Abstract
An original method is presented to study single-colloid interaction with a substrate in liquid environment. Colloids, either in solution or adsorbed on a surface, are fixed by suction against the aperture of a microchanneled atomic force microscopy cantilever. Their adhesion to the substrate is measured, followed by their release via a short overpressure surge. Such colloid exchange procedure allows for 1), the quick variation of differently functionalized colloids within the same experiment; 2), the investigation of long-term interactions by leaving the colloids on a surface for a defined time before detaching them; and 3), the inspection of irreversible interactions. After validation of the method by reproducing literature results obtained with traditional colloidal atomic force microscopy, the serial use of colloids with different surface functionalization was shown on a micropatterned surface. Finally, concanavalin A-coated colloids were allowed to adsorb on human embryonic kidney cells and then detached one by one. The adhesion between cells and colloids was up to 60 nN, whereas individual cells adhered with 20 nN to the glass substrate. A cellular elastic modulus of 0.8 kPa was determined using the attached colloid as indenter.
Introduction
Colloids in the nanometer to micrometer scale are present everywhere in our life, from the ink on this paper, to the milk in the morning and the exhaust fumes of our cars. They are a part of our consumer goods as well as of our waste. In research applications, colloids are used as means of transport (1), detection (2), and measurement (3).
Colloidal probe atomic force microscopy (AFM) was developed in the last two decades (3–5) to investigate the interaction forces between single colloids and a substrate. Either a single, spherical colloid is glued precisely (6) to a tipless AFM cantilever, or the apex of an AFM pyramid is rounded to assume the shape of a half-sphere. Colloidal AFM has since been used to measure forces in the pico- to nanoNewton range: Examples range from hydration forces at the nanoscale (5) to mechanical properties of soft matter such as cells (7) or polymer films (8). Colloidal probes are generally the preferred way to quantify interfacial forces with AFM, for theory requires the contact radius to be much larger than the separation distance (6,9–11), which is not the case for standard AFM pyramidal probes with a tip curvature down to 10 nm.
Yet, colloidal AFM is affected by some inherent limitations: Most colloidal experiments are carried out in buffer and any exchange of the AFM probe results in a waiting period to stabilize the signal drift. Drift is also the reason why contact times above a few minutes between colloid and surface have not been studied. Contamination and degradation of the colloid surface further limit the lifetime of a colloid probe, therefore most data have to be collected using fresh probes. Experiments with living cells, for example, have been limited to three data points per tip (7,11,12). Finally, colloidal probes with a chemical functionalization cannot be simply exchanged during an experiment, making measurements of different biomolecular interactions on the same sample difficult.
In this work, we developed a strategy to overcome these limitations by using fluid force microscopy (FluidFM) (13,14) to manipulate single colloids. The tipless or pyramidal microchanneled FluidFM probes transform the AFM into a force-controlled micro syringe or micropipette (15–17).
A single colloid can be attracted to the aperture at the end of the cantilever (see Fig. 1 A) by negative pressure. It is either grabbed directly from the solution where it is suspended or picked up from the substrate where it was previously adsorbed, as shown schematically in Fig. 1, B and C. After use, for example for a force spectroscopy, it can be released by applying a quick overpressure pulse. Thus, FluidFM offers unprecedented flexibility to investigate single-colloid interactions:
The user has the freedom to exchange the colloid probe anytime, eventually using each time a differently functionalized colloid.
By grasping a fresh colloid for each force spectroscopy curve, the first contact with the substrate can be investigated in statistically relevant numbers. This allows exploring strong repetition-dependent interactions, where the first few contacts bear the most valuable information.
The colloids can be previously spread on the substrate, interact for a defined time and finally be detached. This enables the investigation of long-term interactions, whereas the same cantilever can be used to quickly detach the colloids one by one.
To demonstrate the feasibility of these concepts, we designed the following experiments:
Colloidal FluidFM was validated against literature data for a system where COOH-coated polystyrene beads interact with a clean glass substrate (18).
Versatile exchange of colloids with different chemical functionalization was shown for the biotin-avidin interaction on an opportunely micropatterned surface.
Colloids were adsorbed on a nonconfluent cell layer on the one hand to measure the adhesion between concanavalin A (ConA)-coated colloids and human embryonic kidney (HEK) cells, and on the other hand to determine the elastic modulus of the cells.
Figure 1.
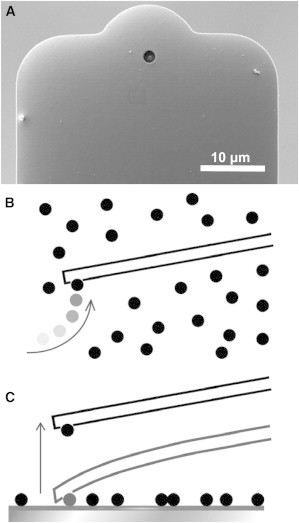
(A) Scanning electron micrograph of a FluidFM tipless microchanneled cantilever with a 2-μm circular opening at the end. A spherical colloid can be attached to the aperture by underpressure in two ways: (B) the colloid is grabbed from a separate reservoir and transferred to the target substrate; and (C) the bead is picked up directly from the substrate. In both cases the colloid is released after use by a short pressure pulse.
Materials and Methods
Characteristics and preparation of FluidFM cantilevers
The FluidFM cantilevers were supplied by Cytosurge (Zürich, Switzerland). For this study, 200-μm-long, 36-μm-wide tipless cantilevers were used with a channel height h of 900 nm and a wall thickness of 300 nm, whereas the circular opening at the end of the cantilever had a diameter do of either 2 μm or 10 μm (14). All cantilevers were already mounted on a CYTOCLIP (Cytosurge). The clip was designed to fit on an AFM probeholder (Nanosurf, Liestal, Switzerland) modified for FluidFM applications.
The spring constant was determined for each cantilever using the Sader method (19) with a script supplied by Nanosurf. It was typically between 0.5 N m−1 and 3 N m−1. The sensitivity was recalibrated each time the medium was changed.
Before an experiment the fresh cantilever was cleaned for 2 min in air plasma (PDC-32G; Harrick Plasma, Ithaca, NY) and then immediately immersed in Poly(L-lysine) (20)-g[3.5]-poly(ethylene glycol) (2) (PLL-g-PEG; SuSoS, Dübendorf, Switzerland) 0.1 mg/mL in 10 mM HEPES + 150 mM NaCl buffer (HEPES2). After a minimum of 45 min of immersion, the cantilever was rinsed with MilliQ water (Merck Millipore, Billerica, MA) and was ready for the experiment.
The over- and underpressure in the FluidFM cantilever was established with a pressure controller (Cytosurge) in a range from −800 mbar to +1000 mbar with 1 mbar resolution and a response time of 200 ms.
Colloidal beads
Polystyrene beads were bought already functionalized (Micromod Partikeltechnologie, Rostock, Germany).Their diameter db was chosen according to the restrictions imposed by the cantilever geometry. The minimal suitable colloid diameter dmin was determined as function of the opening diameter do of the aperture and the height h of the channel (as illustrated in Fig. 2A, where the equation is valid only if db > do and db > 2h):
For the 2-μm opening we used db = 3 μm polystyrene colloids, and for the 10-μm opening we used db = 50 μm. The vertical force Fv fixing a colloid to the cantilever could be calculated using the typical suction p of 750 mbar and the area of the cantilever opening,
Assuming tight sealing between cantilever and bead, this resulted in forces Fv of ∼230 nN for the 2-μm opening and ∼6 μN for the 10-μm opening. The lateral fixation force Fl depends of the torque T created by the suction and of the vertical distance dv ≈ db, which the colloid creates between cantilever and substrate. The torque T can be found through an integration of the circle chord along the diameter of the opening,
For a 3-μm bead, this results in a lateral fixation force Fl of ∼100 nN. However, the potential rotational freedom might lead to higher effective lateral forces before the bead loses its fixation.
Figure 2.
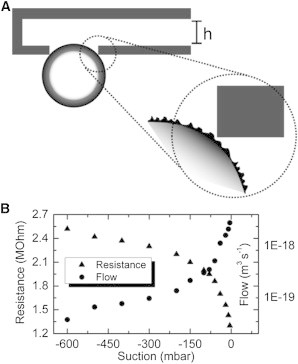
From resistance to flow. The electrical resistance of the FluidFM channel with an attached bead was measured and the leak flow was calculated via simulations. (A) A nanometric average gap due to the contact roughness between bead and cantilever allowed the buffer solution to conduct electrical current between inside and outside (B, triangles). By analyzing the occurring electric and hydrodynamic resistance in a FEM model, the measured electric resistance could be converted into a corresponding volumetric flow (B, circles).
Colloid sealing the cantilever: fluorescence measurement
The fluorescence images were taken with a C9100-13 camera (Hamamatsu Photonics, Tokyo, Japan) on an AXIO OBSERVER microscope with COLIBRI illumination (Carl Zeiss, Jena, Germany). Epifluorescence images were taken while illuminating with a 470-nm LED (423052-9050-000; Carl Zeiss) and using the 38HE filter set (Carl Zeiss).
The leak flow was observed for a cantilever with 2-μm opening with a bare bead attached (MICROMER 01-00-303; Micromod Partikeltechnologie). The channel was filled with 1 mg/mL fluorescein sodium salt (F6377; Sigma-Aldrich, Saint Louis, MO) in PBS buffer (P4417; Sigma-Aldrich). The bare beads were suspended in PBS buffer and then attracted by the cantilever with 150 mbar suction pressure. The fluorescence solution in the cantilever was replaced by buffer within 1 s when suction was applied. The corresponding flow was calculated and measured to be ∼150 fL s−1 mbar−1 as shown in Fig. S1 in the Supporting Material. A soon as the bead blocked the entrance, fluorescein could diffuse back into the channel coming from a reservoir of 5 μL in the CYTOCLIP, only hindered by potential leak flow.
The expected diffusion of fluorescein in the blocked channel was calculated applying Fick’s laws (20) and using the software MATHEMATICA (Wolfram Research, Oxfordshire, UK). The diffusion constant of fluorescein in buffer was assumed to be 2.7 × 10−10 m2 s−1 (21).
Colloid sealing the cantilever: impedance measurement
The microchannel of FluidFM cantilever with 2-μm opening was electrically contacted through the CYTOCLIP with a homemade connector based on a silver chloride-coated silver wire. The electrical ground electrode from the same material was placed in the buffer solution.
The impedance was measured using a PICO 2 (TECELLA, Foothill Ranch, CA) clamping the voltage at 10 mV. A self-written LABVIEW (National Instruments, Austin, Texas) script was used to evaluate and visualize the data.
The dielectric bead (MICROMER 01-00-303; Micromod Partikeltechnologie) was attracted with 150 mbar suction pressure. As soon as the colloid was fixed at the cantilever opening, the suction pressure was swept from 5 to 600 mbar (Pressure Controller; Cytosurge) while measuring the electrical current. The impedance was then calculated with Ohm’s law. The experimental data of the impedance were correlated to an equivalent flow in the microchannel using COMSOL (Comsol Group, Stockholm, Sweden). The assumption was that the electric current past the dielectric bead was due to roughness of the bead and the cantilever as illustrated in Fig. 2 A. This was approximated by an average gap distance g between the two, which was varied 0.01–10 nm. It was found that only the geometry just around the contact zone had any influence on the flow. For each gap distance g the hydrodynamic- and the electrical resistance could be calculated. The hydrodynamic resistance Rh scaled with g−2 and the electric resistance Re scaled with –ln[g], as shown in Fig. S7 and Fig. S8. Simple analytical approximations supported these scaling laws and showed <20% numerical difference to the simulations (see the Supporting Material). The relation between Rh (Pa s m−3) and Re (Ohm) was empirically derived as
Thus, the measured resistance data were converted into an equivalent volumetric flow Q,
where Δp is the applied pressure difference.
Adsorption of COOH-coated beads on glass
Glass substrate preparation
The glass substrate was thoroughly cleaned before the measurement according to a protocol of three cycles of 10-min washing with different liquids in the ultrasonic bath (T 710 DH; Elma Hans Schmidbauer, Singen, Germany) and rinsing with MilliQ water. To begin, we used COBAS CLEANER (Roche, Basel, Switzerland), then isopropanol, and finally MilliQ water. At this point, the substrate was dried with a nitrogen blower, cleaned for 2 min in air plasma, and immersed into 1 mM PBS buffer.
Bead preparation and measurement
One microLiter of COOH-coated beads (MICROMER 30-02-303; Micromod Partikeltechnologie) was mixed into 4 mL of 1 mM PBS. The 3-μm large beads were picked up with a FluidFM cantilever with a 2-μm opening from their stock petri dish (GWSB-5040; WillCo Wells, Amsterdam, Netherlands) and then transferred through air to the petri dish with the cleaned glass substrate. The transfer took <10 s. This two-dish approach was preferred to avoid the noise induced in the photodiode by the floating colloid particles, which disturbed the AFM laser.
Force spectroscopy curves were recorded with a setpoint of 5 nN and a speed of 100 nm s−1. A contact area of ∼5 × 10−16 m2 was calculated using the JKR model (22). With the same attached bead, the contact time was systematically varied 0.1–10 s, repeating each measurement at least 20 times at different points on the substrate. The data were analyzed with SPIP (Image Metrology, Hørsholm, Denmark).
Exchange of beads on micropatterned substrate
Pattern preparation
The clean glass surface was patterned with a microcontact printing protocol from Csucs et al. (23). The polydimethylsiloxane (PDMS) stamp had a chessboard structure, consisting of 50 × 50 μm2 squares with a depth of 2 μm. PLL (20)-g[3.5]-PEG (2)/PEG(3.4)-biotin(50%) (SuSoS) was mixed with PLL (20)-g[3.5]-PEG (2)/FITC (SuSoS) 1:1 in a concentration of 1 mg mL−1 in HEPES2. A drop of this fluorescent and biotinylated solution was then placed on the freshly cleaned stamp and left there for 1 h in the dark. The glass substrate was air-plasma-cleaned for 2 min just before the stamp was pressed on it for 15 s. The glass was then thoroughly rinsed with MilliQ water and backfilled with PLL-g-PEG 0.1 mg mL−1 in HEPES2. After 1 h incubation, the substrate was rinsed again with MilliQ water and immersed in HEPES2.
Bead preparation and measurement
Two types of beads were used: bare (MICROMER 01-00-303; Micromod Partikeltechnologie) or-coated with avidin (MICROMER 01-18-303; Micromod Partikeltechnologie). A quantity of 1 μL of stock bead solution was diluted in 4 mL HEPES2 and then kept in separate petri dishes for each type. A selected colloid was then sucked against the aperture just before use and transferred onto the patterned substrate.
Force spectroscopy curves were sampled with a setpoint of 50 nN and a speed of 750 nm s−1. The contact area of ∼4 × 10−14 m2 could be calculated using the JKR theory (22) in combination with the radius of gyration of the PEG chain (24).
Adhesion of human embryonic kidney cells 293 to concanavalin A beads
Preparation of HEK cells
The human embryonic kidney cells 293(HEK) cells were cultured in a medium containing the following ingredients: 450 mL Dulbecco’s Modified Eagle Medium (DMEM, Cat. No. 41965; Life Technologies, Carlsbad, CA), 50 mL fetal bovine serum (heat-inactivated Cat. No. 10082147; Life Technologies), 10 mL L-glutamine 200 mM (Cat. No. 25030024; Life Technologies), 0.2 mL gentamicin 50 mg/L (Cat No. 15750060; Life Technologies) and 1 mL zeocin 100 mg/mL (Cat. No. R25001; Life Technologies).
When the cells were 60% confluent in a 25 cm2 flask (SPL Life Science, Gyeonggi-do, Korea) they were washed with PBS and then incubated with 1 mL 0.01% trypsin (Cat. No. 25300054; Life Technologies) in PBS for 2 min. At this point, 10 mL of medium was added, and the solution was centrifuged at 1500 revolutions per min for 2 min. The supernatant was removed and 5 mL fresh medium was added. A quantity of 1–3 drops of this concentrated solution were added to a glass-bottom petri dish (WillCo Wells) filled with 3 mL medium. After 18 h of incubation, the cells were ready for the experiments.
Preparation of ConA colloids
Streptavidin-coated polystyrene beads with a diameter of 50 μm were used (MICROMER 01-19-504; Micromod Partikeltechnologie). A protocol adapted from Friedrichs et al. (25) was applied to bind ConA-Biotin (C2272; Sigma-Aldrich) to the beads. First, 100 μL of the pure bead solution was centrifuged at 10,000 revolutions per min for 2 min. The supernatant was removed and 100 μL of ConA-Biotin solved as 0.3 mg mL−1 in PBS was added. After 30 min of incubation at room temperature, the solution was centrifuged again, and the supernatant was replaced with PBS. After repeating the washing step three times, the ConA-functionalized beads were stored at 4°C.
Measurement
A quantity of 10 μL of the ConA colloid solution was added to the petri dish with the adsorbed cells. The beads settled and could bind to the HEK cells for 1 h at 37°C.
To measure the long distance adhesion of HEK cells (longer than the 10-μm piezo course), a home-built AFM was utilized. A linear motor (M-227.10; Physik Instrumente, Auburn, MA) allowed moving the cantilever vertically with a resolution of 4 nm for up to 1 cm. FluidFM cantilever with an opening of 10 μm were used with an underpressure of 750 mbar to grab the colloids. After grabbing, we never observed that the colloid moved or detached from the cantilever.
The force spectroscopy procedure was carried out in an incubator with air heated at 37°C without CO2 control. It was started a few micrometers above the colloid and was ended when either the cell detached from the surface or the colloid from the cell. A speed of 1 μm s−1 was used and the setpoint for the return was set to 10 nN, where a pause of 5 s allowed the bead to properly connect with the cantilever. Although the motor was used to record the retraction curves, the forward curves were still controlled by the piezo because of its better spatial resolution to properly extract the young’s modulus.
The data were analyzed with the Scanning Probe Image Processor (SPIP, NanoScience Instruments, Phoenix, AZ) software for the maximum adhesion as well as the apparent Young’s modulus. Cases where the bead was not located on top of the middle of the cell were discarded for the Young’s modulus calculation, because the bead could have partially touched the glass as well. Such bead-glass contact could indeed be detected in the forward curve by observing an ideally steep slope.
Errors
In this work, results were indicated with the corresponding standard deviation of the statistical data analysis. Nonetheless, each datum was affected by an uncertainty estimated as follows:
—AFM forces and Young’s modulus: 20% relative error introduced by the spring constant.
—Fluorescence intensity measurements: 20% relative error due to background selection.
—Electrical resistance measurements: 5% relative error due to resistance offset selection.
For the simulations, 1% relative error was estimated as consequence of the finite element meshing and the fitting of the results.
Results and Discussion
Strong connection between colloid and cantilever
Typically, colloids are glued to the cantilever for colloidal AFM. The exact alignment of AFM tip and colloid is crucial at this step because it influences both sensitivity and the effective spring constant of the setup. With FluidFM, the colloid is automatically centered on the opening of the cantilever, the energetically most stable position. The fixation strength depends on the suction force, and therefore on the pressure gradient across the bead-cantilever contact. This pressure distribution is directly linked to the leak flow across the bead. With a large leakage, there would be minimal pressure drop, while with a perfectly tight seal the full pressure would hold the bead in place.
Therefore, it was essential to determine whether the beads would seal the cantilever tightly. We investigated the seal between colloid and FluidFM probe using fluorescence and electrical impedance. Both measurements indicated that the leak flow past the bead is in the attoLiter s−1 range. In effect, the applied pressure in the channel dropped exclusively across the bead originating strong fixation forces up to a few μN, which explained the observed mechanical stability of the beads. We also never observed that the applied underpressure had any influence on the measured adhesion forces or on the noise level.
For the leakage measurements, the cantilever was filled with FITC solution while the surrounding medium was plain buffer. The fluorescent channel content was then replaced with buffer by applying a 1 s suction pulse. After a bead blocked the channel opening, the fluorescence partially recovered within minutes while under pressure was still applied (see Fig. S3). A numeric calculation suggested that unhindered FITC diffusion could explain the measured data alone. We concluded that the <0.1 pL s−1 diffusion of the fluorophore in the channel was much greater than the leak flow past the bead.
The leak flow was further quantified by electrical measurements. The impedance was measured through the cantilever channel while a polystyrene micro bead was attached to its opening. Finite element method (FEM) simulations were used to extract a leak flow out of these measurements. For the FEM model, it was assumed that the measured leak current was due to an average gap between colloid and tip caused by nano-roughness of bead and cantilever, as illustrated in Fig. 2 A. The calculated flow was ∼aL s−1 for typical suction pressures and is depicted in Fig. 2 B. This corresponded to an average gap size of <0.1 nm, indicating that, in fact, the majority of the ∼6-μm-long bead-cantilever contact line was tightly sealed. Probably only a few nanometer-sized holes were present due to the surface roughness. Leak current and flow both decreased with increasing suction, which could be due to the elasticity of the polystyrene microbead. Thus, with increasing suction force, more of the roughness-induced nano holes could be sealed. As control of the tip integrity, its opening was blocked by pressing it against dielectric PDMS in buffer solution, resulting in an open circuit as expected.
A tight seal between colloid and cantilever implicates a vertical fixation force up to a few μN depending on the opening radius (see the Materials and Methods). The lateral fixation force does not exceed 100 nN, depending on the bead diameter and the opening radius. With a friction coefficient of 0.1, this results in allowed scanning forces up to 1 μN. However, any lateral force between substrate and bead potentially leads to a rotation of the bead rather than its translation. In fact, we never lost any beads during such experiments.
Validation of the colloidal FluidFM against selected colloidal AFM results from literature
To validate the concept of colloidal FluidFM, a benchmark measurement with glue-attached beads was chosen from Xu and Logan (18), based on a comparable setup. In both cases, the colloid beads were 3 μm in diameter and consisted of polystyrene functionalized with COOH (18). The colloid was brought into contact with a clean glass slide using ∼5 nN of force and different residence times up to 10 s. Fig. 3 shows both the reference data (18) and our measurement (n = 20 contacts per data point).
Figure 3.
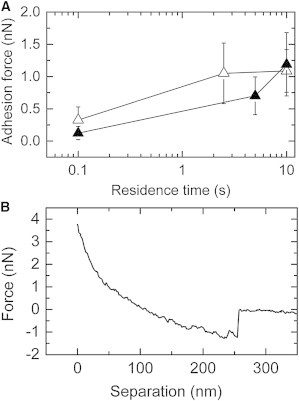
COOH-coated polystyrene beads on glass. Comparison between colloidal FluidFM and standard colloidal AFM. (A) (Solid triangles) Reference data, courtesy of Li-Chong Xu (18). (Open triangles) Data collected in this study. (Bars) Respective standard deviation. (B) A retraction curve measured for a 2.5 s contact with the glass substrate.
The results obtained with standard colloidal AFM and FluidFM measurement agree within the standard deviation. This proves that our setup can be used to gain quantitative data on colloid-substrate interactions.
It has to be kept in mind that two main discrepancy factors cannot be eliminated:
-
1.
The beads in the literature were from a different manufacturer, and thus may differ in surface chemistry (22), although would be nominally the same, and its elasticity as well. Both properties affect the adhesion behavior and the contact area.
-
2.
The probe calibration typically gives an error to the AFM cantilever spring constant of 10% or more (26). This is pronounced for colloidal AFM tips, as the exact position of the colloid influences the effective spring constant (10).
Rapid exchange of beads with different chemical coatings
The lifetime of a functionalized colloid probe can be as short as a few contacts because of contamination (27), because the investigated molecules unfold irreversibly (28) or because they detach from either the colloidal or substrate surfaces (29). Although colloidal probe cantilevers are available for purchase, financial costs are not the only argument against replacing them in large numbers. Each exchanged cantilever has its own spring constant altering the sensitivity of the setup. As a side effect of the cantilever exchange, the system is a drifting one with typical waiting periods of ∼1 h (30).
With the FluidFM we have the possibility to exchange the colloidal probe by applying a positive pressure pulse followed by the attraction of a new colloid from the solution. The colloids can either be spread directly on the surface or are kept in a dedicated reservoir from which they are picked up and transferred onto the substrate.
In this proof-of-principle experiment, we used 3-μm polystyrene beads to measure their adhesion forces on the different chemical domains of a micropatterned surface. We chose the widely used biotin-avidin system to generate a contrast against a PLL-g-PEG background. The avidin-biotin bond is among the strongest noncovalent interactions, and known for its high affinity (31,32). Here we used PLL-g-PEG-biotin on the substrate while the avidin was bound to the bead. In contrast, pure PLL-g-PEG impedes unspecific binding of proteins (24) and was used as passivation. Shown in Fig. 4 A is an epifluorescence image of the chessboard patterned glass substrate as obtained by microcontact printing. Bright squares were functionalized with PLL-g-PEG-biotin and dark squares were coated with repellent PLL-g-PEG. The pattern was then probed with both avidin-coated and plain polystyrene beads. Each type of bead was kept in a separate petri dish. Before measurement, a bead was picked from its source petri dish and transferred through air to the dish containing the substrate pattern. Switching between the bead reservoir and the substrate took typically <10 s. The cantilever remained wet and caused a measurable setpoint drift for less than a minute after exchange of the colloid probe. Stable force spectroscopy curves could be recorded as soon as the substrate pattern was approached again.
Figure 4.
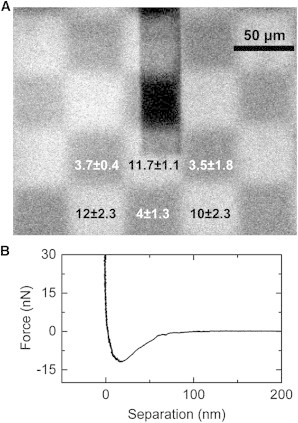
Exchangeable colloids on a micropatterned surface. (A) Fluorescence image of the pattern obtained by microcontact printing: (Bright squares) biotinylated PLL-g-PEG-FITC; (dark areas) covered with bare PLL-g-PEG. The cantilever is visible as shadow due to a high fluorescence background. The average forces on six adjacent squares probed with avidin-coated beads are indicated in [nN] with the respective standard deviation. (B) A typical curve recorded for an avidin-coated bead on PLLgPEG-biotin.
A clear pattern (Fig. 4 A) was visible for the adhesion of the avidin-coated beads, which held to the PLL-g-PEG -biotin regions with 11 ± 1 nN and to the plain PLL-g-PEG regions with 3.7 ± 0.3 nN (2 beads, 12 contacts each). The bare beads were not responsive to the surface pattern and showed no adhesion (<0.3 nN noise, 2 beads, 12 contacts each). This experiment confirms that the exchange of the colloid probe by FluidFM and the transfer from a dedicated reservoir is feasible. It also confirms that force maps can be acquired using colloidal FluidFM. By only switching the colloid instead of the whole cantilever the drift is limited, enabling immediate measurement of adhesion forces.
The measured nonzero adhesion between avidin-coated beads and the PLL-g-PEG layer is attributed to an imperfect PLL-g-PEG coating, which can be a batch-dependent issue. This allowed the avidin molecules to contact the glass substrate directly and adhere on it unspecifically. The unspecific adhesion of avidin beads to clean glass was measured 6 ± 2 nN (15 contacts), which was stronger than on the imperfect PLL-g-PEG layer. In a control experiment, free PLL-g-PEG was added to the buffer to block unspecific interactions on the patterned surface and adhesion was no longer observed. Even the avidin-biotin interaction was blocked, which indicates that the polystyrene/avidin bead was coated by the PLL and thus encased by a PEG shell. This is expected from Dusseiller et al. (33). Also, the avidin-biotin interaction could no longer be measured if free biotin was added to the buffer. This saturated the avidin molecules on the colloid surface, and confirms the specificity of the interaction. Another control experiment measured the adhesion of plain beads on a clean glass substrate to be negligible. This is consistent with the results from Xu and Logan (18), and explains why the plain beads showed zero adhesion even on an imperfect PLL-g-PEG layer and thus with direct access to the glass substrate.
Taking into account the surface density of avidin on the beads and the contact area given by the JKR theory and the thickness of the PLL-g-PEG layer, ∼140 avidin molecules contribute to the adhesion. With a reported binding strength of ∼160 pN per avidin-biotin bond (34), this gives a theoretical maximum adhesion of ∼22 nN. Adhesion curve shape implies, however, that not all connections were broken at the same time, which resulted in a smaller maximum adhesion that scales with the number of available bonds.
Repeated contact between the same avidin-coated bead and the PLL-g-PEG-biotin-coated surface resulted in a decay of contact adhesion. Fig. 5 shows the adhesion normalized by the first contact. The adhesion gradually dropped to <40% of the initial value after only 10 contacts (n = 10 beads). This could be due to PLL backbone being stripped from the surface because the biotin-avidin interaction is stronger than the electrostatic force holding PLL on the glass surface.
Figure 5.
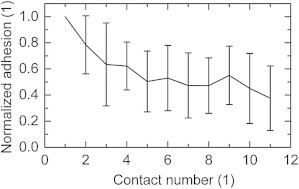
Decay of the maximum adhesion during the first 10 contacts of avidin-coated beads on PLL-g-PEG-biotin. The values are normalized with respect to the first contact adhesion of each measurement (10 beads), whereas the bars indicate the standard deviation. The decaying trend was observed for each bead.
Similar results were reported for biotin tethered to a lipid bilayer by PEG (35) and dioctadecylamine (36). Even for biotin bound to agarose, a drop of adhesion was observed during the first 100 contacts (35). For these and other rapidly decaying systems, colloidal FluidFM could become an efficient alternative.
Application in cell biology measuring adhesion and elastic modulus of HEK cells
Another strategy possible with FluidFM is to spread the colloids directly onto the substrate and then detach them one by one. This enables long interaction times between colloid and substrate, and exploration of many different substrate sites each with a fresh bead in quick succession. We applied this protocol to study the interaction between human embryonic kidney 293 cells (HEK) and concanavalin A (ConA)-coated polystyrene beads.
HEK cells are popular in the field of electrophysiology (37) but only a few studies have tried to quantify their mechanical and adhesion properties (17,38,39), which will represent the reference for our work. ConA is known to bind to the manose residues on the cell membrane strongly enough to be used for single cell force spectroscopy (SCFS) (25,40,41). In standard SCFS experiments, single cells are linked chemically to an AFM tipless cantilever to measure their adhesion to a substrate. Typically, in SFCS, only one cell can be examined per cantilever, as the interaction between cell and ConA cannot be established a second time and thus is irreversible. Additionally, long contact times between the ConA-coated surface and the cell are needed to measure stronger cell-substrate adhesion forces. This either limits the incubation time of the cells to a few minutes, so they cannot spread and adhere too strongly, or it drastically reduces the acquired data points per experiment if fully spread cells are investigated (42). This drawback was recently improved by detaching cells from substrate directly with FluidFM (17).
Using colloidal FluidFM, we designed an experiment combining a long contact time with a quick succession of probed cells. We could assess both the adhesion between cell and substrate as well as between cell and bead. Only one cantilever was utilized per experiment day without any contamination issues. In addition, the apparent elastic modulus of the cells could be obtained from the same experiment by pushing the adsorbed bead against the underlying cell before detaching it, and employing the Hertz model for a spherical indenter.
The HEK cells were cultured on a glass dish for 18 h and were observed to grow in large confluent groups with only few single, double, or triple cells. Then 50-μm beads were added to the dish and were left to sediment and adhere for 60 min. Detachment of the colloids from the cells was performed using a FluidFM cantilever with 10-μm opening and an expected bead fixation force of ∼6 μN. The colloids showed measurable adhesion to the cells only during the first retraction, as expected. A second contact never did originate significant adhesion even if sustained for several minutes (smaller than the 1 nN noise background).
Two different responses could be observed, depending on how many cells were growing together:
-
1.
When picked up from large cell groups, the beads detached from the cells with a maximum adhesion force of ∼60 nN and a minimum force of 10 nN (20 beads).
-
2.
When picking the beads from up to three connected cells, the cells detached from the substrate with the adhesion force linearly dependent of the number of cells. The adhesion force per cell was 20 ± 3 nN (9 cells). Larger cell clusters (4+) could not be detached, which is expected considering the maximum adhesion of a bead to the cells of 60 nN and the individual adhesion of a cell to the substrate of 20 nN.
In Fig. 6, A and B, the detachment of a fully spread cell is shown: As the cantilever retracted from the surface, the cell footprint decreased while the cell elongated and finally detached. The cell remained stuck to the bead after detaching from the surface. A force curve of such a single cell detachment is shown in Fig. 6 C.
Figure 6.
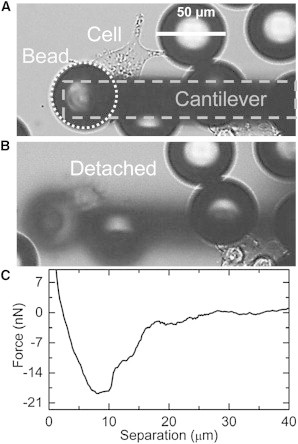
(A) A ConA-coated bead is adsorbed on top of a single HEK cell while the cantilever is visible as shadow. (B) After grabbing the bead and retracting the cantilever, both bead and cell are out of focus. This indicates that the cell is also completely detached from the surface. (C) Retraction curve of a single cell.
The 20 ± 3 nN adhesion per individual cell is slightly lower than reported (33 ± 9 nN (17)) where HEK was directly picked up with FluidFM, which could be due to the shorter incubation time of the cells in our study.
The measured adhesion forces between beads and cells were in the range reported for SCFS (41), where cells are linked to an AFM cantilever using ConA. In contrast, forces between a 10-μm plain latex bead and HEK cells were reported to be 2–3 orders of magnitude smaller (43).
Along with the adhesion strength, the elasticity of the cell is an equally interesting physiological indicator. It can be used to distinguish cancerous from noncancerous cells (44,45), and can reflect changes in the chemical environment (39). Here the apparent elastic modulus of the cells could be extracted from the forward force versus separation curves using the Hertz model (46) for spherical indenters. The Young’s modulus resulted in a value of 0.77 ± 0.68 kPa (15-cell cluster). No relation was found between the Young’s modulus and the size of the cell cluster. The measurement was in agreement with reported values of ∼1 kPa for HEK cells by other groups (38,39).
Conclusion and Outlook
This study shows that FluidFM in combination with colloids can be a reliable and versatile alternative to colloidal probe AFM. It opens the door to the investigation of irreversible and long-term biophysical interactions on a large scale. Several applications can be envisioned based on the option of exchanging the colloid probe and its functionalization at any time, and to observe irreversible interactions.
Spreading the colloids on the substrate allows for long interaction times without sacrificing throughput. This is especially interesting for the study of cell-substrate interactions, where the elastic modulus can also be acquired. Instead of a flat substrate a coated colloid can present the interface of interest to a confluent cell layer. This has not been possible so far, because
-
1.
More than one contact between cell and colloid would already show artifacts of degradation and dirt in colloidal AFM; and
-
2.
Only individual cells could be studied with SCFS. It also allows the study of many different colloid materials and coatings in the same cell condition, by using, for example, fluorescently coded colloids.
The inherent alignment of colloid and cantilever opening should allow working with colloids smaller than ∼0.5 μm, where standard colloidal AFM reaches its limits. Experiments with pyramidal FluidFM probes are being planned and could potentially lead to the investigation of single nanoparticles. With particle radii of <50 nm, the spatial resolution could even approach that of pyramidal AFM tips, although the suction force keeping such small particles in place is expected to be <0.5 nN.
The technique also enables the investigation of nonsolid colloids where gluing of the colloid is not possible due to its liquid or gaseous nature. Investigation of gas bubbles in liquid or between immiscible liquids will be approached in future projects.
The tight seal between colloid and cantilever results in fixation forces strong enough to withstand typical AFM operation. Although only vertical forces have been addressed in this study, lateral forces and thus rotational friction of single colloidal spheres could also be addressed with FluidFM, which has never been reported in literature.
Acknowledgments
We are indebted to Li-Chong Xu (Pennsylvania State University) for providing the raw colloidal AFM data of Xu and Logan (18). Many thanks to Filipe Barata, Michael Kleint, Konstantin Kunz, and Lisa Maier, the students who helped us in the design of the home-built AFM. We also thank Edin Sarajlic (SmartTip, Netherlands), Michael Gabi, Pascal Behr (Cytosurge, Switzerland), and Patrick Frederix (Nanosurf, Switzerland) for their constant support. Finally, we thank Stephen Wheeler for his technical expertise in the LBB workshop.
The development of the colloidal FluidFM was supported by the Swiss innovation promotion agency KTI-CTI (Contract No. 14336.1 PFNM-NM) and by a grant from the Swiss National Science Foundation (SNSF Interdisciplinary Project No. CR23I2-135535).
Supporting Material
References
- 1.Mishra B., Patel B.B., Tiwari S. Colloidal nanocarriers: a review on formulation technology, types and applications toward targeted drug delivery. Nanomedicine. 2010;6:9–24. doi: 10.1016/j.nano.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Wohltjen H., Snow A.W. Colloidal metal-insulator-metal ensemble chemiresistor sensor. Anal. Chem. 1998;70:2856–2859. [Google Scholar]
- 3.Kappl M., Butt H.-J. The colloidal probe technique and its application to adhesion force measurements. Part. Part. Systems Char. 2002;19:129–143. [Google Scholar]
- 4.Ducker W.A., Senden T.J., Pashley R.M. Direct measurement of colloidal forces using an atomic force microscope. Nature. 1991;353:239–241. [Google Scholar]
- 5.Butt H.-J. Measuring electrostatic, van der Waals, and hydration forces in electrolyte solutions with an atomic force microscope. Biophys. J. 1991;60:1438–1444. doi: 10.1016/S0006-3495(91)82180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilal N., Bowen W.R., Ogunbiyi O. A review of atomic force microscopy applied to cell interactions with membranes. Chem. Eng. Res. Des. 2006;84:282–292. [Google Scholar]
- 7.Bartalena G., Loosli Y., Snedeker J.G. Biomaterial surface modifications can dominate cell–substrate mechanics: the impact of PDMS plasma treatment on a quantitative assay of cell stiffness. Soft Matter. 2012;8:673–681. [Google Scholar]
- 8.Schneider A., Francius G., Picart C. Polyelectrolyte multilayers with a tunable Young’s modulus: influence of film stiffness on cell adhesion. Langmuir. 2006;22:1193–1200. doi: 10.1021/la0521802. [DOI] [PubMed] [Google Scholar]
- 9.Pasche S., Textor M., Griesser H.J. Relationship between interfacial forces measured by colloid-probe atomic force microscopy and protein resistance of poly(ethylene glycol)-grafted poly(L-lysine) adlayers on niobia surfaces. Langmuir. 2005;21:6508–6520. doi: 10.1021/la050386x. [DOI] [PubMed] [Google Scholar]
- 10.Hodges C.S. Measuring forces with the AFM: polymeric surfaces in liquids. Adv. Colloid Interface Sci. 2002;99:13–75. doi: 10.1016/s0001-8686(02)00003-9. [DOI] [PubMed] [Google Scholar]
- 11.Mahaffy R.E., Shih C.K., Käs J. Scanning probe-based frequency-dependent microrheology of polymer gels and biological cells. Phys. Rev. Lett. 2000;85:880–883. doi: 10.1103/PhysRevLett.85.880. [DOI] [PubMed] [Google Scholar]
- 12.McNamee C.E., Pyo N., Higashitani K. Atomic force microscopy study of the specific adhesion between a colloid particle and a living melanoma cell: effect of the charge and the hydrophobicity of the particle surface. Biophys. J. 2006;91:1960–1969. doi: 10.1529/biophysj.106.082420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meister A., Gabi M., Zambelli T. FluidFM: combining atomic force microscopy and nanofluidics in a universal liquid delivery system for single cell applications and beyond. Nano Lett. 2009;9:2501–2507. doi: 10.1021/nl901384x. [DOI] [PubMed] [Google Scholar]
- 14.Dörig P., Stiefel P., Zambelli T. Force-controlled spatial manipulation of viable mammalian cells and micro-organisms by means of FluidFM technology. Appl. Phys. Lett. 2010;97:023701. [Google Scholar]
- 15.Stiefel P., Schmidt F.I., Mercer J. Cooperative vaccinia infection demonstrated at the single-cell level using FluidFM. Nano Lett. 2012;12:4219–4227. doi: 10.1021/nl3018109. [DOI] [PubMed] [Google Scholar]
- 16.Guillaume-Gentil O., Potthoff E., Vorholt J.A. Force-controlled fluidic injection into single cell nuclei. Small. 2013;9:1904–1907. doi: 10.1002/smll.201202276. [DOI] [PubMed] [Google Scholar]
- 17.Potthoff E., Guillaume-Gentil O., Vorholt J.A. Rapid and serial quantification of adhesion forces of yeast and mammalian cells. PLoS ONE. 2012;7:e52712. doi: 10.1371/journal.pone.0052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu L.-C., Logan B.E. Adhesion forces between functionalized latex microspheres and protein-coated surfaces evaluated using colloid probe atomic force microscopy. Colloids Surf. B Biointerfaces. 2006;48:84–94. doi: 10.1016/j.colsurfb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Sader J.E., Chon J.W.M., Mulvaney P. Calibration of rectangular atomic force microscope cantilevers. Rev. Sci. Instrum. 1999;70:3967–3969. [Google Scholar]
- 20.Hardt S., Schönfeld F. Springer; New York: 2007. Microfluidic Technologies for Miniaturized Analysis Systems. [Google Scholar]
- 21.Periasamy N., Verkman A.S. Analysis of fluorophore diffusion by continuous distributions of diffusion coefficients: application to photobleaching measurements of multicomponent and anomalous diffusion. Biophys. J. 1998;75:557–567. doi: 10.1016/S0006-3495(98)77545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L.-C., Logan B.E. Interaction forces between colloids and protein-coated surfaces measured using an atomic force microscope. Environ. Sci. Technol. 2005;39:3592–3600. doi: 10.1021/es048377i. [DOI] [PubMed] [Google Scholar]
- 23.Csucs G., Michel R., Danuser G. Microcontact printing of novel co-polymers in combination with proteins for cell-biological applications. Biomaterials. 2003;24:1713–1720. doi: 10.1016/s0142-9612(02)00568-9. [DOI] [PubMed] [Google Scholar]
- 24.Kenausis G.L., Vörös J., Spencer N.D. Poly(l -lysine)-g-poly(ethylene glycol) layers on metal oxide surfaces: attachment mechanism and effects of polymer architecture on resistance to protein adsorption. J. Phys. Chem. B. 2000;104:3298–3309. [Google Scholar]
- 25.Friedrichs J., Helenius J., Muller D.J. Quantifying cellular adhesion to extracellular matrix components by single-cell force spectroscopy. Nat. Protoc. 2010;5:1353–1361. doi: 10.1038/nprot.2010.89. [DOI] [PubMed] [Google Scholar]
- 26.Cumpson P.J., Clifford C.A., Hedley J. Quantitative analytical atomic force microscopy: a cantilever reference device for easy and accurate AFM spring-constant calibration. Meas. Sci. Technol. 2004;15:1337–1346. [Google Scholar]
- 27.Hinterdorfer P., Dufrêne Y.F. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods. 2006;3:347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 28.Tinoco I.J., Jr. Force as a useful variable in reactions: unfolding RNA. Annu. Rev. Biophys. Biomol. Struct. 2004;33:363–385. doi: 10.1146/annurev.biophys.33.110502.140418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hugel T., Seitz M. The study of molecular interactions by AFM force spectroscopy. Macromol. Rapid Commun. 2001;22:989–1016. [Google Scholar]
- 30.Zauscher S., Klingenberg D.J. Friction between cellulose surfaces measured with colloidal probe microscopy. Colloids Surf. A Physicochem. Eng. Asp. 2001;178:213–229. [Google Scholar]
- 31.Florin E.L., Moy V.T., Gaub H.E. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 32.Teulon J.-M., Delcuze Y., Pellequer J.-L. Single and multiple bonds in (strept)avidin-biotin interactions. J. Mol. Recognit. 2011;24:490–502. doi: 10.1002/jmr.1109. [DOI] [PubMed] [Google Scholar]
- 33.Dusseiller M.R., Schlaepfer D., Textor M. An inverted microcontact printing method on topographically structured polystyrene chips for arrayed micro-3-D culturing of single cells. Biomaterials. 2005;26:5917–5925. doi: 10.1016/j.biomaterials.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 34.Lee C.-K., Wang Y.-M., Lin S. Atomic force microscopy: determination of unbinding force, off rate and energy barrier for protein-ligand interaction. Micron. 2007;38:446–461. doi: 10.1016/j.micron.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 35.Wong J., Chilkoti A., Moy V.T. Direct force measurements of the streptavidin-biotin interaction. Biomol. Eng. 1999;16:45–55. doi: 10.1016/s1050-3862(99)00035-2. [DOI] [PubMed] [Google Scholar]
- 36.Leckband D., Müller W., Ringsdorf H. Molecular mechanisms determining the strength of receptor-mediated intermembrane adhesion. Biophys. J. 1995;69:1162–1169. doi: 10.1016/S0006-3495(95)79990-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas P., Smart T.G. HEK293 cell line: a vehicle for the expression of recombinant proteins. J. Pharmacol. Toxicol. Methods. 2005;51:187–200. doi: 10.1016/j.vascn.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 38.Reed J., Frank M., Gimzewski J.K. High throughput cell nanomechanics with mechanical imaging interferometry. Nanotechnology. 2008;19:235101. doi: 10.1088/0957-4484/19/23/235101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuerrier C.M., Benoit M., Grandbois M. Real-time monitoring of angiotensin II-induced contractile response and cytoskeleton remodeling in individual cells by atomic force microscopy. Pflugers Arch. 2009;457:1361–1372. doi: 10.1007/s00424-008-0596-0. [DOI] [PubMed] [Google Scholar]
- 40.Müller D.J., Helenius J., Dufrêne Y.F. Force probing surfaces of living cells to molecular resolution. Nat. Chem. Biol. 2009;5:383–390. doi: 10.1038/nchembio.181. [DOI] [PubMed] [Google Scholar]
- 41.Helenius J., Heisenberg C.-P., Muller D.J. Single-cell force spectroscopy. J. Cell Sci. 2008;121:1785–1791. doi: 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- 42.Weder G., Blondiaux N., Liley M. Use of force spectroscopy to investigate the adhesion of living adherent cells. Langmuir. 2010;26:8180–8186. doi: 10.1021/la904526u. [DOI] [PubMed] [Google Scholar]
- 43.Mescola A., Vella S., Vassalli M. Probing cytoskeleton organization of neuroblastoma cells with single-cell force spectroscopy. J. Mol. Recognit. 2012;25:270–277. doi: 10.1002/jmr.2173. [DOI] [PubMed] [Google Scholar]
- 44.Cross S.E., Kreth J., Gimzewski J.K. Nanomechanical properties of glucans and associated cell-surface adhesion of Streptococcus mutans probed by atomic force microscopy under in situ conditions. Microbiology. 2007;153:3124–3132. doi: 10.1099/mic.0.2007/007625-0. [DOI] [PubMed] [Google Scholar]
- 45.Darling E.M., Zauscher S., Guilak F. A thin-layer model for viscoelastic, stress-relaxation testing of cells using atomic force microscopy: do cell properties reflect metastatic potential? Biophys. J. 2007;92:1784–1791. doi: 10.1529/biophysj.106.083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radmacher M., Tillmann R.W., Gaub H.E. Imaging viscoelasticity by force modulation with the atomic force microscope. Biophys. J. 1993;64:735–742. doi: 10.1016/S0006-3495(93)81433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siekman H.E., Thamsen P.U. 2nd Ed. Springer; Berlin Heidelberg: 2009. Fluid Mechanics for Engineering [Strömungslehre für den Maschinenbau] [Google Scholar]
- 48.Tipler P.A. 4th Ed. W.H. Freeman; New York: 1999. Physics for Scientists and Engineers. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


