Abstract
AIMS
The pathogenesis of obesity remains incompletely understood. Drosophila have conserved neuroendocrine and digestion systems with human and become an excellent system for studying energy homeostasis. Here, we reported a novel obesity Drosophila model, in which expression of human protein, synphilin-1 (SP1), in neurons fosters positive energy balance.
SUBJECTS AND METHODS
To further understand the actions of SP1 in energy balance control, the upstream activation sequence UAS/GAL4 system was used to generate human SP1 transgenic Drosophila. We characterized a human SP1 transgenic Drosophila by assessing SP1 expression, fat lipid deposition, food intake and fly locomotor activity to determine the major behavioral changes and their consequences in the development of the obesity-like phenotype.
RESULTS
Overexpression of SP1 in neurons, but not peripheral cells, increased the body weight of flies compared with that of non-transgenic controls. SP1 increased food intake but did not affect locomotor activity. SP1 increased the levels of triacylglycerol, and the size of fat body cells and lipid droplets, indicating that SP1 increased lipid-fat disposition. Survival studies showed that SP1 transgenic flies were more resistant to food deprivation. SP1 regulated lipin gene expression that may participate in SP1-induced fat deposition and starvation resistance.
CONCLUSION
These studies demonstrate that SP1 expression affects energy homeostasis in ways that enhance positive energy balance and provide a useful obesity model for future pathogenesis and therapeutic studies.
Keywords: Synphilin-1, fat disposition, transgenic Drosophila, lipin
INTRODUCTION
Obesity has become a major health problem worldwide. Although tremendous research investments have been made, the pathogenesis of obesity is still poorly understood and pharmacological treatments are underdeveloped. Identification of novel factors in energy balance pathways may open new avenues for obesity prevention and intervention. Mammalian synphilin-1 (SP1), a 919-amino acid, cytoplasmic protein is expressed in various tissues and is enriched in brain neurons. It is expressed in many regions of the brain, including in multiple hypothalamic nuclei involved in energy balance. Previous reports suggest that SP1 may be involved in neuronal signaling and activity in the brains.1–3 SP1 contains ankyrin-like repeats and a coiled-coil domain suggesting protein–protein interaction functions.1,2,4 SP1 has been shown to interact with α-synuclein, parkin, leucine-rich repeat kinase-2, other ubiquitin ligases and proteasome subunit/regulators, and has implications in Parkinson’s disease pathogeneses related to protein aggregation.1,3–12 Previously we and others have reported that SP1 does not cause neuronal degeneration but has neurotrophic and neuroprotective effects in Parkinson’s disease models.13–15 Our recent studies using a transgenic mouse model suggest that SP1 may be involved in the controls of food intake and body weight.16 In this mouse model, neuronal overexpression of SP1 resulted in phenotypes consistent with the metabolic syndrome: hyperphagia, obesity, hyperinsulinemia, hyperleptinemia and impaired glucose tolerance. To further understand the actions of SP1 in energy balance control, Drosophila models were employed.
Drosophila has become an excellent model for studying metabolism homeostasis and nutrient-sensing pathways.17,18 Drosophila has conserved neuroendocrine and digestion systems that are similar to vertebrates. Drosophila and humans use analogous organs systems and biochemical pathways for maintaining circulating sugar levels, and for sugars and fat storage. Glycogen and fat (most as triacylglycerols (TAGs)) are stored in fly fat bodies, which can be considered the fly equivalent of the liver and white adipose tissue. Pars intercerebralis–corpora cardiac system of insects, the endocrinological equivalent of the hypothalamus–pituitary system of vertebrates, receives information on the internal metabolic status and coordinates the physiological activities of various peripheral organs. Drosophila models have been utilized to investigate multiple aspects of energy homeostasis including food perception, feeding control, energy flux and lipometabolism.17,18 Although the molecular basis of regulating metabolic homeostasis is far from understood, study of how novel proteins function to influence metabolic responses, such as lipid and glucose homeostasis, may open new avenues for understanding the overall controls of energy balance and identify potential novel therapeutics.
To further investigate the actions of SP1 in energy balance, we generated human SP1 transgenic Drosophila models using the upstream activation sequence UAS/GAL4 system to induce SP1 expression in various tissues in both larvae and adult flies. There is no ortholog of human SP1 gene in Drosophila. Thus, transgenic Drosophila is an ideal system to study the actions of human SP1. In this study, we investigated the effects of human SP1 expression in various fly neurons that regulate energy balance, including fruitless-gal4, dopaminergic, serotonergic and pan neurons. We also assessed the effects of human SP1 expression in fat body and insulin-like peptide secretory cells in flies. We measured fly fat deposition, body weight and food intake to assess the actions of SP1.
MATERIALS AND METHODS
SP1 transgenic Drosophila
The UAS/GAL4 system was used to generated human SP1 transgenic Drosophila. Human SP1 cDNA was cloned into pUASTattB vector containing attB site and sequence insulators for high expression. Site-specific integration of attB plasmids containing SP1 gene into fly germ line(attP40) that contains attP lading sites was carried out by coinjection with phiC31-integrase RNA as previously described.19,20 SP1 expression levels were examined by anti-SP1 western blot analysis. Various GAL4 driver flies were obtained from Bloomington Stock Center (Bloomington, IN, USA) to express UAS-attB-synphilin-1 (UAS-SP1) in various tissues as listed in Table 1. Fruitless-Gal4(ref. 21) was a gift from Dr Bader Al-Anzi at Division of Biology, California Institute of Technology (Pasadena, CA, USA). TPH-Gal4 was from Dr Youngseok Lee at Johns Hopkins University School of Medicine, Baltimore, MD. Drosophila were grown in regular fly cultured vials (25 ml volume) on standard cornmeal medium. There were 30 flies in each of vial. All the fly vials were cultured in a Micoprocessor Controlled Low Temperature Illuminated Incubator (Thermo Scientific, Dubuque, IA, USA) at 25 °C and humidity at 55% with a 12-h light/12-h dark cycle. Weight of larvae and adult flies was measured to 0.0001 mg using a Sartorius microbalance ME5 (Sartorius Corporation, Edgewood, NY, USA).
Table 1.
Tissue-specific GAL4 driver flies
| GAL4 drivers | Targeting tissues |
| Dilp2–GAL4 | IPCs |
| ppl-GAL4 | Fat body |
| Fruitless-GAL4 (Fru-GAL4) |
IPCs, mushroom body, the subesophageal ganglion, and the ventral ganglion |
| ddc- GAL4 | Predominately dopaminergic neuron |
| TPH-GAL4 | Serotonergic neuron |
| TH-GAL4 | Dopaminergic neuron |
| Elav-GAL4 | Pan-neuron |
| Actin5c-GAL4 | Ubiquitous expression |
Abbreviation: IPC, insulin-producing cells.
Western blot analysis
Flies were homogenized in lysis buffer (50 mM Tris • HCl, pH 7.5/1 mM EGTA/0.5 M NaCl/1% Triton X-100/1 mM DTT with protease inhibitors) as described previously.22 The resulting homogenates were subjected to Bradford protein assays to ensure equal protein loading and resolved on 4–12% SDS/NuPAGE Bis-Tris gels and transferred onto PVDF membranes (Invitrogen, Grand Island, NY, USA). The membranes were blocked in buffer (pH 7.4, 10mM Tris HCl/ 150 mM NaCl/0.1% Tween 20) containing 4% nonfat milk and then probed with various antibodies. Proteins were detected by using enhanced chemiluminescence reagents (NEN, Boston, MA, USA). Anti-SP1 rabbit polyclonal antibodies were made against human SP1 (30–543 aa fragment).4 Anti α-tublin antibody was from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA).
TAG and glycogen measurement
For TAG assays, flies were homogenized in phosphate-buffered saline (PBS) with 1% Triton-X and immediately incubated at 70 °C for 10 min. Heat-treated homogenates were incubated with Free Glycerol Reagent (Sigma, St Louis, MO, USA) for 5 min at 37 °C. Samples were assayed using a BioTek Synergy (Winooski, VT, USA) HT microplate spectrophotometer at 540 nm. TAG was determined by subtracting the amount of free glycerol in the PBS-treated sample from the total glycerol present in the sample treated with triglyceride reagent. TAG levels were normalized to protein levels. Data were analyzed using a Student’s t-test.
For glycogen assays, flies were prepared in an identical manner as described for the TAG assays, except that flies were homogenized in PBS. Heat-treated homogenates (20 µl) were transferred to each of three wells of a 96-well plate. Into one well was added 150 µl PBS, into the second was added 150 µl of glucose reagent (Sigma) and into the third was added 150 µl of glucose reagent and 0.7 U of amyloglucosidase (Sigma). The plate was incubated at 37 °C for 1 h. The color intensity of each sample was measured using a BioTek Synergy HT microplate reader at 340 nm. Glucose and glycogen were determined using a standard curve. Total glycogen was normalized to protein levels.
Buoyancy-based density assay (floating assay)
Floating assays were performed as previously described23 with a slight modification based on the principle that the fatter larvae float better in a higher density solution. Briefly, 10–30 larvae in each experimental group at wandering stage were put in 10 ml of 9% sucrose (Fisher Scientific, PA, USA), PBS solution. Larvae were placed in the sucrose solution, gently mixed and, at a 3 minutes time point, the larvae floating at the surface of solution were counted. The percentage of larvae that were floating was calculated. The experiments were repeated at least three times.
Fat body staining assay
Phalloidin staining of fat body tissue was carried out according to the manufacturers’ protocols.24 Briefly, fat bodies were dissected from third instar larvae and fixed with 3.7% formaldehyde for 10 min. After blocking with 5% normal goat serum for 2 h, samples were incubated with Alexa Fluor 568 phalloidin (Invitrogen) and mounted in Vectashield with DAPI. The images were captured under a fluorescent Leica DMRB microscope (Buffalo Grove, IL, USA). NIH ImageJ software (La Jolla, CA, USA) was used to analyze cell area, represented as pixels per cell.
Nile red staining for lipid droplets
Fat bodies were dissected from third instar larvae, fixed with 3.7% formaldehyde. Samples were incubated in the buffer (0.00002% Nile red and 75% glycerol) for 5 min before mounting with a coverslip. Slides were imaged using a fluorescent Leica DMRB microscope at × 40 magnification with an excitation wavelength of 515–560 nm and an emission of 590 nm or excitation: 450–500 nm, emission: 528 nm.
Capillary feeder assay
Capillary feeder (CAFÉ) assay was performed as described previously.25 Briefly, a Drosophila wide vial with 15 ml 1% agar covered with a foam plug was used to provide humid condition. Through the foam plug was inserted a truncated 200-µl pipette tip which held 5 µl disposable glass capillary tube containing 5% sugar solution. The experiments were conducted for 3 days under a 12-h light/12-h dark cycle in a room kept at 25 °C and 70% humidity. Each experiment included an identical CAFE chamber without flies to determine evaporative losses (typically <10% of ingested volumes), which were subtracted from experimental readings.25
Climbing assay and actometer test
Climbing assay (negative geotaxis assay) was used to determine locomotor activity as described previously.22 Cohorts of 60 flies from each genotype were subjected to the assay at 10 days of age. The tested flies were age-matched, randomly selected, anesthetized and placed in a vertical plastic column (length, 25 cm; diameter, 1.5 cm). After a 30-min recovery from CO2 exposure, flies were gently tapped to the bottom of the column. We counted and calculated the percentage of flies that could climb to or above the median line of the cylinder in 10 s.
For actometer test, cohorts of 20 flies from each genotype were subjected to the actometer assay at 10 days of age. A single fly was placed in a small tube with food at one end and was monitored for 3 days under standard conditions of 12-h light and 12-h darkness intervals. Activity was recorded on the computer every time the fly crossed an infrared beam (locomotion actograms), and the data were grouped into 5-min bins as described previously.22
Starvation resistance assay
For starvation experiments, adult flies at 10 days of age were fed with 1% agar in 12-h light/12-h dark cycle at 25 °C. Survival was monitored daily through the fly lifetime. Periodically, flies were transferred to fresh vials to prevent fungal growth. Each experiment was performed in triplicate. Survival data were analyzed by using Kaplan–Meier survival curves.
Real-time reverse transcription PCR for lipin gene expression
Flies at 10 days of age were transferred to vials containing only water. After starvation of 24 or 48 h, total RNA was extracted from whole flies using Trizol reagent (Invitrogen). DmLpinA and DmLpinK were analyzed by one-step quantitative reverse transcriptase (qRT)-PCR using a HotStart-IT SYBR green qRT-PCR kit (product no. 75770; Affymetrix, Cleveland, OH, USA) and a Bio-Rad CFX96 real-time PCR detection system. The rp49 gene was used as a normalized gene with primers with the sequences 5′-CGGATCGATA TGCTAAGCTGT-3′ and 5′-GCGCTTGTTCGATCCGTA-3′. Primers for DmLpinA were 5′-ATCCCACGTCCCTGATATCG-3′ and 5′-TTCATCTTGGTTGGTTAGCAGG-3′. Primers for DmLpinK were 5′-CGGTAGATCGGTAAATCGC-3′ and 5′-TGGAC CCGCTGCTTCTGAG-3′. Comparative quantification was carried out using the standard curve method according to the manufacturer’s protocol.
Data analysis
Quantitative data were expressed as arithmetic means ± s.e.m. The differences among groups were statistically analyzed by analyses of variance followed by post hoc test (Tukey’s range test). A P-value of < 0.05 was considered significant.
RESULTS
Expression of human SP1 increased fly body weight
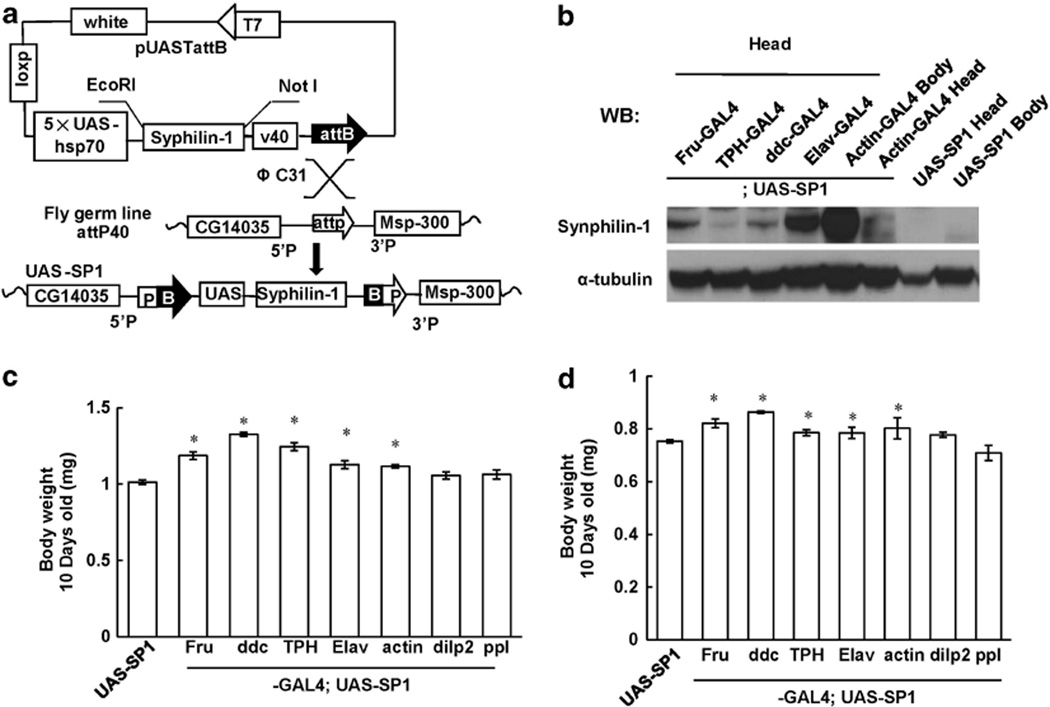
To generate human SP1 transgenic flies, UAS/GAL4 system was employed.26,27 In this system, the yeast GAL4 transcription factor binds specifically to the UAS. Thus, UAS-linked transgenes can be expressed in specific cell types under the control of a given promoter (promoter-GAL4). This recently developed targeted integration method was utilized to overcome the transgene position effect and to obtain high expression levels.19,20 The principle of this method is that using a gypsy retrovirus insulator reliably produces highly expressed transgenes in a variety of tissues. To be effective, gypsy-flanked transgenes must be targeted to specific loci such as attP to achieve the highest levels of induction, and used in conjunction with neutral core promoters to ensure tight basal regulation. Human SP1 gene was cloned into the UAS-attB vector containing the attB site (Figure 1a). The resulting construct was coinjected with phiC31-integrase RNA to fly germ line(attP40) containing attP landing sites as previously described.19,20 The UAS-SP1 fly line was established by genetic mapping.
Figure 1.
Synphilin-1 increases adult fly body weight. (a) Schematic of site-specific integration between the attB and attP sequences to generate UAS-SP1 fly line. Human synphilin-1 cDNA with a myc tag was cloned into vector pUASTattB containing attB site. Site-specific integration of the resulting construct into attP40 flies at attP lading site was carried out by coinjection with phiC31-integrase RNA. (b) Western blot analysis showing human synphilin-1 protein expression in various fly tissues under indicated tissue-specific GAL4 promoter drivers using anti-synphilin-1 antibodies. (c, d) Human synphilin-1 was expressed by combination of the pUASTattB-synphilin-1(UAS-SP1) with Fru-GAL4, ddc-GAL4, TPH-GAL4 or Elav-GAL4 driver in Fru-GAL4 neurons, dopaminergic and seronergic neurons, and all neurons, respectively. The body weight of flies was measured at 10 days of age. There were 70 flies in each group. (c) female; (d) male. Data are means ± s.e.m. Significant differences between SP1 transgenic flies and non-transgenic control mice as indicated, *P < 0.05 by analyses of variance.
To express human SP1 in fly tissues, various neuronal and endocrine tissue GAL4 driver flies were used including Dilp2-GAL4 (leading to expression in insulin producing cells, IPCs), Fruitless-GAL4 (leading to expression in IPCs, mushroom body, the subesophageal ganglion and the ventral ganglion neurons), ddc-GAL4 (leading to expression predominantly in dopaminergic neuron), TPH-GAL4 (leading to expression in serotonergic neurons), Elav-GAL4 (leading to expression in pan neurons) and actin5c-GAL4 (leading to ubiquitous expression in all tissues). Western blot analysis showed that human SP1 was expressed well using these GAL4 driver fly lines (Figure 1b). Expression of human SP1 resulted in significantly increased body weight in adult flies at 10 days of age using all above drivers except Dilp2-GAL4 and fat body-GAL4, indicating that expression of SP1 in IPCs and fat body cells was not sufficient to affect body weight (Figures 1c and d). Using the same tissue GAL 4 promoter, female SP1 transgenic flies had greater body weight increases than male adult flies, consistent with the overall sex difference in adult body weights.28,29 Most interestingly, expression of SP1 in dopaminergic neurons by ddc-GAL4 driver, and in serotonergic neuron by TPH-GAL4 driver produced greater effects on body weight than pan neuron or ubiquitous expression. Thus, most of experiments in this study used the ddc-GAL4 promoter for expression in dopaminergic neurons.
SP1 increased fat storage in transgenic flies
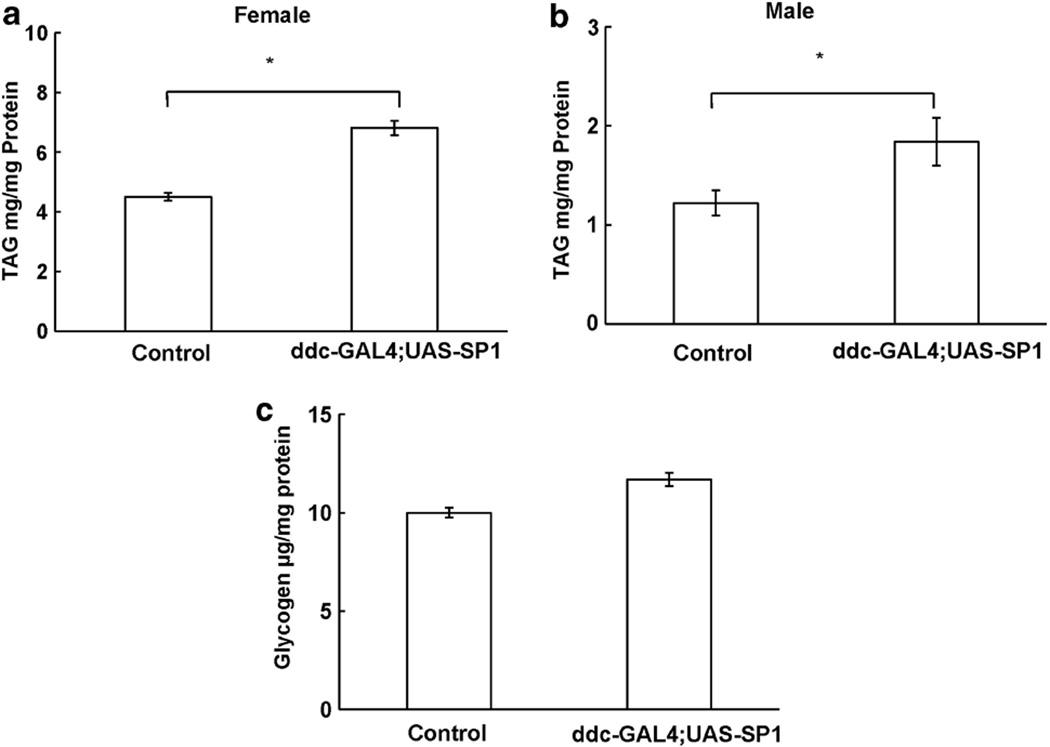
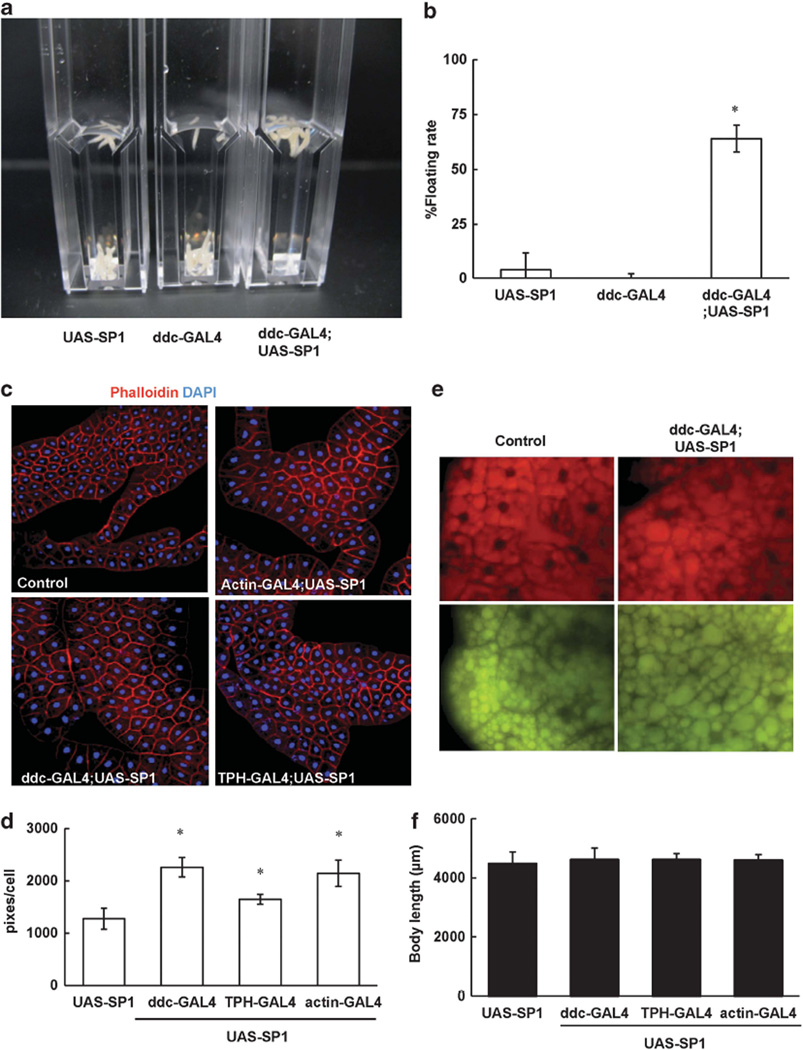
TAG and glycogen are the major intracellular forms of stored energy in flies.17,18 In adult flies, expression of human SP1 in dopaminergic neurons significantly increased TAG content (Figures 2a and b) but did not alter glycogen content (Figure 2c). To further assess whether expression of SP1 in dopaminergic neurons increased fat deposit in the development stage, we employed a floating assay to indirectly detect body fat content in Drosophila larvae at the wandering stage. The principle of this assay is that larvae with higher fat content float better in 9% sucrose solution than do the lean ones, similar to the previously described assay.23 When larvae were in a 9% sucrose/ PBS solution, 94% of the non-transgenic larvae sank to the bottom of vials. In contrast, about 71% of the ddc-GAL4;UAS-SP1 larvae floated at the surface of the solution (Figure 3).
Figure 2.
Synphilin-1 increased TAG and glycogens in adult flies. The homogenates from adult non-transgenic and ddc-GAL4;UAS-SP1 transgenic flies at 10 days of age were assayed for TAG (a, b) or glycogen (c). Triacylglycerol and glycogen levels were normalized to protein levels, and are presented relative to the level in non-transgenic control flies (pUASTattb-synphilin-1). Error bars represent s.e.m., independent samples of 40 animals each group; *P < 0.05 by analyses of variance vs non-transgenic control flies.
Figure 3.
Synphilin-1 increased fat storage in the third larvae stage. (a, b) SP1 increased fat storage in the third instar larvae using a buoyancy-based density assay. The non-transgenic and SP1 larvae were immersed in the 9% sucrose solution in plastic cuvettes and photographed after reaching equilibrium. (a) Images of floating assays. (b) For the indicated genotypes, mean floatation scores (% floating larvae) were calculated from three independent replicates, for each using ~ 20 larvae submerged in sucrose. Error bars represent s.e.m. *P < 0.05 by analyses of variance (ANOVA). Significant differences between SP1 transgenic flies and non-transgenic controls (UAS-SP1 or ddc-GAL4) as indicated. (c–f) Synphilin-1 was expressed using various GAL4 drivers as indicated. (c) Representative images of fat body cells from wandering third instar larvae in each experimental group stained with phalloidin (red) and DAPI (blue) for nuclei. (d) Quantitation of fat body cell size was carried out with Image-J software. Significant differences between SP1 and non-transgenic control larvae as indicated, *P < 0.05 by ANOVA. (e) Representative fluorescent microscopy images of lipid droplets in fat body cells from third instar larvae in each experimental group stained with nile red. Top: excitation: 515–560 nm, emission: 590 nm; bottom: excitation: 450–500 nm, emission: 528 nm. (f) The body size of the third instar larvae was measured. Data are means ± s.e.m. There were 10 larvae in each group. Significant differences between SP1 and non-transgenic control larvae as indicated, *P < 0.05 by ANOVA.
The Drosophila fat body is not only the equivalent of adipose tissue but also serves major liver-like functions in energy homeostasis,17,18 as this mammalian organ is absent in insects. The fat body operates as the executive organ of energy storage and mobilization in energy balance control in response to humoral signals of the brain neuroendocrine system. The larval fat body consists of a defined number of large polyploid cells building a single contiguous organ sheet, which stretches throughout the body cavity and responds to nutrient excess by hypertrophy. These polyploidy cells are largely composed of lipid droplet (filled trophocytes). Fat body cell size was strikingly increased in larvae expressing human SP1 in dopaminergic neurons compared with non-transgenic controls (Figures 3c and d). The cell size of fat bodies in the larvae of ddc-GAL;UAS-SP1 flies increased roughly twofold. Moreover, the size of lipid droplet was also increased in these SP1 transgenic larvae compared with non-transgenic controls (Figure 3e). However, the body length of the larvae was not altered by SP1 expression (Figure 3f).
SP1 increased food intake but did not affect locomotor activity
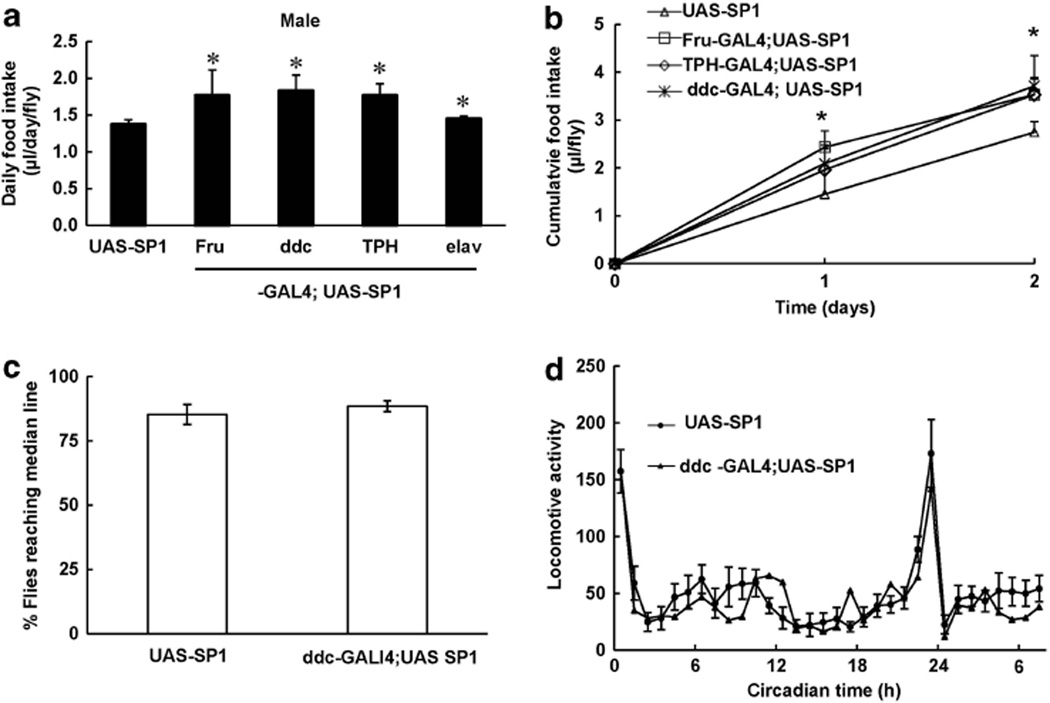
To assess whether SP1 expression alters caloric intake in flies, CAFÉ assays were employed to measure adult fly food intake in all the transgenic lines that had increased body weight. Neuronal expression of SP1 significantly increased food intake in transgenic lines relative to non-transgenic control flies (Figures 4a and b). To observe fly locomotor activity as a surrogate for energy expenditure, climbing assays and actometer measurements were employed. There were no differences at any age on climbing assays between ddc-Gal4;UAS-SP1 and non-transgenic control flies (Figure 4c). To further assess whether SP1 alters fly overall activity, we used an actometer assay to assess the movement pattern in 10-day-old flies during a 12-h dark/12-h light cycle. The flies displayed two peaks of activity. Expression of SP1 in dopaminergic neurons did not affect times, or frequency at which peak activity occurred or the overall level of locomotor activity (Figure 4d).
Figure 4.
Synphilin-1 increased food intake in adult flies. (a, b) CAFÉ assay was used to monitor fly food intake at 5 days of age for 1–5 days. The food intake of each fly was recorded daily. Data are means ± s.e.m. (a) Daily food intake; (b) 48 h cumulative food intake. Significant differences between SP1 transgenic flies and non-transgenic control mice as indicated, *P < 0.05 by analyses of variance. (c, d) Cohorts of 60 flies from each group at 10 days of age were subjected to climbing assay (c) and the actometer assay (d) to measure locomotor activity. Shown are representative data from three separate experiments.
SP1 transgenic flies were resistant to food deprivation and decreased lipin gene expression
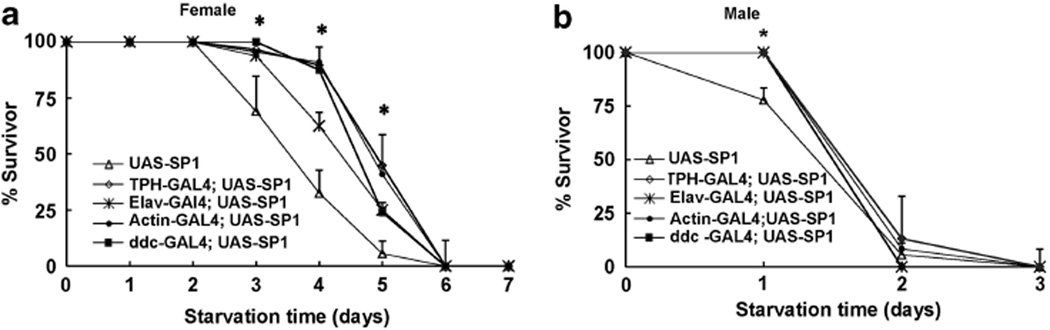
To further characterize the obese phenotypes of SP1 transgenic flies, a starvation-resistance assay was performed. Overall, SP1 expression that resulted in increased body weight also improved survival in response to starvation (Figure 5). This effect was much more evident in female than in male flies. This is possibly due to male flies having less overall fat storage than female flies (Figures 2a and b). At 5-day starvation, 97% of female non-transgenic flies were dead. In contrast, at 5-day starvation, there were still 50% SP1 transgenic female flies were alive.
Figure 5.
Synphilin-1 transgenic flies were resistance to food deprivation. Cohort of 60 flies from each group at 10 days of age were subjected to starvation resistance assays. Flies were kept at 25 °C, and dead flies were counted and removed from the vials daily. Each experiment was performed in triplicate. (a) female; (b) male. Survival data were analyzed by Kaplan–Meier log-rank survival analysis. *P < 0.05, statistically significant differences between non-transgenic and SP1 transgenic flies.
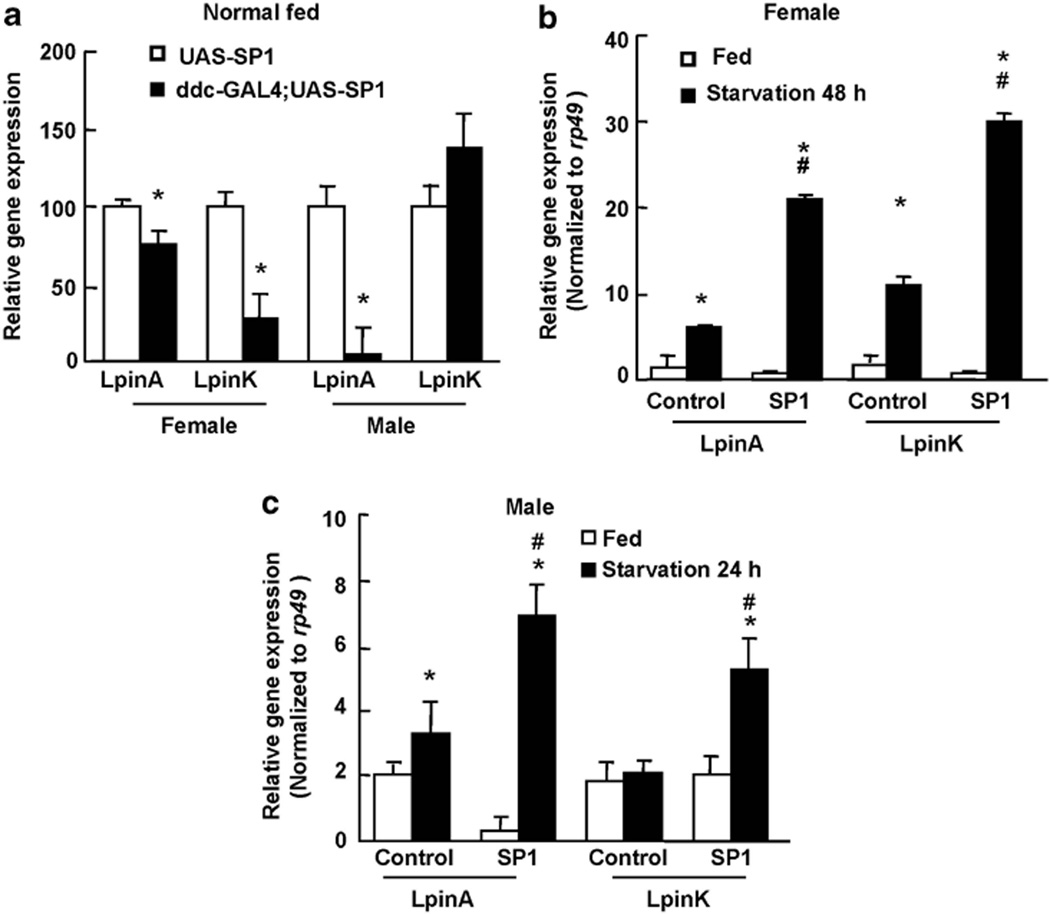
Previous reports suggest that Lipin promotes fat storage and is upregulated in response to food deprivation.30–33 In Drosophila, there is only one Lipin ortholog (CG8709, here named DmLpin) expressing at least three isoforms (DmLpinA, DmLpinK and DmLpinJ). The highest levels of lipin are found in the fat body, where DmLpinA and DmLpinK are expressed. DmLpinK is the most abundant isoform in the central nervous system. DmLpinJ is the predominant isoform in the testis.30–32 To assess the effect of SP1 on lipin gene expression, we performed real-time reverse transcription PCR to detect the mRNA levels of DmLpinA and DmLpinK in ddc-GAL4;UAS-SP1 transgenic flies. The mRNA levels of DmLpinA and DmLpinK significantly decreased in ddc-GAL;UAS-SP1 transgenic flies at 10 days of age (when the body weight was significantly increased) compared with non-transgenic control flies in the fed state (Figure 6a). Starvation increased both DmLpinA and DmLpinK mRNA in non-transgenic and SP1 transgenic flies (Figure 6b). However, upregulation of both DmLpinA and DmLpinK by starvation in SP1 transgenic flies occurred to a greater degree than in non-transgenic control flies (Figure 6b).
Figure 6.
Synphilin-1 regulates lipin gene expression. Cohort of 60 flies from each group at 10 days of age were food deprived for 24h (male) or 48 h (female). The total RNA were extracted and subjected to real-time RT-PCR to detect the mRNA level of DmLpinA and DmLpinK. (a) Flies at normal fed condition. *P < 0.05 by ANOVA, statistically significant differences between non-transgenic and SP1 transgenic flies. (b, c) Flies at starvation conditions. (b) Female; (c) male. *P < 0.05 by ANOVA, statistically significant differences between normal fed and starvation group. #P < 0.05 by ANOVA, statistically significant differences between non-transgenic and SP1 transgenic flies.
DISCUSSION
In this study, we characterized the metabolic actions of human SP1 by generating transgenic Drosophila expressing human SP1 under various promoters resulting in various tissue-specific expression, including fruitless-GAL4, dopaminergic, serotonergic and pan neurons, and fat body and insulin-like peptide secretory cells. Expression of human SP1 in neurons but not peripheral cells significantly increased fly body weight and food intake compared with non-transgenic controls. Moreover, expression of SP1 in dopaminergic neurons increased TAG content, fat body and lipid droplet size. SP1 transgenic flies were more resistant to starvation challenge and more sensitive in regulating lipin gene expression compared with non-transgenic control flies. These findings not only provide new insight into the actions of SP1 in regulating energy balance but also provide a unique model for the study of the pathogenesis of obesity and its potential treatments.
Given there is no endogenous SP1 homology protein in flies, human SP1 was specifically expressed in various neuronal and endocrine tissues under various driver promoters to assess its actions. Our results demonstrate that expression of SP1 in neuronal tissues is more likely to increase body weight rather than expression in endocrine peripheral cells. In contrast, expression of human LRRK2 with the same approach as used for the expression of SP1 in flies does not alter body weight but induces Parkinson’s disease-like phenotypes.22 Furthermore, expression of SP1 in dopaminergic (by ddc-GAL4 or TH-GAL4 driver) or serotonergic (by TPH-GAL4 driver) neurons increased body weight and fat deposition to a greater degree than either pan neuron or ubiquitous expression. Expression of SP1 in dopamine neurons increased fat deposition in both larvae and adulthood. In adult ddc-GAL4;UAS-SP1 flies, TAG content, one of major indicators of fat storage, was significantly increased. In larvae, SP1 increased the size of fat body cells and lipid droplet. Furthermore, the larvae of ddc-GAL4;UAS-SP1 transgenic flies displayed high floating rate than non-transgenic flies in sucrose solution due to SP1- induced the high fat deposition. The fly fat body plays a critical role in Drosophila metabolism and expresses more than 7 000 genes (http://www.Flyatlas.org/) that regulate metabolic homeostasis as described.34 The fat body is the equivalent of mammalian adipose and acts like mammalian liver in the regulation of energy balance and the detoxification of xenobiotics.17,18 The fat body acts as a central organ of energy storage and is largely composed of lipid droplets. The cytosolic lipid droplets not only store neutral lipid but also are dynamic organelles involved in important metabolic reactions including fatty acid and sterol biosynthesis, fatty acid activation and lipolysis.35 Our findings suggest that overexpression of SP1 in dopaminergic neurons in flies may act as a positive energy balance regulator and has a critical role in fat body and lipid droplet-associated energy storage.
Expression of SP1 in neurons in flies significantly increased food intake compared with non-transgenic flies. However, SP1 expression did not alter locomotor activity, suggesting that it is the SP1-induced hyperphagia that leads to body weight gain and increased fat deposition. These results are consistent with our previous findings in a human SP1 transgenic mouse model,16 in which expression of human SP1 in neurons increased food intake, body weight and body fat. Pair-feeding SP1 transgenic mice to the amount of food that non-transgenic mouse consumed for 10 weeks normalized the obesity phenotypes, demonstrating that the obesity is secondary to increase in food intake.
Lipid droplets have a central role in lipid homeostasis by mediating the transient storage of fatty acids in the form of triglyceride, while preventing high levels of toxic lipid intermediates or oxidized lipids that mediate cellular lipotoxicity. Lipin proteins have been found to have crucial roles in the regulation of lipid homeostasis. Most if not all mammalian and non-mammalian cells convert excess fatty acid into triglyceride for storage within lipid droplets.35,36 During fasting, adipose-tissue lipolysis provides a convenient source of fatty acid fuel for energy demands in non-adipose tissues. Most interestingly, our results showed that SP1 expression resulted in increased lipid droplets in the fat body and greater resistance to starvation than was found in non-transgenic flies. Expression of SP1 in dopaminergic neurons decreased lipin expression in the basal fed condition and resulted in starvation-induced upregulation of lipin expression. These findings are consistent with previous reports that lipin transcript levels are upregulated under starvation conditions.33,37 Expression of SP1 in dopamine neurons regulating lipin gene expression is most likely secondary to the increased feeding and weight gain. This provides a possible explanation for SP1 transgenic flies’ enhanced starvation resistance after the cessation of food intake. In the normal feeding condition, ddc-GAL4;UAS-SP1 flies displayed downregulation of lipin expression thereby leading to fat synthesis and storage; in the starvation condition, lipin gene was upregulated by SP1 expression leading to more-efficient energy use through improved salvage of fatty acids released during lipolysis (by recycling to TAG). Previous mammalian studies of the role of lipin during starvation in adipose tissue support this explanation.38 In mammalian liver, starvation also leads to lipin 1 upregulation.39 There, lipin activates genes of the fatty acid β-oxidation pathway, bolstering the capacity of the liver to process the increasing supply of free fatty acids. However, the pathways mediating how expression of SP1 in neurons in flies regulate lipid droplet biogenesis and lipin gene expression require further investigation. Our recent studies in mice demonstrated that food deprivation significantly increased endogenous SP1 mRNA expression in important hypothalamic nuclei involved in the controls of food intake and energy balance (paraventricular nucleus and arcuate nucleus) in normal mice,16 suggesting that endogenous SP1 expression is regulated by energy intake status. Together with SP1-induced hyperphagia in mice and flies, it suggests that SP1 may alter various aspects of neuronal signaling in the brains to increase food intake leading to increased fat deposition and alerations in lipin gene expression.
In summary, we demonstrate that expression of human SP1 in fly neurons induced obesity-like phenotypes including increases in body weight, body fat (TAG content), food intake and starvation resistance. These phenotypes were most obvious in transgenic flies when SP1 was expressed in dopaminergic neurons. SP1 increased the size of the fat body and lipid droplets, downregulated lipin gene expression and increased the larvae floating rate in sucrose. Our findings demonstrate that SP1 expression positively regulates energy homeostasis. These results also provide a useful novel model for further understanding the pathogenesis of obesity and for the development of novel therapeutics for obesity prevention and intervention.
ACKNOWLEDGEMENTS
We thank Dr Craig Montell for helpful discussion. We also thank Drs Bader Al-Anzi and Dr Youngseok Lee for kindly providing us the GAL4 driver flies. This work was supported by the National Institutes of Health, Grants: DK083410 to WWS, and the Paul R McHugh Chair for Motivated Behavior to THM.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Ribeiro CS, Carneiro K, Ross CA, Menezes JR, Engelender S. Synphilin-1 is developmentally localized to synaptic terminals, and its association with synaptic vesicles is modulated by alpha-synuclein. J Biol Chem. 2002;277:23927–23933. doi: 10.1074/jbc.M201115200. [DOI] [PubMed] [Google Scholar]
- 2.Nagano Y, Yamashita H, Takahashi T, Kishida S, Nakamura T, Iseki E, et al. Siah-1 facilitates ubiquitination and degradation of synphilin-1. J Biol Chem. 2003;278:51504–51514. doi: 10.1074/jbc.M306347200. [DOI] [PubMed] [Google Scholar]
- 3.Wakabayashi K, Engelender S, Yoshimoto M, Tsuji S, Ross CA, Takahashi H. Synphilin-1 is present in Lewy bodies in Parkinson’s disease. Ann Neurol. 2000;47:521–523. [PubMed] [Google Scholar]
- 4.Engelender S, Kaminsky Z, Guo X, Sharp AH, Amaravi RK, Kleiderlein JJ, et al. Synphilin-1 associates with alpha-synuclein and promotes the formation of cytosolic inclusions. Nat Genet. 1999;22:110–114. doi: 10.1038/8820. [DOI] [PubMed] [Google Scholar]
- 5.Smith WW, Margolis RL, Li X, Troncoso JC, Lee MK, Dawson VL, et al. Alpha-synuclein phosphorylation enhances eosinophilic cytoplasmic inclusion formation in SH-SY5Y cells. J Neurosci. 2005;25:5544–5552. doi: 10.1523/JNEUROSCI.0482-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung KK, Zhang Y, Lim KL, Tanaka Y, Huang H, Gao J, et al. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat Med. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- 7.Szargel R, Rott R, Engelender S. Synphilin-1 isoforms in Parkinson’s disease: regulation by phosphorylation and ubiquitylation. Cell Mol Life Sci. 2008;65:80–88. doi: 10.1007/s00018-007-7343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Farrell C, Murphy DD, Petrucelli L, Singleton AB, Hussey J, Farrer M, et al. Transfected synphilin-1 forms cytoplasmic inclusions in HEK293 cells. Brain Res Mol Brain Res. 2001;97:94–102. doi: 10.1016/s0169-328x(01)00292-3. [DOI] [PubMed] [Google Scholar]
- 9.Lee G, Tanaka M, Park K, Lee SS, Kim YM, Junn E, et al. Casein kinase II-mediated phosphorylation regulates alpha-synuclein/synphilin-1 interaction and inclusion body formation. J Biol Chem. 2004;279:6834–6839. doi: 10.1074/jbc.M312760200. [DOI] [PubMed] [Google Scholar]
- 10.Avraham E, Szargel R, Eyal A, Rott R, Engelender S. Glycogen synthase kinase 3beta modulates synphilin-1 ubiquitylation and cellular inclusion formation by SIAH: implications for proteasomal function and Lewy body formation. J Biol Chem. 2005;280:42877–42886. doi: 10.1074/jbc.M505608200. [DOI] [PubMed] [Google Scholar]
- 11.Marx FP, Soehn AS, Berg D, Melle C, Schiesling C, Lang M, et al. The proteasomal subunit S6 ATPase is a novel synphilin-1 interacting protein--implications for Parkinson’s disease. FASEB J. 2007;21:1759–1767. doi: 10.1096/fj.06-6734com. [DOI] [PubMed] [Google Scholar]
- 12.Alvarez-Castelao B, Castano JG. Synphilin-1 inhibits alpha-synuclein degradation by the proteasome. Cell Mol Life Sci. 2011;15:2643–2654. doi: 10.1007/s00018-010-0592-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith WW, Liu Z, Liang Y, Masuda N, Swing DA, Jenkins NA, et al. Synphilin-1 attenuates neuronal degeneration in the A53T {alpha}-synuclein transgenic mouse model. Hum Mol Genet. 2010;19:2087–2098. doi: 10.1093/hmg/ddq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Liu Z, Tamashiro K, Shi B, Rudnicki DD, Ross CA, et al. Synphilin-1 exhibits trophic and protective effects against Rotenone toxicity. Neuroscience. 2010;165:455–462. doi: 10.1016/j.neuroscience.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giaime E, Sunyach C, Herrant M, Grosso S, Auberger P, McLean PJ, et al. Caspase-3-derived C-terminal product of synphilin-1 displays antiapoptotic function via modulation of the p53-dependent cell death pathway. J Biol Chem. 2006;281:11515–11522. doi: 10.1074/jbc.M508619200. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Tamashiro KL, Liu Z, Bello NT, Wang X, Aja S, et al. A novel obesity model: synphilin-1-induced hyperphagia and obesity in mice. Int J Obes (Lond) 2011 doi: 10.1038/ijo.2011.235. e-pub ahead of print 13 December 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker KD, Thummel CS. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leopold P, Perrimon N. Drosophila and the genetics of the internal milieu. Nature. 2007;450:186–188. doi: 10.1038/nature06286. [DOI] [PubMed] [Google Scholar]
- 19.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni JQ, Markstein M, Binari R, Pfeiffer B, Liu LP, Villalta C, et al. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5:49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Anzi B, Sapin V, Waters C, Zinn K, Wyman RJ, Benzer S. Obesity-blocking neurons in Drosophila. Neuron. 2009;63:329–341. doi: 10.1016/j.neuron.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Wang X, Yu Y, Li X, Wang T, Jiang H, et al. A Drosophila model for LRRK2-linked parkinsonism. Proc Natl Acad Sci USA. 2008;105:2693–2698. doi: 10.1073/pnas.0708452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reis T, Van Gilst MR, Hariharan IK. A buoyancy-based screen of Drosophila larvae for fat-storage mutants reveals a role for Sir2 in coupling fat storage to nutrient availability. PLoS Genet. 2010;6:e1001206. doi: 10.1371/journal.pgen.1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Liu J, Li CR, Momen B, Kohanski RA, Pick L. Deletion of Drosophila insulin-like peptides causes growth defects and metabolic abnormalities. Proc Natl Acad Sci USA. 2009;106:19617–19622. doi: 10.1073/pnas.0905083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, et al. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci USA. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Yang D, Sushchky S, Liu Z, Smith WW. Models for LRRK2-linked Parkinsonism. Parkinsons Dis. 2011;2011:942412. doi: 10.4061/2011/942412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markow TA. Perspective: female remating, operational sex ratio, and the arena of sexual selection in Drosophila species. Evolution. 2002;56:1725–1734. doi: 10.1111/j.0014-3820.2002.tb00186.x. [DOI] [PubMed] [Google Scholar]
- 29.DiAngelo JR, Birnbaum MJ. Regulation of fat cell mass by insulin in Drosophila melanogaster. Mol Cell Biol. 2009;29:6341–6352. doi: 10.1128/MCB.00675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Csaki LS, Reue K. Lipins: multifunctional lipid metabolism proteins. Annu Rev Nutr. 2010;30:257–272. doi: 10.1146/annurev.nutr.012809.104729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris TE, Finck BN. Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol Metab. 2011;22:226–233. doi: 10.1016/j.tem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reue K. The lipin family: mutations and metabolism. Curr Opin Lipidol. 2009;20:165–170. doi: 10.1097/MOL.0b013e32832adee5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ugrankar R, Liu Y, Provaznik J, Schmitt S, Lehmann M. Lipin is a central regulator of adipose tissue development and function in Drosophila melanogaster. Mol Cell Biol. 2011;31:1646–1656. doi: 10.1128/MCB.01335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ku¨hnlein RP. Energy homeostasis regulation in Drosophila: a lipocentric perspective. In: Meyerhof W, Beisiegel U, Joost HG, editors. Sensory and Metabolic Control of Energy Balance. Berlin, Germany: Springer Berlin Heidelberg; 2010. pp. 159–173. [Google Scholar]
- 35.Goodman JM. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J Lipid Res. 2009;50:2148–2156. doi: 10.1194/jlr.R001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beller M, Thiel K, Thul PJ, Jackle H. Lipid droplets: a dynamic organelle moves into focus. FEBS Lett. 2010;584:2176–2182. doi: 10.1016/j.febslet.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 37.Harbison ST, Chang S, Kamdar KP, Mackay TF. Quantitative genomics of starvation stress resistance in Drosophila. Genome Biol. 2005;6:R36. doi: 10.1186/gb-2005-6-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reue K, Brindley DN. Thematic review series: glycerolipids. multiple roles for lipins/phosphatidate phosphatase enzymes in lipid metabolism. J Lipid Res. 2008;49:2493–2503. doi: 10.1194/jlr.R800019-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]