Abstract
Understanding the surface properties of the human opportunistic pathogen Aspergillus fumigatus conidia is essential given the important role they play during the fungal interactions with the human host. Although chitin synthases with myosin motor-like domain (CSM) play a major role in cell wall biosynthesis, the extent to which deletion of the CSM genes alter the surface structural and biophysical-biological properties of conidia is not fully characterized. We used three complementary atomic force microscopy techniques—i.e., structural imaging, chemical force microscopy with hydrophobic tips, and single-molecule force spectroscopy with lectin tips—to gain detailed insights into the nanoscale surface properties (ultrastructure, hydrophobicity) and polysaccharide composition of the wild-type and the chitin synthase mutant (ΔcsmA, ΔcsmB, and ΔcsmA/csmB) conidia of A. fumigatus. Wild-type conidia were covered with a highly hydrophobic layer of rodlet nanostructures. By contrast, the surface of the ΔcsmA mutant was almost completely devoid of rodlets, leading to loss of hydrophobicity and exposure of mannan and chitin polysaccharides. The ΔcsmB and ΔcsmA/csmB mutants showed a different behavior, i.e., the surfaces featured poorly organized rodlet layers, yet with a low hydrophobicity and substantial amounts of exposed mannan and chitin at the surface. As the rodlet layer is important for masking recognition of immunogenic fungal cell wall components by innate immune cells, disappearance of rodlet layers in all three chitin synthase mutant conidia was associated with an activation of human dendritic cells. These nanoscale analyses emphasize the important and distinct roles that the CSMA and CSMB genes play in modulating the surface properties and immune interactions of A. fumigatus and demonstrate the power of atomic force microscopy in fungal genetic studies for assessing the phenotypic characteristics of mutants altered in cell surface organization.
Introduction
The human opportunistic pathogen Aspergillus fumigatus causes several important respiratory diseases such as allergic bronchopulmonary and invasive aspergillosis (1,2). Understanding the molecular bases of A. fumigatus diseases requires a detailed knowledge of the fungal cell wall composition and the cell surface structure as these play key roles in pathogen-host interactions (3). Particularly important are the cell wall properties of aerial asexual spores (i.e., conidia) because these are the forms of the fungus invading immunocompromised patients and developing within the lungs. The outermost cell wall layer of A. fumigatus conidia is characterized by the presence of clustered nanofibrils, called rodlets, composed of hydrophobins, a family of small, hydrophobic proteins characterized by the conserved eight cysteine residues (4,5). The hydrophobic character of this outer layer allows the conidia to be easily dispersed into the air and to favor adhesion of the fungus to surfaces. Inner cell wall layers are essentially composed of polysaccharides, such as α-glucans, galactomannan, β-glucans, and chitin (6,7). Chitin, which accounts for 4–15% of the A. fumigatus cell wall depending on the morphotypes (8), is one of the two structural polysaccharides that provide cell shape and cell wall stiffness (7). In A. fumigatus, chitin synthesis, which occurs at the plasma membrane, involves eight chitin synthase (CHS) enzymes. Two of those with a myosin motor-like domain (AfCSMA and AfCSMB) have been recently shown to play a role not only in chitin synthesis but also on the entire conidial cell wall architecture (8). Conidia from ΔcsmA mutant showed strongly reduced amounts of chitin, whereas ΔcsmB and ΔcsmA/ΔcsmB mutants did not show any modification of the chitin content in their cell wall but all showed a modification of the overall cell wall integrity and permeability. Yet, the impact of these genetic modifications on the organization, properties, and biological interactions of the cell wall remains poorly understood.
In this study, we used atomic force microscopy (AFM) in three different modes—imaging, chemical force microscopy (CFM), and single-molecule force spectroscopy (SMFS)—to probe the ultrastructure, hydrophobicity, and polysaccharide properties (nature and distribution) of ΔcsmA, ΔcsmB, and ΔcsmA/csmB mutant conidia from A. fumigatus, and we compared these properties with those of the wild-type (WT) conidia. The results show that deletion of the CSMA and CSMB leads to major changes in the conidial nanoscale surface properties, and illustrate the power of AFM techniques in the high-resolution phenotypic characterization of fungal mutants with altered cell surface organization. These changes were associated with modifications in the immunogenic properties of the conidia.
Materials and Methods
Microorganisms and cultures
We used CEA17ΔakuBKU80 (wild-type), ΔcsmA, ΔcsmB, and ΔcsmA/csmB strains of A. fumigatus generated as described (8). Briefly, conidia were harvested from 10-day-old culture grown at 25°C on 2% malt extract agar containing 6% KCl. They were rinsed 3 times in Tween water (0.05%), 5 times in deionized water, and then 3 times with sodium acetate buffer (pH 4.75), and resuspended in 10 mL buffer to a concentration of ∼106 conidia per mL.
AFM
AFM measurements were performed in aqueous solution (acetate buffer for imaging, deionized water for CFM, and acetate buffer supplemented with 1 mM Ca2+ and 1 mM Mn2+ for SMFS) using a Multimode V AFM (Bruker AXS, Santa Barbara, CA), and, unless stated otherwise, using oxide-sharpened microfabricated Si3N4 cantilevers with a nominal spring constant of ∼0.01 N/m (Microlevers, Bruker AXS). Cells were immobilized, by mechanical trapping, into porous polycarbonate membranes (Millipore). After filtering a concentrated cell suspension, the filter was gently rinsed with buffer, carefully cut, attached to a steel sample puck using a small piece of double faced adhesive tape, and the mounted sample was transferred into the AFM liquid cell while avoiding dewetting. The sensitivity of the cantilevers was calibrated on glass substrates, and their spring constants measured using the thermal noise method (Picoforce, Bruker AXS). Data processing was performed using the commercial Nanoscope Analysis software (Bruker AXS) and the MATLAB software (The MathWorks, Natick, MA). For each strain, the structural images shown were obtained for multiple conidia from independent cultures and analyzed with different tips, and were representative of the entire conidial population.
For quantifying cell surface hydrophobicity by means of CFM (9), hydrophobic tips were prepared by immersing gold-coated cantilevers (OMCL-TR4, Olympus, Tokyo, Japan; nominal spring constant ∼0.02 N/m) for 12 h in 1 mM solutions of HS(CH2)11CH3 in ethanol and then rinsed with ethanol. Single cells were first localized and imaged with a silicon nitride tip. The tip was then changed with a hydrophobic tip to record adhesion force maps.
For single-molecule imaging, AFM tips were functionalized with Concavalin A (ConA) or wheat germ agglutinin (WGA) lectins (Sigma) using polyethylene glycol (PEG)-benzaldehyde linkers as described by Wildling et al. (10). Cantilevers were washed with chloroform and ethanol, placed in an ultraviolet-ozone-cleaner for 30 min, immersed overnight into an ethanolamine solution (3.3 g ethanolamine into 6 mL of dimenthyl sulfoxide (DMSO)), and then washed 3 times with DMSO and 2 times with ethanol, and dried with N2. The ethanolamine-coated cantilevers were immersed for 2 h in a solution prepared by mixing 1 mg Acetal-PEG-NHS dissolved in 0.5 mL chloroform with 10 μL triethylamine, and then washed with chloroform and dried with N2. Cantilevers were further immersed for 5 min in a 1% citric acid solution, washed in MilliQ water, and then covered with a 200 μL droplet of a phosphate buffered saline (PBS) solution containing lectins (0.2 mg/mL) to which 2 μL of a 1 M NaCNBH3 solution were added. After 50 min, cantilevers were incubated with 5 μL of a 1 M ethanolamine solution to passivate unreacted aldehyde groups, and then washed with and stored in buffer. Single cells were first localized and imaged with a silicon nitride tip. The tip was then changed with a lectin-tip to record adhesion force maps. Force curves were analyzed one by one to determine whether they featured specific adhesion events. These events were plotted as bright pixels, brighter colors meaning larger adhesion values.
Extraction and characterization of the conidial surface proteins
Conidia were washed twice with Tween (0.05%)-water and incubated in 0.5 M NaCl at room temperature for 90 min. The supernatant obtained was dialyzed against water, concentrated, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12.5% gel. One set of the gel was subjected to silver staining and proteins on another set of the gel was subjected to Western blotting using peroxidase-conjugated ConA and revealed using the ECL kit as per manufacturer’s instructions (Invitrogen).
WGA-fluorescein isothiocyanate (FITC) labeling
Conidia were washed extensively with PBS and incubated for 30 min in darkness in a solution containing 5 μg/ml WGA-FITC in 50 mM NaHCO3. After washing in the same buffer, conidia were observed under fluorescent microscopy. At least 300 conidia were checked at different fields under bright field microscopy and the same fields were counted for fluorescent FITC-labeled conidia; the ratio of FITC-labeled conidia × 100/conidia counted under bright field microscopy was expressed as the percentage of WGA-FITC-labeled conidia.
Stimulation of human dendritic cells
Isolations of the peripheral blood monocytes from the health donors’ blood, their differentiation into dendritic cells (DC), and the stimulation experiments with A. fumigatus conidia were performed as described earlier (11,12). The phenotype of DC was determined by flow cytometry analysis of surface markers by using fluorochrome-conjugated monoclonal antibodies to CD83, CD80, CD86, HLA-DR, and CD40. The values are presented as either percentage DC positive for indicated markers or intensity of expression of the molecules depicted as mean fluorescence intensity. The significance of differences between series of results was assessed using the one-way ANOVA and comparison between sets of results was assessed using Tukey’s posttest. Values of p < 0.05 were considered as statistically correlated (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001). All statistical analyses were performed using Prism 5 software (GraphPad softwares).
Results and Discussion
CSMA and CSMB genes modulate cell surface architecture
We first investigated the surface structure of WT and mutant conidia using high-resolution AFM imaging. Cells were immobilized on polymer membranes, a method which allows live cells to be imaged by AFM without using any drying or fixation procedure (13). Fig. 1, a–c, and Fig. S1, a and c in the Supporting Material, show typical deflection images recorded at low, medium, and high resolution for the surface of A. fumigatus WT conidia. Because height images could not reveal the fine ultrastructure of the specimens, deflection images were preferred. Deflection images revealed the presence of typical hydrophobin fasciculated rodlets, several hundred nm in length and 10 nm in width, consistent with earlier electron microscopy (14) and AFM (15) observations. By contrast, ΔcsmA conidia (Fig. 1, d–f, and Fig. S1, e and g), were devoid of homogeneous rodlet layers but presented an amorphous, granular surface (Fig. 1 f, label A). On close examination, however, some residual parts of the rodlet layer could be seen (Fig. 1 f, label R). Furthermore, tip-induced alterations were seen on the cell edge, close to the polymer membrane where sample curvature is the highest. As these features were not seen on WT cells, this further confirms that the cell wall architecture of the mutant is altered and more fragile.
Figure 1.

AFM imaging reveals that deletion of the CSMA and CSMB genes leads to major remodeling of the cell wall architecture. AFM deflection images of the surface of WT (a–c), ΔcsmA (d–f), ΔcsmB (g–i), and ΔcsmA/csmB (j–l) conidia recorded in acetate buffer at low (a, d, g, j), medium (b, e, h, k), and high (c, f, i, l) resolution. Labels R and A” indicate regions made of rodlets and amorphous material. The arrow indicates artifacts resulting from the interaction between the tip and loosely bond material. For each strain, images are representative of those obtained for at least 15 cells from three independent cultures, and analyzed with three different tips (see also Fig. S1 for additional examples).
Analysis of ΔcsmB conidia (Fig. 1, g–i, and Fig. S1, i and k), showed the presence of rodlets (Fig. 1 i, label R) but the layer seemed to be less compact and less organized than the WT. In addition, amorphous material was frequently spotted over the surface (Fig. 1 i, label A). Finally, ΔcsmA/csmB double mutant conidia (Fig. 1, j–l, and Fig. S1, m and o) featured a heterogenic population with the majority being globally similar to the ΔcsmB mutant. In addition, many conidia of the double mutant showed rodlet regions that were easily damaged by the scanning tip (Fig. 1 l, arrow), suggesting that the hydrophobin layer was synthesized but somewhat disorganized, thus mechanically fragile.
Structural changes in the mutants correlate with differences in cell surface hydrophobicity
Because hydrophobins are known to convey strong hydrophobic properties to the conidia (16), we then asked whether the observed structural changes correlate with differences in hydrophobicity. To this end, we used CFM with hydrophobic (CH3) tips (9,15,17) to map and quantify the nanoscale hydrophobic character of WT and mutant A. fumigatus conidia. Fig. 2 shows a set of CFM data collected on a single WT cell in pure water (no buffer was used because it is known to interfere with the measurements). Force-distance curves recorded across the cell surface revealed large adhesion forces (Fig. 2 b), with a magnitude of 2196 ± 148 pN (mean ± standard deviation σ, n = 1024) (Fig. 2 d). Adhesion maps recorded over 0.5 μm2 (Fig. 2 c) displayed a homogeneous distribution of hydrophobic forces, in agreement with previous work (15) and with the presence of the outermost surface layer of hydrophobins. Comparison with the data obtained on reference surfaces indicated that the conidial surface has a marked hydrophobic character (9), thus indeed consistent with hydrophobins (Fig. 3, a–c, and Fig. S1, a–d).
Figure 2.

CFM demonstrates that rodlet layers on WT conidia are uniformly hydrophobic. (a) Deflection image of a WT conidial surface recorded in water with a silicon nitride tip. (b–d). Representative force-distance curves (b), adhesion force map (z-range: 4 nN) (c), and adhesion force histogram (n = 512). (d) Recorded on the same cell with a hydrophobic tip (see also Fig. S1 for additional examples).
Figure 3.
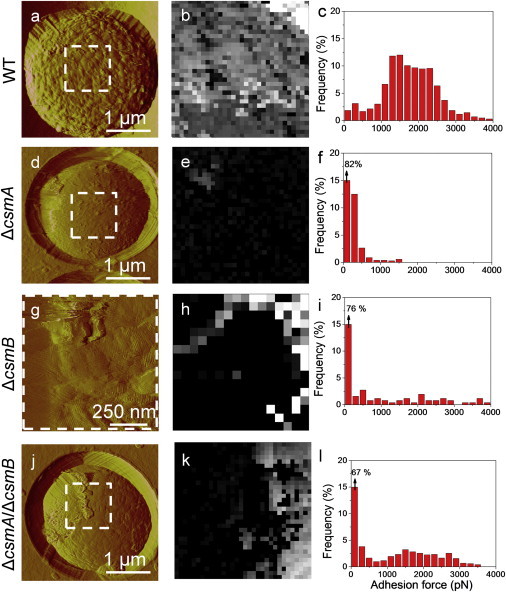
Structural changes in the mutants correlate with differences in cell surface hydrophobicity. Deflection images (a, d, g, j) recorded in water with silicon nitride tips, adhesion force maps (z -range: 4 nN) (b, e, h, k), and adhesion force histograms (n = 512) (c, f, i, l) recorded with hydrophobic tips on the surface of WT (a–c), ΔcsmA (d–f), ΔcsmB (g–i), and ΔcsmA/csmB (j–l) conidia (see Fig. S1).
Notably, we found that structural changes in ΔcsmA conidia (Fig. 1, d–f), were correlated with profound modifications of the cell surface hydrophobicity (Fig. 3, d–f, and Fig. S1, e–h). Force-distance curves and force maps recorded with the hydrophobic tip showed adhesion forces of only 362 ± 59 pN (mean ± σ, n = 1024), uniformly distributed across the surface, indicating that the conidial surface of the ΔcsmA mutant is hydrophilic (9). From these data, we conclude that the ΔcsmA mutation leads to the exposure of a hydrophilic amorphous layer on the cell surface. Because hydrophobins could still be extracted by hydrogen fluoride treatment from ΔcsmA mutant conidia (8), we suggest that rodlets are still present but hidden by this amorphous material. For the ΔcsmB (Fig. 3, g–i, and Fig. S1, i–l) and the ΔcsmA/csmB double (Fig. 3, j–l, and Fig. S1, m–p) mutants, we observed that the nanoscale structural heterogeneities were to some extent correlated with differences in hydrophobicity, the surface displaying contrasted hydrophobic and hydrophilic characters. Note that in the present work structural and adhesion maps could not be truly correlated (overlapped) as they were obtained with two different tips (silicon nitride and hydrophobic tips).
Chitin synthase mutations lead to the exposure of glycans
To elucidate the nature of the outer layer covering the rodlet structure, we next analyzed the occurrence and distribution of two of the cell wall polysaccharides, i.e., mannans and chitin, using SFMS with tips functionalized with the ConA and WGA lectins, respectively (18,19). Fig. 4 and Fig. S2 show the adhesion force maps, the adhesion force histograms, and representative force curves obtained for WT conidia with either ConA- or WGA-tips. Adhesive events were rarely detected (2–8%) and randomly distributed over the surface. The mean adhesion forces, in the 50–250 pN range, are consistent with values expected for the rupture of single (∼50 pN) or multiple lectin-sugar complexes (18,19). Supporting this view, force-distance curves (Fig. 4 c) showed well-defined single force peaks with short rupture distances (10–150 nm) that are typical of specific molecular recognition events. The mean rupture distances were quite short indicating that the detected residues are associated with macromolecules that are either short or not flexible like bacterial exopolysaccharides (19) or yeast mannoproteins (18).
Figure 4.
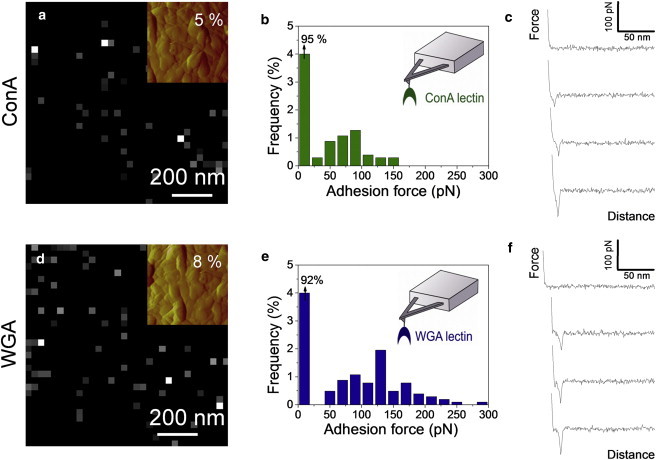
Mapping single polysaccharide residues on WT conidia using SMFS. Adhesion force maps (z-range: 300 pN; values in the top right corners correspond to the percentage of adhesive events) (a and d), adhesion force histograms (n = 512) (b and e) and representative force-distance curves (c and f) recorded on WT conidia in acetate buffer supplemented with Ca2+and Mn2+, using AFM tips bearing ConA (a–c) and WGA (d–f) lectins. The insets in panels a and d show deflection images of the areas corresponding to the adhesion maps. See Fig. S2 for an independent set of data.
Both ConA- and WGA-positive molecules were clearly more abundant (12–25% depending on the cell investigated) on the surface of the ΔcsmA mutant (Fig. 5, a and b, and Fig. S3), consistent with the absence of rodlets. Adhesion forces were well defined and in the 50 pN range (see Fig. 5, g and h), thus typical of lectin-carbohydrate bonds (18,19). Interestingly, ConA- and WGA-positive molecules were essentially detected on smooth, featureless areas lacking the rodlets. Proteins were also present on the surface of the conidia of all csm mutants and could be extracted using 0.5 M NaCl. These surface proteins, which were present in low amount (20 fg /mutant conidium), were negative upon Western blotting using peroxidase conjugated lectins (data not shown). Accordingly, these results suggested that ConA- and WGA-positive molecules corresponded most probably to mannan and chitin and not to surface glycoproteins. These polysaccharides were hardly exposed on the surface of WT conidia, which is fully consistent with our structural (rodlet-coated cell surface) and chemical (very hydrophobic cell surface) data. The ΔcsmB and ΔcsmA/csmB mutants yielded results that were somewhat similar to the ΔcsmA mutant, i.e., 11–27% of mannose and N-acetylglucosamine-rich residues cell wall components (mannan and chitin, respectively) randomly distributed over the surface (Fig. 5, c–h, Fig. S4, and Fig. S5). This may seem surprising because in contrast to ΔcsmA, the surface of the conidia of ΔcsmB and ΔcsmA/csmB conidia was less homogenously covered with amorphous material. However, rodlet patterns were often fuzzy and hardly visualized, suggesting they were loosely structured and possibly mixed with underlying/extruding carbohydrates.
Figure 5.

Chitin synthase mutations lead to the exposure of glycans. Adhesion force maps (z-range: 300 pN; the values correspond to the percentage of adhesive events) recorded on the surface of ΔcsmA (a and b), ΔcsmB (c and d), and ΔcsmA/csmB (e and f) conidia, using AFM tips bearing ConA (a, c, e) and WGA (b, d, f) lectins. The insets show deflection images of the areas corresponding to the adhesion maps. (g and h). Historgams of the mean (± SD) adhesion force measured on each mutant with ConA (g) and WGA (h) tips. See Figs. S3–S5 for independent sets of data.
These AFM data were confirmed by an analysis of the labeling of the conidia by FITC conjugated WGA. Labeling showed heterogeneous but increased labeling of the csm mutant conidia compared to WT (the percent of conidia labeled with WGA-FITC were 51 ± 3, 44 ± 2, and 45 ± 9 for ΔcsmA, ΔcsmB, and ΔcsmA/csmB, respectively, compared to 11 ± 2 for the WT). Moreover, these fluorescence observations were in agreement with the AFM data that showed heterogeneous conidial populations in the csm mutants.
Chitin synthase mutant conidia are immunostimulatory
DC play an important role in orchestrating the immune response to invading pathogens (20,21). In steady state DC rest in immature phenotype and express low level of class II antigen presenting molecule HLA-DR and costimulatory molecules CD80, CD86, and CD40. When DC encounter pathogens, the interaction of pattern recognition receptors on DC with pathogen-associated molecular patterns leads to maturation and activation of DC. The maturation of DC is associated with a high level of surface expression of CD83 and antigen peptide-loaded HLA-DR that interacts with T cell receptor-CD3 complex on T cells. In addition, mature DC show an increased expression of costimulatory molecules. CD80 and CD86 interact with CD28 and CD40 with CD154 on T cells to provide stimuli for T cell activation and proliferation. The surface rodlet layer on dormant conidia of A. fumigatus masks the immunogenic components of the conidia and hence imparts immunological inertness to air-borne conidia (12). In contrast to DC stimulated with WT conidia DC stimulated with csm mutant conidia expressed significantly high levels of costimulatory molecules CD80, CD86, CD40, antigen presenting molecule HLA-DR and terminal maturation marker CD83 (Fig. 6). These results correlate with our previous results that show that chemical or genetic removal of the rodlet layer exposes the immunogenic motifs that stimulate innate cells such as DC (12). In line with these previous observations, and the present data showing poorly organized rodlet layer on the surfaces of the ΔcsmA, ΔcsmB, and ΔcsmA/csmB mutants, our results indicate that chitin synthase plays an important role in preventing immune recognition of airborne conidia by organizing the outer surface layer of the conidia. The lack of the significant differences among three mutants (ΔcsmA, ΔcsmB, and ΔcsmA/csmB) in their ability to stimulate DC was in agreement with the data obtained with AFM using tips functionalized with the ConA and WGA lectins.
Figure 6.

Chitin synthase mutant conidia stimulate human dendritic cells. The expression of CD83, CD86 (both expressed as % positive cells) (a and b); CD80, CD86, HLA-DR, and CD40 (all expressed as mean fluorescence intensities) (c–f, respectively) on DC that were cultured with cytokines alone or cytokines plus WT conidia, ΔcsmA, ΔcsmB, or ΔcsmA/csmB conidia for 48 h. Data (mean ± SE) from six independent donors. Statistical significance as determined by ANOVA test is indicated (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001).
Conclusion
Knowledge of the surface properties of A. fumigatus chitin synthase mutants appears critical to our understanding of the cell wall synthesis and organization of this pathogen and has some direct impact on the immunological properties of this microorganism (21). We have shown that AFM-based imaging, CFM, and SMFS are valuable tools to study the nanoscale surface properties (ultrastructure, hydrophobicity) and polysaccharide composition of A. fumigatus chitin synthase mutants. Our results emphasize the important and distinct roles that the CSMA and CSMB genes play in modulating the cell wall architecture and properties of A. fumigatus. Compared to WT conidia, ΔcsmA conidia were profoundly altered: the surface was entirely devoid of rodlets, leading to a loss of hydrophobicity and to the exposure of mannan/chitin polysaccharides. Whereas, ΔcsmB and ΔcsmA/csmB showed somewhat different behaviors: loosely structured rodlet layers were present on the conidial surfaces, yet with a low hydrophobicity, smooth and rough patches of amorphous component, and exposure of polysaccharides. In agreement with the AFM observations, all three mutant conidia (ΔcsmA, ΔcsmB, and ΔcsmA/csmB) showed an increase in the surface exposure of immunostimulatory cell wall polysaccharides otherwise hidden by the surface rodlet layer in the WT conidia.
Taken together, our AFM experiments show that disruption of all CSM genes leads to a very dramatic change in the conidial cell surface organization resulting from unexpected compensatory reactions that were not obvious based only on biochemical data (8) The AFM results indicated that the three-dimensional organization of the cell wall polymers has been extensively and unpredictedly changed by the CSM deletion resulting in the appearance of new molecules on the surface structure of the conidial cell wall. AFM analyses suggested that the loosely structured rodlets allow passage of the molecules that are exposed on the conidial surface. These observations emphasize that AFM is of great help to complement biochemical analysis of the fungal cell wall because the later deals with the overall conidial cell wall composition, whereas AFM analyze the outermost cell surface. Spatial resolution (lateral and vertical) is a unique advantage of AFM over biochemical analysis of the cell wall. In addition, AFM is advantageous over fluorescent labeling as AFM tips functionalized with specific markers enables the precise localization and the quantification of surface exposed cell wall components (6,12,22). Understanding of the fungal surface-specific properties is critical during host-microbial interaction as the fungal surface is the first component confronting with the host immune system.
Acknowledgments
Work at the Université catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the Région Wallonne, the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté française de Belgique (Concerted Research Action). Work at the Institut Pasteur and Institut National de la Santé et de la Recherche Médicale, Paris were supported by the ANR grant ANR-10-BLAN-1309 HYDROPHOBIN, European Community’s Seventh Framework Programme [FP7/2007-2013] under Grant Agreement No: 260338 ALLFUN and European Science Foundation grant Fuminomics RNP06-132. Y.F.D. and D.A. are Senior Research Associate and Postdoctoral Researcher of the FRS-FNRS.
Contributor Information
Jean-Paul Latgé, Email: Jean-paul.latge@pasteur.fr.
Yves F. Dufrêne, Email: Yves.Dufrene@uclouvain.be.
Supporting Material
References
- 1.Chai L.Y.A., Hsu L.Y. Recent advances in invasive pulmonary aspergillosis. Curr. Opin. Pulm. Med. 2011;17:160–166. doi: 10.1097/MCP.0b013e328343eb49. [DOI] [PubMed] [Google Scholar]
- 2.Mahdavinia M., Grammer L.C. Management of allergic bronchopulmonary aspergillosis: a review and update. Ther. Adv. Respir. Dis. 2012;6:173–187. doi: 10.1177/1753465812443094. [DOI] [PubMed] [Google Scholar]
- 3.Annaix V., Bouchara J.P., Tronchin G. Specific binding of human fibrinogen fragment D to Aspergillus fumigatus conidia. Infect. Immun. 1992;60:1747–1755. doi: 10.1128/iai.60.5.1747-1755.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beever R.E., Dempsey G.P. Function of rodlets on the surface of fungal spores. Nature. 1978;272:608–610. doi: 10.1038/272608a0. [DOI] [PubMed] [Google Scholar]
- 5.Cole G.T., Sekiya T., Kasai R. Surface ultrastructure and chemical composition of the cell walls of conidial fungi. Exp. Mycol. 1979;3:132–156. [Google Scholar]
- 6.Latgé J.P. Tasting the fungal cell wall. Cell. Microbiol. 2010;12:863–872. doi: 10.1111/j.1462-5822.2010.01474.x. [DOI] [PubMed] [Google Scholar]
- 7.Latgé J.P., Mouyna I., Nierman W. Specific molecular features in the organization and biosynthesis of the cell wall of Aspergillus fumigatus. Med. Mycol. 2005;43(Suppl 1):S15–S22. doi: 10.1080/13693780400029155. [DOI] [PubMed] [Google Scholar]
- 8.Jiménez-Ortigosa C., Aimanianda V., Latgé J.P. Chitin synthases with a myosin motor-like domain control the resistance of Aspergillus fumigatus to echinocandins. Antimicrob. Agents Chemother. 2012;56:6121–6131. doi: 10.1128/AAC.00752-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alsteens D., Dague E., Dufrêne Y.F. Direct measurement of hydrophobic forces on cell surfaces using AFM. Langmuir. 2007;23:11977–11979. doi: 10.1021/la702765c. [DOI] [PubMed] [Google Scholar]
- 10.Wildling L., Unterauer B., Gruber H.J. Linking of sensor molecules with amino groups to amino-functionalized AFM tips. Bioconjug. Chem. 2011;22:1239–1248. doi: 10.1021/bc200099t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bansal K., Sinha A.Y., Bayry J. Src homology 3-interacting domain of Rv1917c of Mycobacterium tuberculosis induces selective maturation of human dendritic cells by regulating PI3K-MAPK-NF-kappaB signaling and drives Th2 immune responses. J. Biol. Chem. 2010;285:36511–36522. doi: 10.1074/jbc.M110.158055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aimanianda V., Bayry J., Latgé J.P. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature. 2009;460:1117–1121. doi: 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- 13.Dufrêne Y.F. Atomic force microscopy and chemical force microscopy of microbial cells. Nat. Protoc. 2008;3:1132–1138. doi: 10.1038/nprot.2008.101. [DOI] [PubMed] [Google Scholar]
- 14.Paris S., Debeaupuis J.P., Latgé J.P. Conidial hydrophobins of Aspergillus fumigatus. Appl. Environ. Microbiol. 2003;69:1581–1588. doi: 10.1128/AEM.69.3.1581-1588.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dague E., Alsteens D., Dufrêne Y.F. Chemical force microscopy of single live cells. Nano Lett. 2007;7:3026–3030. doi: 10.1021/nl071476k. [DOI] [PubMed] [Google Scholar]
- 16.Bayry J., Aimanianda V., Latgé J.P. Hydrophobins—unique fungal proteins. PLoS Pathog. 2012;8:e1002700. doi: 10.1371/journal.ppat.1002700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dague E., Alsteens D., Dufrêne Y.F. High-resolution cell surface dynamics of germinating Aspergillus fumigatus conidia. Biophys. J. 2008;94:656–660. doi: 10.1529/biophysj.107.116491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alsteens D., Dupres V., Dufrêne Y.F. Structure, cell wall elasticity and polysaccharide properties of living yeast cells, as probed by AFM. Nanotechnology. 2008;19:384005. doi: 10.1088/0957-4484/19/38/384005. [DOI] [PubMed] [Google Scholar]
- 19.Francius G., Lebeer S., Dufrêne Y.F. Detection, localization, and conformational analysis of single polysaccharide molecules on live bacteria. ACS Nano. 2008;2:1921–1929. doi: 10.1021/nn800341b. [DOI] [PubMed] [Google Scholar]
- 20.Cheng S.C., Joosten L.A.B., Netea M.G. Interplay between Candida albicans and the mammalian innate host defense. Infect. Immun. 2012;80:1304–1313. doi: 10.1128/IAI.06146-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romani L. Immunity to fungal infections. Nat. Rev. Immunol. 2011;11:275–288. doi: 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- 22.Roy R.M., Paes H.C., Klein B.S. Complement component 3C3 and C3a receptor are required in chitin-dependent allergic sensitization to Aspergillus fumigatus but dispensable in chitin-induced innate allergic inflammation. mBio. 2013;4:e00162-13. doi: 10.1128/mBio.00162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


