge. Tolerance was defined as the absence of a clinical response to a double-blind, placebo-controlled food challenge or cow's milk exposure.
METHOD:
The genomic DNA of the 50 patients and 224 healthy controls (HCs) was used to investigate five IL10 gene polymorphisms (-3575A/T, -2849A/G, -2763A/C, -1082G/A, -592C/A) and one TGFβ1 polymorphism (-509C/T).
RESULTS:
Among the five IL10 polymorphisms analyzed, homozygosis for the G allele at the -1082 position was significantly higher in the patients compared with the healthy controls (p = 0.027) and in the persistent cow's milk allergy group compared with the healthy controls (p = 0.001).
CONCLUSIONS:
Homozygosis for the G allele at the IL10 -1082G/A polymorphism is associated with the persistent form of cow's milk allergy.
Keywords: Food Hypersensitivity, Cow's Milk Allergy, Clinical Outcome, Gene Polymorphism, Interleukin 10, Transforming Growth Factor β1
INTRODUCTION
The prevalence of food allergy has been increasing over the last decades. Many reports indicate that new eating habits associated with epigenetic factors may be responsible for this change worldwide (1,2).
Sensitization, the first step in food allergy, is characterized by allergen-specific IgE production along with a Th2 immune response, inflammatory cytokine release and the onset of signs and symptoms (3). Despite the daily ingestion of many proteins in the diet, only a few are associated with clinical manifestations of allergy: cow's milk (CM), soy, eggs, peanuts, wheat, nuts, fish and crustaceans (4,5). CM allergy (CMA) is one of the leading causes of food allergy in childhood, affecting 2.5% of children under the age of 3 and 6% of children under the age of 4 (4). CM proteins, such as casein, beta-lactoglobulin and alpha-lactalbumin, may be involved in long-term immunological learning (6). Although most patients overcome the allergy, the persistence of CMA has been reported (7). CMA prognosis is usually good in patients developing tolerance up to 3 years of age, although recent studies have shown more severe outcomes in patients with IgE-mediated CMA (5). The persistence of CMA has also been associated with sensitization to casein and the occurrence of anaphylaxis (6).
Predictive markers of persistent CMA have been extensively studied to identify groups eligible for new treatments targeted to induce tolerance, such as immunotherapy with food allergens associated with bacterial products or the use of genetically modified allergens. A number of regulatory cells play a key role in the immune response to food, including (CD4+CD25+) lymphocytes (Treg), Th3 cells and T regulatory type 1 cells (Tr1), which produce immunoregulatory cytokines, such as transforming growth factor β1 (TGFβ1) and interleukin 10 (IL10). A reduction in TGFβ1 production due to the inadequate stimulation of innate immunity by intestinal microflora may contribute to the failure of oral tolerance (8). Several studies have associated low TGFβ1 levels with sensitization to food (8,9). The deletion/anergy of reactive T cells against specific antigens associated with the expanding population of regulatory T cells and the production of IL10 are mechanisms involved in oral tolerance (3).
The present study aimed to investigate whether the presence of single nucleotide polymorphisms (SNPs) in such immunoregulatory genes as IL10 and TGFβ1 are associated with demographic and clinical data in order to better characterize the group of children at a high risk of evolving persistent CMA.
MATERIALS AND METHODS
Ethics statement
Written informed consent was obtained from all participants, and the study was approved by the Internal Review Board of the University of São Paulo (CAPpesq n°1107/07, 03/17/2008). Permission to use DNA samples from HCs was also granted strictly according to the Brazilian Ethical Committee guidelines (CAPpesq n°560/00, 08/09/2002).
Study population
This study enrolled 50 children older than 5 years of age with IgE-mediated CMA who were followed up at the Child Institute, General Hospital, Faculdade de Medicina da Universidade de São Paulo, Brazil. The children were diagnosed according to the following criteria: anaphylaxis triggered by CM or immediate clinical reactivity induced by CM during a double-blind, placebo-controlled food challenge (DBPCFC). The patient group was compared with 224 HCs recruited from bone marrow donors at the same hospital. These individuals were clinically evaluated to exclude any type of allergy.
DNA preparation
Blood samples were obtained from the CMA patients. Genomic DNA was extracted according to the DTAB/CTAB method (10).
SNP selection and genotyping
The choice of SNPs was made on the basis of the SNPs' role in cytokine production (11-13).
SNPs located at the promoter region of the IL10 gene or at the -3575 (rs1800890), -2849 (rs6703630), -2763 (rs6693899) or -592 (rs1800872) position were tested by polymerase chain reaction (PCR)-RFLP according to previously described protocols (14,15).
Briefly, PCR was performed in a final volume of 25 μL containing 100 ng of genomic DNA, 40 μM dNTP, 0.2 U of Taq polymerase, 1.5 mM MgCl2 and 0.25 pM of each primer. In the case of the -592 SNP, 2.0 mM MgCl2 and 0.5 pM of each primer was used. The PCR conditions were an initial denaturation step at 95°C for 5 min, followed by 35 cycles at 95°C for 20 s; annealing temperatures of 62°C (-3575), 60°C (-2849 and -2763) or 56°C (-592) for 30 s; and an extension step at 72°C for 20 s, ending with 7 min at 72°C. An aliquot of 10 μL of the PCR product was digested for 3 h at 37°C with 2.5 U of Apo I (-3575), 2.5 U of Alw I (-2849), 5 U of Dde I (-2763) or 3.0 U of Rsa I (all enzymes from New England Biolabs, Ipswich, MA) in a total volume of 20 μL. Digestion products were separated by electrophoresis in 4% agarose gels, stained with ethidium bromide and visualized under UV light.
The IL10 -1082 polymorphism (rs1800896) was genotyped using an allele-specific PCR (ASPCR). The specific primer S1 or S2 paired with the common primer C were used to amplify a 258 bp fragment (16). The housekeeping hGH gene was used as an internal control, yielding a 480 bp amplification product. PCR was performed using the same conditions as for the IL10 -592 polymorphism.
The TGFβ1 -509 polymorphism (rs1800469) was typed by PCR-RFLP with primers described elsewhere (17). Briefly, PCR-RFLP was performed in a final volume of 25 μL containing 100 ng of genomic DNA, 40 μM dNTP, 0.2 U of Taq polymerase, 1.5 mM MgCl2 and 0.25 pM of each primer, using the same protocol as for IL10 -592. An aliquot of 20 μL of the PCR product was digested for 3 h at 37°C with 3 U of Dde I (New England Biolabs) in a total volume of 40 μL. Digestion products were separated by electrophoresis in a 4% agarose gel, stained with ethidium bromide and visualized under UV light.
Statistics
Considering a type I error to be 0.05 and a relative risk of 2.5, defined according to our significant results, the power was estimated to detect statistically significant differences for the SNP frequencies in the two studied groups (CMA and HC) EPI Center version 4.1 (Extreme Networks, Inc., Santa Clara, California, USA). The values obtained were 76% and 67% for IL10 -1082 G/A and TGFβ1 -509C/T, respectively, and 66% for IL10 -1082 G/A considering the CMA group (persistent, n = 36), indicating that our sample size was adequate to avoid false-negative association.
Genotype and allele frequencies were calculated by direct gene counting. Comparisons between the CMA and HC groups were performed by the x2 test along with calculation of the odds ratio (OR) and 95% confidence interval (CI). Statistical comparisons were performed using Fisher's exact test whenever a value in the contingency table was below five and to perform genotype analysis using GraphPad Prism version 5.0 software. The p values were corrected for the number of different alleles tested (pc). Statistical significance was defined as a p value less than 0.05. Haplotype frequency, Hardy-Weinberg expectation (HWE) and linkage disequilibrium (LD) analyses were performed using Arlequin 3.1 software (18).
A logistic regression model was applied, considering the development of tolerance as a dichotomous variable, and continuous and qualitative parameters in univariate analysis were considered to be independent variables. Parameters with p<0.2 (sex, anaphylaxis, the presence of other allergic diseases, IgE serum levels and specific IgE) were initially included in the multivariable analysis, and only those parameters with p<0.05 were considered to be significant. These analyses were performed considering genotypes and allele carriage (the number of individuals having the allele on either one or both chromosomes). IgE levels (BN100; Siemens Healthcare Diagnostics, Marburg, Germany) were categorized into two groups: normal and elevated, according to the reference interval defined for each age by the manufacturer.
RESULTS
Clinical features of and laboratory findings for the children with CMA
The 50 studied patients were divided into two groups according to their clinical evolution: 36 patients were classified with persistent CMA (group P) and 14 were classified with tolerant CMA (group T). The onset of disease occurred very early in both groups, with no difference in the median age, although signs and symptoms appeared later in the persistent CMA group (15 vs 2 days). Familial atopy was slightly more frequent in the persistent CMA group compared with the tolerant CMA group (72% vs 57%). Anaphylaxis was associated with the persistent CMA group (p = 0.004). Similar trends were observed regarding the presence of wheezing (p = 0.018) and other allergies, such as asthma, rhinitis or dermatitis (p = 0.031) (Table 1).
Table 1.
Clinical and laboratory characteristics of patients with persistent or tolerant, IgE-mediated cow's milk allergy.
| Tolerant | Persistent | p-value | |
| (n = 14) | (n = 36) | ||
| Sex F/M | 8/6 | 13/23 | ns |
| Age of onset (days) | |||
| Median | 135 | 120 | ns |
| Range | 2-330 | 15-600 | |
| Diagnosis (days) | |||
| Median | 195 | 180 | ns |
| Range | 20-900 | 15-690 | |
| Follow-up (months) | |||
| Median | 48 | 46 | ns |
| Range | 11-124 | 5-164 | |
| Family atopy | 8 | 26 | ns |
| Anaphylaxis | 3 | 24 | 0.004 |
| Manifestation | |||
| Cutaneous | 13 | 34 | ns |
| Gastrointestinal | 8 | 24 | ns |
| Respiratory | 7 | 26 | 0.018 |
| Wheezing | 1 | 27 | |
| Other food allergies | 3 | 12 | ns |
| Other allergiesa | 6 | 27 | 0.031 |
| IgE (IU/mL) | |||
| Median | 186.5 | 473 | ns |
| Range | 9.3-10,180 | 43-6,740 | |
| Specific IgE (>0.35/total) | ns | ||
| CM | 14/14 | 33/35 | ns |
| Alpha | 8/10 | 20/23 | ns |
| Beta | 9/11 | 21/23 | ns |
| Casein | 10/11 | 21/24 | ns |
ns = not significant; aasthma, rhinitis or dermatitis.
Association of TGFβ1 and IL10 gene polymorphisms with CMA
All six genotyped SNPs are in HWE. The five IL10 polymorphisms were in strong LD (data not shown). We observed a statistically significant difference in both the allele and the genotype distributions of the IL10 -1082 G/A polymorphism when CMA patients were compared with HCs. Homozygosis for the G allele was higher in the CMA group compared with the HC group (19 vs 12%; p = 0.027), but this difference was not maintained after Bonferroni correction (pc = 0.054) or in patients with persistent CMA compared with HCs (24 vs 12%; p = 0.001; pc = 0.002). The genotype containing the G allele (AG plus GG vs AA) was significantly more frequent in the persistent CMA group compared with the tolerant CMA group (OR = 6.77, 95% CI 1.59-28.73, p = 0.006). The presence of the G allele conferred a twofold risk of developing persistent CMA (OR = 2.43, 95% CI 1.45-4.08, p<0.001), and the risk was even higher in the persistent CMA group compared with the tolerant CMA group (OR = 3.24, 95% CI 1.20-8.76, p = 0.017) (Table 2).
Table 2.
Genotype and allele frequency of IL10 -1082 A/G polymorphism in children with cow's milk allergy and in healthy controls (HCs).
| lL10 -1082A/G | T (n = 13) | p-value (n = 34) | Total (n = 47) | HC (n = 217) | p-value | pc | x2 | OR | 95% CI |
| Genotype | |||||||||
| AA | 7 (54) | 5 (14) | 12 (26) | 100 (46) | |||||
| AG | 5 (38) | 21 (62) | 26 (55) | 91 (42) | 0.032 | ns | 6.90 | ||
| GG | 1 (8) | 8 (24) | 9 (19) | 26 (12) | |||||
| Genotype comparisons | |||||||||
| AA vs GG | |||||||||
| CMA vs HC | 0.027 | ns | 4.90 | 2.89 | 1.10-7.58 | ||||
| T vs HC | ns | ||||||||
| P vs HC | 0.001 | 0.002 | 10.67 | 6.15 | 1.86-20.39 | ||||
| P vs T | ns | ||||||||
| AG and GG vs AA | |||||||||
| CMA vs HC | 0.001 | 0.002 | 6.68 | 2.49 | 1.23-5.06 | ||||
| T vs HC | ns | ||||||||
| P vs HC | <0.001 | <0.001 | 11.89 | 4.96 | 1.85-13.29 | ||||
| P vs T | 0.006 | 0.016 | 7.58 | 6.77 | 1.59-28.73 | ||||
| Allele | |||||||||
| A | 19 (73) | 31 (46) | 50 (53) | 291 (67) | |||||
| G | 7(27) | 37 (54) | 44 (47) | 143 (33) | |||||
| Allele comparison A vs G | |||||||||
| CMA vs HC | 0.011 | 0.022 | 6.49 | 1.74 | 1.14-2.81 | ||||
| T vs HC | ns | ||||||||
| P vs HC | <0.001 | <0.001 | 11.77 | 2.43 | 1.45-4.08 | ||||
| P vs T | 0.017 | 0.034 | 5.71 | 3.24 | 1.20-8.76 |
HC = healthy control; n = number of individuals; x2$ = Chi-square; OR = odds ratio; ns = not significant.
The presence of the CC genotype in the TGFβ1 -509C/T promoter region showed an initial association with persistent allergy compared with the genotype in tolerant patients (14% vs 42%; p = 0.048) that was not maintained after correction (pc = 0.096).
The remaining studied polymorphisms (IL10 -3575, IL10 -2849, IL10 -2763 and IL10 -592) showed no association with CMA (Supplementary Table 1).
Supplementary Table 1.
Genotype frequency of TGFb1 -509T/C, IL10 -3575A/T, IL10 -2849 A/G, IL10 -2763 C/A and IL10 - 592A/C, comparing children with cow's milk allergy with healthy individuals and comparing the tolerant form with the persistent form in CMA patients.
| CMA | ||||||||
| HC | CMA | x2 | p-value | Tolerant | Persistent | x2 | p-value | |
| TFGB1-509 | ||||||||
| CC | 36 (0.38) | 17(0.34) | 2 (0.14) | 15 (0.42) | 6.06 | 0.0484 | ||
| CT | 39 (0.41) | 19 (030) | 0.89 | 0.6419 | 9 (0.64) | 10 (0.27) | ||
| TT | 20 (0.21) | 14 (0.38) | 3 (0.33) | 11 (0.31) | ||||
| TOTAL | 95 | 50 | 14 | 36 | ||||
| IL10 -3575 | ||||||||
| AA | 13 (0.06) | 5 (0.22) | 2 (0.14) | 3 (0.09) | 0.47 | 0.7906 | ||
| AT | 77 (0.35) | 20 (0.28) | 2.83 | 0.2428 | 5 (0.36) | 15 (0.44) | ||
| TT | 134 (0.60) | 23 (0.50) | 7 (0.50) | 16 (0.47) | ||||
| TOTAL | 224 | 49 | 14 | 34 | ||||
| IL10-2849 | ||||||||
| AA | 12 (0.05) | 1 (0.02) | 1 (0.07) | 0 (0.00) | 4.52 | 0.1045 | ||
| AG | 52 (0.23) | 19 (0.39) | 5.54 | 0.0626 | 3 (0.21) | 16 (0.46) | ||
| GG | 160 (0.71) | 29 (0.59) | 10 (0.71) | 19 (0.54) | ||||
| TOTAL | 224 | 49 | 14 | 35 | ||||
| IL10-2763 | ||||||||
| AA | 21 (0.09) | 3 (0.06) | 1 (0.07) | 2 (0.06) | 0.40 | 0.8188 | ||
| AC | 66 (0.29) | 22 (0.44) | 4.16 | 0.1251 | 7 (0.50) | 15 (0.42) | ||
| CC | 138 (0.62) | 25 (0.50) | 6 (0.43) | 19 (0.52) | ||||
| TOTAL | 224 | 50 | 14 | 36 | ||||
| IL10-592 | ||||||||
| AA | 32 (0.14) | 7 (0.15) | 2 (0.18) | 5 (0.14) | 1.02 | 0.5944 | ||
| AC | 95 (0.42) | 24 (0.51) | 1.33 | 0.5154 | 8 (0.62) | 16 (0.44) | ||
| CC | 97 (0.43) | 16 (0.34) | 3 (0.23) | 13 (0.36) | ||||
| TOTAL | 224 | 47 | 13 | 36 | ||||
HC = healthy control; CMA = cow's milk allergy; x2 = Chi-square.
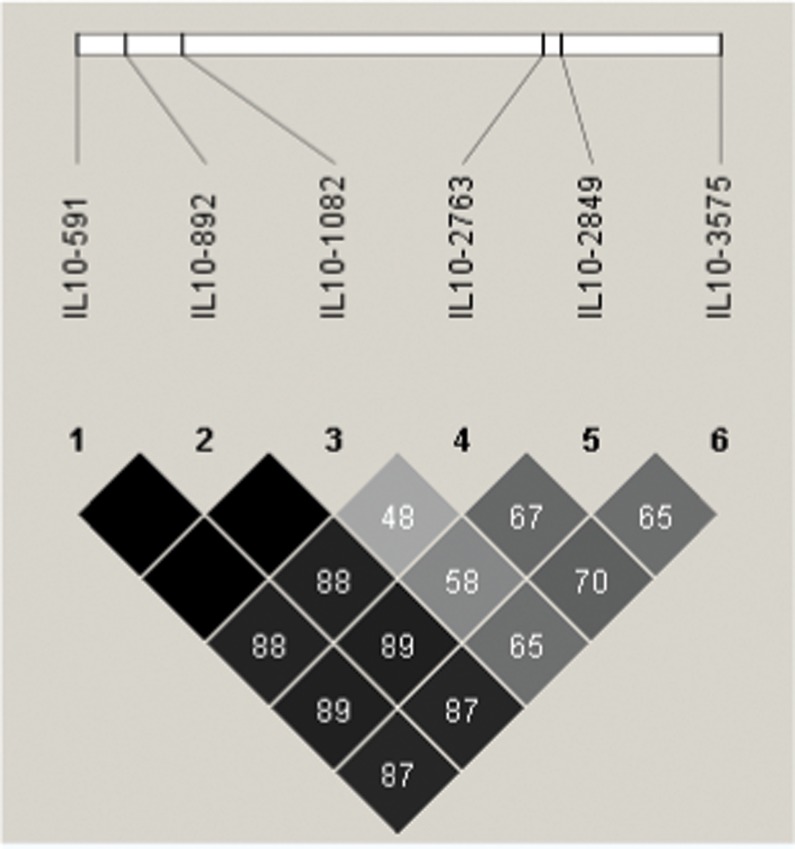
Due the LD observed at the studied IL10 polymorphisms (Figure 1), we analyzed the haplotypes containing these SNPs, except for -892, which was excluded from this study due to a strong association with -592 (r2 = 0.99). Three different SNP combinations showed an association with CMA. Curiously, the IL10 -3575A, IL10 -2849A, IL10 -2763C, IL10 -1082G and IL10 -592C haplotypes, strongly associated with CMA compared with HCs (p<0.001; OR = 4.71, 95% CI, 1.86-11.9 (Table 3), were completely absent in the tolerant CMA group (data not shown).
Figure 1.
Linkage disequilibrium (LD) map of the IL10 SNPs rs1800872, 1800896, 6693899, 3703630 and 1800890. An LD map based on the controls was drawn using Haploview version 4.1. Triangular heat map display of the LD index. Pairs of the IL10 SNP alleles in strong LD (D′>0.75) are highlighted in gray or black.
Table 3.
IL10 (-3575A/T, -2849A/G, -2763A/C, -1082A/G, -592A/C) haplotype frequency in children with cow’s milk allergy (CMA) and in healthy controls (HCs).
| IL10 -3575 | IL10 -2849 | IL10 -2763 | IL10 -1082 | IL10 -592 | HC | CMA | x2 | p-value | OR | 95% CI |
| n = 434 (%) | n = 90 (%) | |||||||||
| A | A | A | G | C | 37 (8) | 2 (3) | ns | |||
| A | A | C | G | C | 10 (2) | 9 (10) | 12.63 | <0.001 | 4.71 | 1.86-11.9 |
| A | G | A | A | C | 8 (2) | 4 (5) | ns | |||
| A | G | A | G | C | 29 (7) | 1 (1) | ns | |||
| T | G | A | A | C | 4 (1) | 6 (7) | 0.027* | 7.67 | 2.12-27.81 | |
| T | G | C | A | A | 136 (31) | 25 (27) | ns | |||
| T | G | C | A | C | 116 (27) | 9 (10) | 11.48 | <0.001 | 0.31 | 0.16-0.63 |
| T | G | C | G | A | 13 (3) | 5 (5) | ns | |||
| T | G | C | T | G | 39 (9) | 10 (12) | ns | |||
| Other** | 43 (10) | 18 (20) | ||||||||
HC = healthy control; x2 = Chi-square; OR = odds ratio;
*Fisher's exact test;
**frequencies less than 5%. ns = not significant; n = sample size, referring to the number of chromosomes.
Association of polymorphisms with clinical features
Several demographic and clinical variables, such as sex, anaphylaxis, the presence of other allergic diseases and total and specific IgE serum levels were analyzed together with the six SNPs studied, comparing tolerant with persistent CMA patients by means of a logistic regression model. The results confirmed that the G allele carriage (AG plus GG) at the IL10 -1082A/G polymorphism and homozygosis for the C allele at the TGFβ1 -509 T/C polymorphism were associated with persistent CMA (p = 0.019 and p = 0.047, respectively).
DISCUSSION
In this study, we analyzed the main clinical characteristics of 50 children with CMA and investigated whether the IgE-mediated persistent form could be at least partially related to the presence of SNPs in the promoter region of the IL10 and TGFβ1 genes, leading to the altered expression of these inflammation markers, which are involved in the gastrointestinal immune response and the development of food tolerance.
To our knowledge, this is the first Brazilian study of children with CMA that has attempted to associate genetic aspects with IgE-mediated persistent disease. TGFβ1 has a fundamental role in intestinal homeostasis and oral tolerance to low antigen doses. In the gastrointestinal tract, this protein is secreted by lymphocytes, contributing to IgA secretion. Yamagiwa et al. (2001) showed that TGFβ1 may also contribute to the expansion of Treg (8). Moreover, a reduction in TGFβ1 production by the intestinal mucosal cells of children with food allergy has been described (9). A TGFβ1 gene polymorphism at the -509 position is associated with low cytokine levels caused by the modification of the Yin Yang 1 transcription factor consensus binding site when the allele C is present. Our findings are consistent with homozygosis of the C allele (low producer) being associated with the persistent form of CMA, but this result must be confirmed by further studies. Several studies have associated reduced levels of TGFβ1 sensitization with foods (8,9). The mechanism of oral tolerance involves the deletion or anergy of T cells reactive with specific antigens associated with the expanding population of regulatory T cells following the production of TGFβ1 and IL10 (3).
One of IL10's main roles is the induction of oral tolerance, which has been evidenced by the association of IL10 levels with oral tolerance (19). Children with CMA who develop tolerance have a high regulatory T cell (CD4+ CD25+) count and a reduced proliferative response to beta-lactoglobulin in vitro compared with children with clinically active disease (20). Negoro et al. found an association of the A allele at the IL10 -627 position with the low expression of this cytokine, IgE levels and the severity of food allergy (20). Different studies have associated IL10 polymorphisms with asthma phenotypes in children, atopic dermatitis, the number of circulating eosinophils and IgE levels (21,22). Our data showed the association of the G allele at the IL10 -1082A/G position with CMA. The reactivity of T cells in children with CMA is associated with IL10 production (23). Previous data showed an association of the GG genotype with an exacerbation of asthma in Costa Rican children exposed to dust mite allergens (24). A recent study showed a functional role for this polymorphism, as the presence of the A allele is associated with susceptibility to and the outcome of sepsis (13). George et al. demonstrated that the A allele at the IL10-1082A/G position is associated with lower IL10 expression (25). Conversely, Giordani et al. observed increased IL10 expression in the presence of the allele G at -1082 position (26). A recent study shows that polymorphisms in the IL10 gene may modify its effect in asthmatics' airways (27). Corroborating findings were observed in an Egyptian study (28), demonstrating the important role of IL10 in allergic processes.
Different SNP combinations in the IL10 gene may impact cytokine production (29) and thus the outcome of immunological diseases. In the present study, we observed an association of IL10 haplotypes with CMA and with the persistent form of the disease. However, the ethnic heterogeneity of our population might be a bias due to the observed difference in IL10 polymorphism between Brazilian and African-derived populations (30). Increased expression levels of IL10 imply the greater control of inflammatory diseases but can also lead to more severe infectious conditions in which the effector response, rich in IFN and TNFα, is important. Haplotypes of polymorphisms in the promoter region of IL10 have been associated with increased levels of IgE in atopic dermatitis (31) and celiac disease (32). The interpretation of our haplotype analysis of polymorphisms in the IL10 gene is difficult due the number of analyzed individuals. Nevertheless, the power results showed that our sample size was adequate to analyze the IL10 -1092 A/G results individually. Of note, greater knowledge of the production of IL10 was linked to different haplotypes as biological relevant changes in the promoter region that usually alter the anchoring of different transcription factors, leading to differences in gene transcription.
In summary, our data have shown that the IL10 gene is associated with the persistent form of CMA in this population of Brazilian children. Further studies, including a study of blood cytokine levels, must be performed to confirm the functional role of these cytokines in CMA.
ACKNOWLEDGMENTS
The authors are very grateful to Anna Carla Goldberg for contributions regarding the control group and for helpful discussions. We also thank Dr. Ulysses Dória Filho for his contribution to the statistical analysis. This study was supported by the “Conselho Nacional de Pesquisa - CNPq” (grant N0 475707/2007-9 - Edital - MCT/CNPq 15/2007) to CMAJ and Process 308105/2012-5 to CMAJ.
Footnotes
No potential conflict of interest was reported.
REFERENCES
- 1.Nowak-Wegrzyn A, Sampson HA. Adverse reactions to foods. Med Clin North Am. 2006;90(1):97–127. doi: 10.1016/j.mcna.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Tan TH, Ellis JA, Saffery R, Allen KJ. The role of genetics and environment in the rise of childhood food allergy. Clin Exp Allergy. 2012;42(1):20–9. doi: 10.1111/j.1365-2222.2011.03823.x. [DOI] [PubMed] [Google Scholar]
- 3.Crittenden RG, Bennett LE. Cow's milk allergy: a complex disorder. J Am Coll Nutr. 2005;24:(6 Suppl):582S–91S. doi: 10.1080/07315724.2005.10719507. [DOI] [PubMed] [Google Scholar]
- 4.Sampson HA. Food allergy. J Allergy Clin Immunol. 2003;111(2 Suppl):S540–547. doi: 10.1067/mai.2003.134. [DOI] [PubMed] [Google Scholar]
- 5.Skripak JM, Matsui EC, Mudd K, Wood RA. The natural history of IgE-mediated cow's milk allergy. J Allergy Clin Immunol. 2007;120(5):1172–7. doi: 10.1016/j.jaci.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 6.Bjorksten B. Genetic and environmental risk factors for the development of food allergy. Curr Opin Allergy Clin Immunol. 2005;5(3):249–53. doi: 10.1097/01.all.0000168790.82206.17. [DOI] [PubMed] [Google Scholar]
- 7.Vanto T, Helppila S, Juntunen-Backman K, Kalimo K, Klemola T, Korpela R, et al. Prediction of the development of tolerance to milk in children with cow's milk hypersensitivity. J Pediatr. 2004;144(2):218–22. doi: 10.1016/j.jpeds.2003.10.063. [DOI] [PubMed] [Google Scholar]
- 8.Yamagiwa S, Gray JD, Hashimoto S, Horwitz DA. A role for TGF-beta in the generation and expansion of CD4+CD25+regulatory T cells from human peripheral blood. J Immunol. 2001;166(12):7282–9. doi: 10.4049/jimmunol.166.12.7282. [DOI] [PubMed] [Google Scholar]
- 9.Perez-Machado MA, Ashwood P, Thomson MA, Latcham F, Sim R, Walker-Smith JA, et al. Reduced transforming growth factor-beta1-producing T cells in the duodenal mucosa of children with food allergy. Eur J Immunol. 2003;33(8):2307–15. doi: 10.1002/eji.200323308. [DOI] [PubMed] [Google Scholar]
- 10.Bignon JD, Viña MF. 12th IHWC Class II Reference Protocol. In: Fauchet R, Charron D, editors. Technical Handbook of 12th International Histompatibility Workshop. Paris: HLA et Médécine; 1995. [Google Scholar]
- 11.Gibson AW, Edberg JC, Wu J, Westendorp RG, Huizinga TW, Kimberly RP. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol. 2001;166(6):3915–22. doi: 10.4049/jimmunol.166.6.3915. [DOI] [PubMed] [Google Scholar]
- 12.Silverman ES, Palmer LJ, Subramaniam V, Hallock A, Mathew S, Vallone J, et al. Transforming growth factor-beta1 promoter polymorphism C-509T is associated with asthma. Am J Respir Crit Care Med. 2004;169(2):214–9. doi: 10.1164/rccm.200307-973OC. [DOI] [PubMed] [Google Scholar]
- 13.Stanilova SA. Functional relevance of IL-10 promoter polymorphisms for sepsis development. Crit Care. 2010;14(1):119. doi: 10.1186/cc8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moraes MO, Santos AR, Schonkeren JJ, Vanderborght PR, Ottenhoff TH, Moraes ME, et al. Interleukin-10 promoter haplotypes are differently distributed in the Brazilian versus the Dutch population. Immunogenetics. 2003;54(12):896–9. doi: 10.1007/s00251-003-0543-3. [DOI] [PubMed] [Google Scholar]
- 15.Lech-Maranda E, Baseggio L, Bienvenu J, Charlot C, Berger F, Rigal D, et al. Interleukin-10 gene promoter polymorphisms influence the clinical outcome of diffuse large B-cell lymphoma. Blood. 2004;103(9):3529–34. doi: 10.1182/blood-2003-06-1850. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Hei P, Deng L, Lin J. Interleukin-10 gene promoter polymorphisms and their protein production in peritoneal fluid in patients with endometriosis. Mol Hum Reprod. 2007;13(2):135–40. doi: 10.1093/molehr/gal106. [DOI] [PubMed] [Google Scholar]
- 17.Pulleyn LJ, Newton R, Adcock IM, Barnes PJ. TGFβeta1 allele association with asthma severity. Hum Genet. 2001;109(6):623–7. doi: 10.1007/s00439-001-0617-y. [DOI] [PubMed] [Google Scholar]
- 18.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 19.Battaglia M, Gianfrani C, Gregori S, Roncarolo MG. IL-10-producing T regulatory type 1 cells and oral tolerance. Ann N Y Acad Sci. 2004;1029:142–53. doi: 10.1196/annals.1309.031. [DOI] [PubMed] [Google Scholar]
- 20.Negoro T, Orihara K, Irahara T, Nishiyama H, Hagiwara K, Nishida R, et al. Influence of SNPs in cytokine-related genes on the severity of food allergy and atopic eczema in children. Pediatr Allergy Immunol. 2006;17(8):583–90. doi: 10.1111/j.1399-3038.2006.00463.x. [DOI] [PubMed] [Google Scholar]
- 21.Lyon H, Lange C, Lake S, Silverman EK, Randolph AG, Kwiatkowski D, et al. IL10 gene polymorphisms are associated with asthma phenotypes in children. Genet Epidemiol. 2004;26(2):155–65. doi: 10.1002/gepi.10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sohn MH, Song JS, Kim KW, Kim ES, Kim KE, Lee JM. Association of interleukin-10 gene promoter polymorphism in children with atopic dermatitis. J Pediatr. 2007;150(1):106–8. doi: 10.1016/j.jpeds.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 23.Tiemessen MM, Van Ieperen-Van Dijk AG, Bruijnzeel-Koomen CA, Garssen J, Knol EF, Van Hoffen E. Cow's milk-specific T-cell reactivity of children with and without persistent cow's milk allergy: key role for IL-10. J Allergy Clin Immunol. 2004;113(5):932–9. doi: 10.1016/j.jaci.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 24.Hunninghake GM, Soto-Quiros ME, Lasky-Su J, Avila L, Ly NP, Liang C, et al. Dust mite exposure modifies the effect of functional IL10 polymorphisms on allergy and asthma exacerbations. J Allergy Clin Immunol. 1991;87(1 Pt 1):93–8. doi: 10.1016/j.jaci.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.George S, Ruan XZ, Navarrete C, Turner D, Reynard M, Sweny P, et al. Renovascular disease is associated with low producer genotypes of the anti-inflammatory cytokine interleukin-10. Tissue Antigens. 2004;63(5):470–5. doi: 10.1111/j.0001-2815.2004.00183.x. [DOI] [PubMed] [Google Scholar]
- 26.Giordani L, Bruzzi P, Lasalandra C, Quaranta M, Schittulli F, Della Ragione F, et al. Association of breast cancer and polymorphisms of interleukin-10 and tumor necrosis factor-alpha genes. Clin Chem. 2003;49(10):1664–7. doi: 10.1373/49.10.1664. [DOI] [PubMed] [Google Scholar]
- 27.Kim KW, Lee KE, Hong JY, Kim MN, Heo WI, Sohn MH, et al. Involvement of IL-10 gene promoter polymorphisms in the susceptibility for childhood asthma. Lung. 2011;189(5):417–23. doi: 10.1007/s00408-011-9312-5. [DOI] [PubMed] [Google Scholar]
- 28.Hussein YM, Shalaby SM, Mohamed RH, Hassan TH. Association between genes encoding components of the IL-10/IL-0 receptor pathway and asthma in children. Ann Allergy Asthma Immunol. 2011;106:474–80. doi: 10.1016/j.anai.2011.02.021. [DOI] [PubMed] [Google Scholar]
- 29.Eskdale J, Keijsers V, Huizinga T, Gallagher G. Microsatellite alleles and single nucleotide polymorphisms (SNP) combine to form four major haplotype families at the human interleukin-10 (IL-10) locus. Genes Immun. 1999;1(2):151–5. doi: 10.1038/sj.gene.6363656. [DOI] [PubMed] [Google Scholar]
- 30.Visentainer JE, Sell AM, da Silva GC, Cavichioli AD, Franceschi DS, Lieber SR, et al. TNF, IFNG, IL6, IL10 and TGFβ1 gene polymorphisms in South and Southeast Brazil. Int J Immunogenet. 2008;35(4-5):287–93. doi: 10.1111/j.1744-313X.2008.00778.x. [DOI] [PubMed] [Google Scholar]
- 31.Shin HD, Park BL, Kim LH, Kim JS, Kim JW. Interleukin-10 haplotype associated with total serum IgE in atopic dermatitis patients. Allergy. 2005;60(9):1146–51. doi: 10.1111/j.1398-9995.2005.00839.x. [DOI] [PubMed] [Google Scholar]
- 32.Nunez C, Alecsandru D, Varade J, Polanco I, Maluenda C, Fernández-Arquero M, et al. Interleukin-10 haplotypes in Celiac Disease in the Spanish population. BMC Med Genet. 2006;7:32. doi: 10.1186/1471-2350-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]