Abstract
In spite of our increased understanding of how genomes are dysregulated in cancer and a plethora of molecular diagnostic tools, the front line and ‘gold standard’ detection of cancer remains the pathologist’s detection of gross changes in cellular and tissue structure, most strikingly nuclear dis-organization. In fact, for over 140 years it has been noted that nuclear morphology is often disrupted in cancer. Even today, nuclear morphology measures include nuclear size, shape, DNA content (ploidy) and ‘chromatin organization’. Given the importance of nuclear shape to diagnoses of cancer phenotypes, it is surprising and frustrating that we currently lack a detailed understanding to explain these changes and how they might arise and relate to molecular events in the cell. It is an implicit hypothesis that perturbation of chromatin and epigenetic signatures may lead to alterations in nuclear structure (or vice versa) and that these perturbations lie at the heart of cancer genesis. In this review, we attempt to synthesize research leading to our current understanding on how chromatin interactions at the nuclear lamina, epigenetic modulation and gene regulation may intersect in cancer and offer a perspective on critical experiments that would help clarify how nuclear architecture may contribute to the cancerous phenotype. We also discuss the historical understanding of nuclear structure in normal cells and as a diagnostic in cancer.
Keywords: Chromatin, Chromosome, Nuclear lamina, Histone, DNA methylation, Lamina Associated Domain, Epigenetics, Fluorescence in situ hybridization (FISH), Hi-C, DamID, ChIP, Cancer, Development
The earliest described genetic abnormality in cancer was abnormal chromatin, described by Teodor Boveri in 1914 [1]. While he emphasized that chromosomes were altered in mitosis and number, for which he is called the father of cancer genetics, his book also refers to a fuzziness of chromatin. One can see this for oneself in his microscopic plates archived at University of Wurzburg. In fairness, he did not mean what we do in describing chromatin, which was described first by Heintz in the 1920s, yet hints of modern chromatin biology go back a century [2].
As indicated above, one of the most noted changes in cancer cells is abnormal nuclear morphology. In fact, the specific morphological changes displayed by nuclei are often used by pathologists to grade and specify cancer type and stage. The hallmark of cancer is genome dysregulation and, of course, genes can become activated or repressed by a variety of mechanisms involving both local and global changes. However, it remains unclear how genome dysregulation and perturbations in nuclear architecture, both so evidently displayed in cancer, are related. One can think of gene regulation as occurring at different levels. The first level of gene activation/repression occurs when a specific transcriptional regulator becomes available at a discreet developmental time point or via signaling in response to stimuli (hormones, nutrients, ligands, etc.). Another layer of regulation occurs at the level of local chromatin modulation in a process that promotes (or prevents) recruitment of transcription factors/complexes to cis elements within a particular locus or regulatory element. The specific local chromatin environment is a consequence of altering the post-translational modifications of histone tails, DNA methylation patterns and/or nucleosome positioning. In cancerous cells, these local chromatin modifications, methylation signatures and gene expression profiles are perturbed and the intersection of these processes in understanding cancer phenotypes is a continuing area of robust investigation. However, the role that overall three dimensional nuclear structure plays in the disease process is not well understood. This ‘higher order’ level of chromatin regulation occurs at a more global level, involving changes in nuclear localization, associations or larger chromatin regions with repressive compartments, such as the nuclear periphery or pericentromeric heterochromatin, and large-scale changes in DNA structure, such as the formation of DNA loops and/or locus contraction. Most studies to date on the role that nuclear structure plays in gene regulation have been carried out in developmental systems or specific disease models, such as Hutchinson–Gilford Progeria (HGPS) early aging syndrome, that have a clear link to disruption of nuclear morphology by a mutation in a site protein coding gene (e.g. Lamin A in HGPS).
Despite the extraordinarily long history of microscopic evidence linking abnormal large-scale chromatin structure to cancer, remarkably little has been done toward understanding the molecular basis of this relationship. The reasons such large-scale molecular chromatin analyses have not been fully applied to the study of cancer are two-fold: (1) it is unclear what the key component(s) are involved the loss of genome structure and (2) the aneuploidy present in cancerous cells is problematic in dissecting the role higher order chromatin structure and scaffolding plays in gene regulation and onset of disease. In this review, we attempt to synthesize research leading to our current understanding on how chromatin interactions at the nuclear lamina, epigenetic modulation and gene regulation may intersect in cancer and offer a perspective on critical experiments that would help clarify how nuclear architecture may contribute to the cancerous phenotype. We discuss the historical understanding of nuclear structure in normal cells and as a diagnostic in cancer, our understanding of epigenetic perturbation in cancer and, finally, how nuclear structure and epigenetics of cancer may be related.
1. Historical perspective of nuclear structure and pathology
The eukaryotic nucleus is now recognized as a highly organized and orchestrated organelle and this structural framework is quite often disrupted cancerous cells. In fact, this disruption is a common diagnostic tool used by pathologists in identifying cancerous cells in an otherwise normal cell population [3]. While much progress has been made in the past few decades on the gene regulatory networks, epigenetic modifications and signaling pathways perturbed by or leading to cancerous phenotypes, less progress has been made in determining the role that nuclear architecture plays in the neoplastic and disease process.
That chromatin is organized in the nucleus is not a new idea. While Carl Rabl (1853–1917) was the first to propose the seminal concept of higher order chromosomal organization (Rabl configuration of chromosomes), Theodor Boveri (1862–1915) was the first to use the term “chromosome territory” (CT). In his 1909 publication, Boveri described chromatin movements and organization in three observational hypotheses [4,5]: first, chromosome territory (CT) arrangements are stably maintained during interphase. Second, this stability is lost during prometaphase and there are greater movements of CTs. Finally, while mother and daughter nuclei do not share similar CT proximity patterns, the daughter nuclei do exhibit symmetry with each other and the general radial CT positioning between mother/daughter nuclei is maintained. Although Boveri was able to make great strides in understanding of nuclear dynamics (although the meaning of chromatin itself came later with Heintz), he was reliant on fixed materials and inferior microscopic instrumentation. The most compelling evidence for CTs did not arrive until the 1970s and 1980s, when the ability to study individual chromosomes and loci in the intact nucleus became possible. These studies confirmed much of Boveri’s findings, including the idea of CTs and general nuclear organization. Functional assays of nuclear architecture have only occurred in the past decade or two, described in more detail later in this review [4].
In spite of our increased understanding of how genomes are dysregulated in cancer and a plethora of molecular diagnostic tools, the front line and ‘gold standard’ detection of cancer remains the pathologist’s detection of gross changes in cellular and tissue structure, most strikingly nuclear dis-organization [3,6,7]. In fact, for over 140 years it has been noted that nuclear morphology is often disrupted in cancer. In the 1860s, Lionel S. Beale of King’s College Hospital examined unstained sputum from a patient with cancer of the pharynx and observed nuclear morphology variations in the cancerous cells [8]. George Papanicolaou developed a stain that enables visualization of many cytoplasmic and nuclear structural features of cells in the 1930s, and applied the stain to cervical cells to test for cancer – the so-called ‘Pap test’ [9]. Even today, nuclear morphology measures include nuclear size, shape, DNA content (ploidy) and ‘chromatin organization’. Given the importance of nuclear shape to diagnoses of cancer phenotypes, it is surprising and frustrating that we currently lack a detailed understanding to explain these changes and how they might arise and relate to molecular events in the cell. Of course, nuclear size and shape is determined by the dynamic nucleoskeleton components and interacting chromatin and RNA. As such, it is an implicit hypothesis that perturbation of chromatin and epigenetic signatures may lead to alterations in nuclear structure (or vice versa) and that these perturbations lie at the heart of cancer genesis.
2. Nuclear scaffolding at the INM – form and function
At the protein level, the nuclear periphery in mammalian cells is comprised of a unique set of inner nuclear membrane (INM) proteins and the nuclear lamins [10,11]. Mammalian INM proteins include lamin B receptor (LBR), lamina associated peptide 2 (Lap2) and Emerin (among many others). These and other INM proteins, including proteins extending into the cytoplasm (e.g. the SUN domain proteins) interact with the nuclear lamina, which is made up of a filamentous meshwork of proteins: lamins A/C and B. Many of the INM proteins (e.g. LBR, Emerin, and Lap2) are able to interact with transcriptional repressors as well as signaling molecules. LBR has been shown to complex with heterochromatin protein 1α (HP1α) and nucleosomes through the core histones H3 and H4. Emerin and Lap2β, which both contain a LEM (Lap2β, Emerin, MAN1) domain, interact with Barrier to Auto-integration Factor (BAF), germ-cell-less (GCL), retinoblastoma protein (Rb) and HDAC3 [12–17]. Finally, the lamins themselves have been implicated in interactions with chromatin. Given the sub-localization of these scaffolding/regulatory proteins in the nuclear volume, much recent work has focused on the role these proteins play in gene regulation.
Chromatin itself is organized into structural domains likely by association with distinct nuclear compartments enriched in regulatory or structural proteins, such as the INM/lamina proteins described above [11,18–23]. Growing evidence suggests that gene activity is modulated by interactions with these sub-nuclear compartments. For instance, late replicating genes and gene-poor chromosomes tend to be located at the nuclear periphery, while early replicating genes and gene-rich chromosomes are more centrally disposed, suggesting that many inactive genes are located at the periphery of the nucleus [24,25]. However, the nuclear periphery has been shown to function in both gene silencing and activation [19,26]. These studies, while quite informative, provide only circumstantial evidence into the potential function of nuclear structure on genome function.
In order to describe more functional assays, let’s focus on one example of a well-studied locus that undergoes changes in nuclear scaffolding and positioning, the Immunoglobulin Heavy chain locus (IgH). It has been shown in murine cells, using 3D-ImmunoFISH, that germ-line (not recombined) immunoglobulin heavy chain loci (IgH) are preferentially localized to the nuclear lamina in hematopoietic progenitors, T lineage cells and non-B cells (such as fibroblasts) but centrally positioned in pro-B cells, where they are active [27–30] (for a graphical description of techniques used in nuclear structure analyses, see Fig. 1). Additional studies have correlated the transcriptional activation of many other mammalian genes with their repositioning away from the nuclear periphery, including muscle specific genes and adipose genes, leading to the hypothesis that the nuclear periphery may be a repressive compartment [31–34]. Intriguingly, it was also demonstrated that localization of the IgH locus to the nuclear periphery is not just cytological, but reflects real molecular contact over a large region with components of the INM/lamina compartment in a Lamin Associated Domain (LAD – see below) [35,36]. Upon locus activation, using chromosome conformation capture (3C) analyses, the IgH locus has been demonstrated to undergo long range compaction dependent upon the scaffolding and regulatory protein CTCF [37]. More recently, using Hi-C (a newer chromosome conformation capture technique) followed by deep sequencing in lymphoid progenitors and pro-B cells, has shown differential interactome map of the IgH locus and other developmentally regulated loci during B cell development [38]. Most intriguingly, the IgH locus has been shown to be in close three dimensional proximity to a common lymphomic translocation partner, Myc, at dynamically regulated transcription factory – again, when it is not scaffolded to the INM/lamina [39]. Thus, the IgH locus provides insight into how a developmentally regulated locus changes its nuclear positioning concomitant with changes in its transcriptional activity and proclivity to translocation.
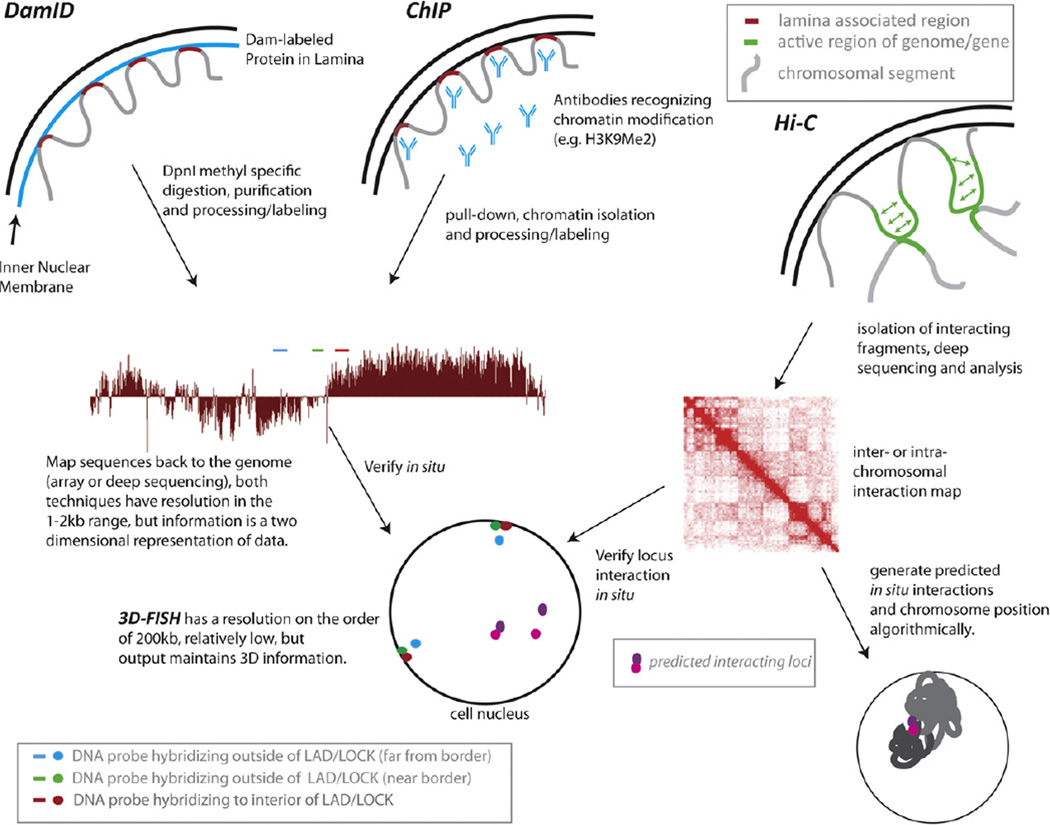
Fig. 1.
Assays used in the study of nuclear architecture. Four types of assays and how they intersect are depicted in the figure above: 3D-FISH, DamID, ChIP and Hi-C. Three dimensional fluorescence in situ hybridization (3D-FISH) has been used for many years to detect the relative positioning of chromosomes and/or specific loci in a single cell nucleus. The advantage of this technique is that relative 3D position is maintained and it is in a single cell. However, the technique is limited by both its sensitivity (relatively large probes are commonly used) and its resolution, which is dictated by microscopic limitations. On a wide-filed microscope, the limit of resolution is 200 nm, which translates to hundreds of kilobases. More recent genome-wide associations with nuclear compartments (specifically, the nuclear lamina) have been detected using DNA Adenine Methyltransferase Identification (DamID). The Dam enzyme from bacteria is fused in frame with a protein of interest (e.g. a nuclear lamina protein, indicated in light blue in nucleus at left) and any chromatin that comes in contact with this protein will be methylated on “A” residues, enabling identification of the associated regions. A methyl specific restriction enzyme DpnI which recognizes GAmeTC or followed by ligation mediated PCR enriches for the associated sequences (shown in red, associated with the nuclear lamina). The resulting DNA can be processed wither for tiled array or deep sequencing. Similarly, recently identified chromatin domains that are implicated in chromatin scaffolding, such as H3K9Me2 LOCKs, can be isolated using chromatin immunoprecipitation (ChIP). Specific antibodies (indicated in light blue, middle nucleus) to the chromatin modification are used to pull down either cross-linked or native chromatin and the associated DNA is isolated and processed for either tiled array or deep sequencing analyses. For both DamID and structural domain ChIP, it is customary to verify several regions of interaction using 3D-FISH to support claims of inferred nuclear structure. As indicated, regions that are not associated with a LAD by DamID (molecular resolution), may cytologically appear to be associated with the nuclear periphery (green probe) due to the inherent resolution of the FISH technique. Hybridization of probes that are interior to a LAD or far away from a LAD border (red and blue, respectively) accurately reflects the molecular nuclear associations. More recently, Hi-C, a derivative of chromosome conformation capture, has been used to identify interacting regions of the genome irrespective of a priori knowledge of protein players or nuclear sub-compartment. In this assay, the genome is crosslinked, digested with a restriction enzyme and subjected to self-ligation. The protocol and subsequent analysis allows for detection of interacting regions – both inter- and intra-chromosomal. The technique is somewhat limited in resolution (tens to hundreds of kilobases), but has the advantage of uncovering genome-wide associations. Perhaps more intriguingly, the technique allows for modeling of the interactome and chromosomal positioning for the entire genome. Hi-C and its predecessors have mostly been used to identify disparate loci that interact with one another with a frequency greater than expected by random chance. Again, these locus specific interactions are usually verified using 3D-FISH analysis and can reveal mono-allelic interactions (purple and pink probes).
To get at general functionality of a specific nuclear subcompartment, several groups recruited genes ectopically to the nuclear periphery and demonstrated that, at least for some DNA segments, recruitment to the nuclear periphery leads to gene repression [35,40,41]. These studies were done in both murine and human cell lines. In both cases, this repression was accompanied by loss of histone H4 acetylation and accumulation of histone H3K27 methylation. Intriguingly, the repression observed was able to spread from the original attachment site to adjacent endogenous loci. Thus, association of a gene with the nuclear periphery can lead to repression of that locus and affect nearby genomic regions. In addition to these artificial systems, a few groups have begun to utilize a technique called DAM-ID to probe genome-wide contacts between DNA and the nuclear lamina (Fig. 1). Again, using the IgH locus as a model, the association with the INM/lamina is robust in non-B lineage cells and occurs over the entire 3 Mb of the murine locus. Additional studies in both mouse and human cells have demonstrated that, on a genome-wide level, these Lamina Associated Domains (LADs) are on the order of 100 Kb to a few megabases in size and comprise mostly inactive chromatin [35,36,42–44]. Interestingly, many developmentally regulated genes reside in LADs and have been demonstrated to dissociate from LADs just prior to or upon activation (unpublished data and [36]). A recent study postulates that the association of chromatin with the nuclear lamina can be either constitutive (cLADs) across cell types, or facultative and dynamic (fLADs) [45]. This is in agreement with previous reports and our own unpublished data. Most intriguingly, LAD domains have been correlated histone H3K9 di-methylation, a facultative heterochromatin mark and DNA methylation BLOCKs (see below). Again, the IgH locus (as our ‘test case’ representative) also undergoes changes in DNA methylation and H3K9-di-methylation, in agreement with the genome-wide data trends. In addition, H3K27 tri-methylation appears to be increased at the borders of the LADs, indicating a potential role for this modification on the establishment or maintenance of the boundary regions. The boundary regions of Lamina Associated Domains have been bioinformatically assessed to be enriched for CTCF binding sites and to be flanked by active gene promoters. It is unclear, however, what and how exactly these boundaries are established and maintained, as CTCF binding by ChIP-seq analyses do not show a strong border enrichment, and indeed, note CTCF binding sites internal to LADs [44]. Finally, using genome-wide chromosome conformation capture techniques, the regions associated with the nuclear lamina, which appear to be comprised of repressed or inactive regions of the genome, are remarkably absent from the interactome maps generated by genome-wide chromosome conformation capture studies (5C and Hi-C), indicating that one function of scaffolding at the INM/lamina is to prevent inter- and intra-chromosomal interactions (such as IgH-Myc, described above) at the lamina associated domains [46,47]
3. Lessons from studies on aging
In addition to potentially regulating normal developmental processes, one of the most well studied functions of nuclear lamina is the role LMNA plays in aging/senescence. At the molecular level, aging can be characterized by global changes in chromatin structure, organization and function. In aged cells several key chromatin proteins, including the higher-order structural heterochromatin protein HP1, are lost [48–50]. Similarly, in the premature aging disorder Hutchinson-Gilford Progeria Syndrome (HGPS), caused by a cryptic splice site mutation in lamin A (progerin) which prevents its proper processing and removal of a c-terminal farnesyl group, the same chromatin proteins are also lost. In fact, in HGPS patient cells, all peripheral heterochromatin is lost [51]. In normally aged cells, a similar non-processed variant of Lamin A (pre-Lamin A) accumulates and likely leads to similar consequences as progerin [52]. Aberrant forms of LMNA likely bring about structural changes which would increase the susceptibility of the genome to damage leading either to senescence, apoptosis or cancer [53,54]. This is supported by findings that implicate the A-type lamins in cell cycle regulation, DNA Damage Response and control of tumor suppressor genes such as Rb (retinoblastoma) and BRCA1 (gonzalo). In addition, physiologically aged and prematurely aged genomes also undergo global changes in epigenetic modifications. Global DNA methylation is reduced in many aged mammalian tissues and several histone modifications are affected during aging [55,56]. In agreement with the observed morphological changes in chromatin (i.e. peripheral heterochromatin loss), the heterochromatin-associated trimethylation of histone H3K9 and histone H3K27 are also abrogated in aged and prematurely aged cells [57]. Interestingly, Histone H3K27 methylation (a facultative heterochromatin mark) is also observed at the edges of Lamina Associated Domains and has been postulated to be important for maintaining or establishing the boundaries of these domains (see preliminary data and [42]). Given these extensive changes in histone modifications, not surprisingly, aged cells show dramatic and global misregulation of gene expression. However, it remains unclear how nuclear architecture and compartmentalization of blocks of chromatin at the INM changes during the aging or cancer processes. Given the importance of LMNA in maintaining nuclear morphology and the role the INM likely plays in scaffolding and regulating large chromatin domains, this is a large hole in our knowledge.
Similar to the aging process, cancer is associated with global changes in the epigenome as well as the transcriptome [55,56]. One can think of both aging and cancer as diseases of development [55]. Generally speaking, methylation patterns are dysregulated in cancer as are other chromatin modifications [58]. It has been known for some time that Lamin A is not present in some hematological malignancies and it is now understood that the promoter region of the LMNA/C gene is hypermethylated in these diseases [16,59]. More recently, while normal B and T cells express Lamin A, leukemia and lymphoma cells do not nor do some normal progenitor cells or early B/T cell intermediaries [58]. Moreover, risk of recurrent colon cancer has been found to be associated with loss of LMNA [60]. Loss of LMNA could simply reflect a less differentiated phenotype, as embryonic stem cells and some hematopoietic precursors do not express lamin A. Alternatively, loss of this key regulator of tumor suppressor pathways and chromatin organization (both scaffolding and chromatin modifications) could itself contribute to the neoplastic process. Interestingly, loss of LMNA in mouse cell lines leads to loss of heterochromatin and irregularly shaped nuclei, similar to the progerin phenotype. Moreover, in leukemia and lymphoma cells it has recently been reported that there is dysregulation of histone H3K27 methylation. This chromatin mark and the modifier that introduces it, EZH2, is necessary for proper B cell development and, as mentioned above, has been found to be enriched in the border regions of the Lamina Associated Domains. Interestingly, this heterochromatic mark is lost in HGPS as well as normal physiological aging. In addition, histone H3K9 methylation (di-, tri-) also appears to be perturbed on a genome-wide basis in some leukemias [61]. Thus, aging and cancer, especially leukemia and lymphoma, appear to share many common features of a perturbed nuclear architecture.
4. Methylation blocks and chromatin LOCKs in cancer
Several groups have approached higher order chromatin organization from the perspective of large regions of posttranslational histone modification, particularly affecting lysine residues on histone tails, or LOCKs, for Large Organized Chromatin K-Modifications. These were first observed at H3K9me2 over ~1/3 of the genome. Although most of these regions seem identical across tissues, some show qualitative and/or quantitative differences among tissue types [62]. In contrast to the LADs, their magnitude also appears to increase between stem cells and somatic cells [62]. In addition, like LADs, there is a clear correlation between H3K9me2 and repression and loss of LOCKs over developmentally regulated genes upon differentiation. More recently, these large LOCK domains were investigated more closely and small ‘euchromatin islands’ over a subset of promoters and transcription start sites were identified [63]. It is tempting to speculate that such euchromatin islands may contribute to poise some loci for transcription.
Bing Ren’s laboratory described large blocks of H3K9Me3 and H3K27Me3 that expand in human lung fibroblasts compared with human embryonic stem cells [64]. The same group found large H3K9me3 and H4K20me3 blocks at the olfactory gene receptor clusters that were tissue specific, enriched in mouse olfactory epithelium but not in liver [65]. All of these regions roughly correspond to LADs described above. However, there is considerable nuance that may be critical for gene regulation, as the dynamics of variation in development have received less attention than the existence of these regions, per se. It is also not clear why different laboratories detect LOCKs with different antibodies. There may be overlap in domains, or the antibodies may suffer from incomplete specificity. However there is little doubt that such regions exist. It is also important to bear in mind that not all tissues show significant LOCKs for H3K9Me2, e.g. brain cells [62,66].
A most intriguing discovery in the last year is the connection between DNA methylation and histone methylation in large regions corresponding to LOCKs and LADs. Hansen et al. [67] performed whole genome bisulfite sequencing of three colorectal adenocarcinomas and three normal mucosal samples matched to the same patients. Approximately one-third of the genome showed substantial DNA demethylation of large genomic regions, tens to hundreds of megabases in size. These regions contained one-third of single copy genes, and they corresponded to LOCKs and LADs, establishing a link between altered DNA methylation and large-scale chromatin decondensation in cancer. Some genes within these regions were also activated functionally in the tumors, and they showed hypervariability of expression. These included several matrix metalloproteinases, which are important in tumor invasion and metastasis, and this hypervariability could provide a mechanism for the tumor cell heterogeneity that is the ultimate reason for drug resistance and tumor progression [67]. A subsequent study confirmed the observation of domain hypomethylation linked to LADs/LOCKs [68]
At present, comparatively little analysis has been performed to compare LOCKs or LADS between cancer and normal cells. The first report of LOCKs described their loss in several tumor cell lines [62]. Recently, McDonald et al. [69] examined both DNA methylation and LOCKs in mouse hepatocytes induced by TGF-β to undergo epithelial-mesenchymal transition [69]. While DNA methylation did not change on short term exposure, LOCKs were reduced quantitatively and globally, and the heterochromatin was restored 24 h after TGF-β withdrawal. Inhibition of demethylation using siRNA or chemical inhibitors of lysine demethylation abrogated downstream effects of EMT including chemoresistance and increased cell motility [69]. These data suggest that LOCKs/LADs/blocks mediate cellular plasticity during normal development or injury repair, and that the same process is hijacked in cancer leading to increased tumor cell plasticity (Fig. 2).
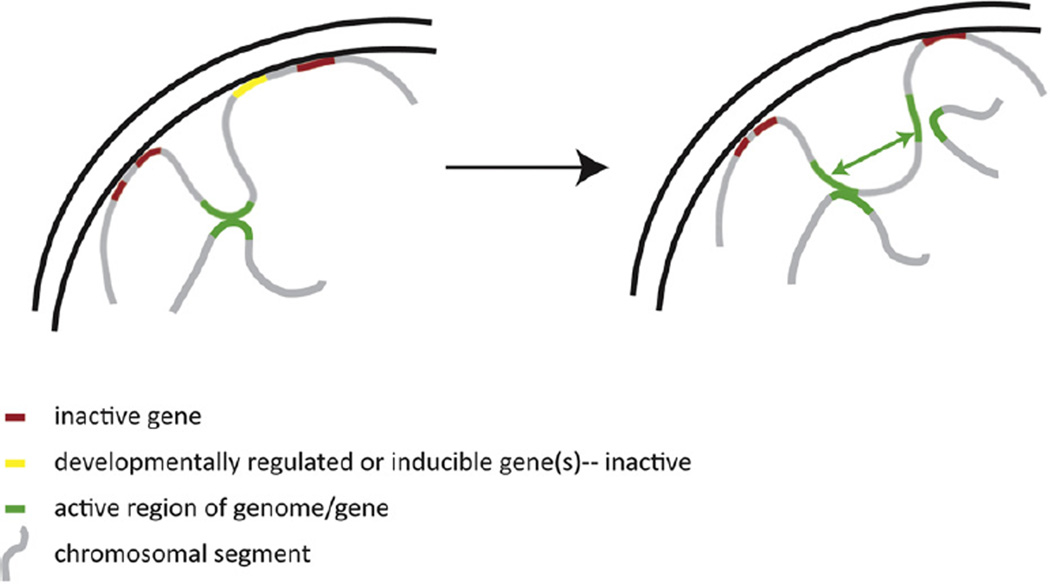
Fig. 2.
LOCKs/LADs potentially regulate gene activation and genome interactions. Depicted above are two cellular states in which a developmentally regulated gene (yellow) is scaffolded at the nuclear lamina when it is transcriptionally silent. Recent evidence has correlated movement away from the nuclear lamina and loss of H3K9Me2 with poising a gene for activation. In this model, not only does the structural association with the nuclear lamina (LAD organization) change, but so too the interaction with other genomic loci (green arrows). Recent evidence has demonstrated that the majority of genome-wide inter-chromosomal interactions are between ‘active’ regions of the genome (green). Thus, in this model, reorganization at the nuclear lamina and LOCKs are concomitant with changes in the nuclear interactome and possibly inter-dependent. These correlated changes have been demonstrated to some degree in developmental systems. We posit that, in cancerous cells, perturbations in nuclear morphology, architecture and chromatin state (both LOCKs and blocks) likely reflect real differences in not only scaffolding, but also in genome wide interactions and coordinated gene regulation (or dysregulation).
5. Future directions
Currently, very few analyses probing aberrations in nuclear scaffolding and interactions at the nuclear lamina have been undertaken in normal versus cancerous cells. Given the obvious nuclear structure abnormalities present in cancer, this, at first glance, seems astonishing. However, genome-wide three-dimensional assays, such as Dam-ID and 5C/Hi-C are still relatively new and the analyses tricky, dependent upon relatively large numbers of cells and reliant on mapping back to the reference genome. Aneuploidy and translocations (known and unknown) are likely to confound such studies. However, much is likely to be gleaned by studying ‘well-behaved’ and well documented cancer cell lines for these types of studies and they should be undertaken. Analysis of LOCKS (H3K9 di-methylation) patterns in such cells will also potentially yield insight into perturbations of nuclear architecture; however, we still do not know if these mostly overlapping features (LADs and LOCKS) are interdependent or if they can be uncoupled. One recent study in C. elegans indicates that localization to the nuclear periphery is dependent upon H3K9 di- and tri-methylation, although this is apparently not the case in murine embryonic stem cells or in certain regions of the mammalian genome [66,70]. Studies into the mechanistic interdependence of these features are, therefore, paramount. In addition, studies investigating the actual genome-wide overlap between LADs and LOCKS in the same cell need to be undertaken. Of equal importance to the epigenetic and genome-wide studies, are avenues of research focused on identifying proteins required for establishing or maintaining associations at the INM. Finally, more targeted and more detailed time and/or cell-type resolved analyses of individual chromosomal domains, as exemplified by studies on the Immunoglobulin Heavy Chain locus, will continue to be key to our understanding of what the functional consequences are of perturbing nuclear architecture.
References
- 1.Boveri T. Zur Frage der Entstehung maligner Tumoren. G. Fischer. 1914. [Google Scholar]
- 2.Heitz E. Heterochromatin, chromocentren, chromomeren. Berichte der Deutschen Botanischen Gesellschaft. 1929;47:274–284. [Google Scholar]
- 3.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nature Reviews Cancer. 2004;4(9):677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 4.Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950 to 1980. Part III. Chromosome territories and the functional nuclear architecture: experiments and m. European Journal of Histochemistry. 2006;50(4):2009. [PubMed] [Google Scholar]
- 5.Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part I. The rise of chromosome territories. European Journal of Histochemistry. 2006;50(3):2009. [PubMed] [Google Scholar]
- 6.Coffey DS. Nuclear matrix proteins as proteomic markers of preneoplastic and cancer lesions: commentary re: G. Brunagel et al., nuclear matrix protein alterations associated with colon cancer metastasis to the liver. Clinical Cancer Research 2002; 8, 3039–3045. Clinical Cancer Research. 2002;8(10):3031–3033. [PubMed] [Google Scholar]
- 7.Nickerson JA, Blencowe BJ, Penman S. The architectural organization of nuclear metabolism. International Review of Cytology. 1995;162A:67–123. doi: 10.1016/s0074-7696(08)61229-2. [DOI] [PubMed] [Google Scholar]
- 8.Long SR, Cohen MB. Classics in cytology. VI: The early cytologic discoveries of Lionel S. Beale. Diagnostic Cytopathology. 1993;9(5):595–598. doi: 10.1002/dc.2840090525. [DOI] [PubMed] [Google Scholar]
- 9.Papanicolaou GN, Traut HF. The diagnostic value of vaginal smears in carcinoma of the uterus. 1941. Archives of Pathology and Laboratory Medicine. 1997;121(3):211–224. [PubMed] [Google Scholar]
- 10.Verstraeten VL, et al. The nuclear envelope: a key structure in cellular integrity and gene expression. Current Medicinal Chemistry. 2007;14(11):1231–1248. doi: 10.2174/092986707780598032. [DOI] [PubMed] [Google Scholar]
- 11.Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nature Reviews Molecular Cell Biology. 2011;12(11):695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- 12.Kourmouli N, et al. Dynamic associations of heterochromatin protein 1 with the nuclear envelope. EMBO Journal. 2000;19(23):6558–6568. doi: 10.1093/emboj/19.23.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holaska JM, et al. Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. Journal of Biological Chemistry. 2003;278(9):6969–6975. doi: 10.1074/jbc.M208811200. [DOI] [PubMed] [Google Scholar]
- 14.Holaska JM, Wilson KL. An emerin proteome: purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry. 2007;46(30):8897–8908. doi: 10.1021/bi602636m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cramer CL, et al. Bioproduction of human enzymes in transgenic tobacco. Annals of the New York Academy of Sciences. 1996;792:62–71. doi: 10.1111/j.1749-6632.1996.tb32492.x. [DOI] [PubMed] [Google Scholar]
- 16.Bjornsson HT, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA: the Journal of the American Medical Association. 2008;299(24):2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somech R, et al. Nuclear envelopathies – raising the nuclear veil. Pediatric Research. 2005;57(5 Pt 2):8R–15R. doi: 10.1203/01.PDR.0000159566.54287.6C. [DOI] [PubMed] [Google Scholar]
- 18.Dernburg AF, Misteli T. Nuclear architecture – an island no more. Developmental Cell. 2007;12(3):329–334. doi: 10.1016/j.devcel.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 19.Taddei A. Active genes at the nuclear pore complex. Current Opinion in Cell Biology. 2007;19(3):305–310. doi: 10.1016/j.ceb.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Albiez H, et al. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Research. 2006;14(7):707–733. doi: 10.1007/s10577-006-1086-x. [DOI] [PubMed] [Google Scholar]
- 21.Woodcock CL. Chromatin architecture. Current Opinion in Structural Biology. 2006;16(2):213–220. doi: 10.1016/j.sbi.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert N, Gilchrist S, Bickmore WA. Chromatin organization in the mammalian nucleus. International Review of Cytology. 2005;242:283–336. doi: 10.1016/S0074-7696(04)42007-5. [DOI] [PubMed] [Google Scholar]
- 23.Boggs BA, et al. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nature Genetics. 2002;30(1):73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- 24.Cremer T, et al. Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Critical Reviews in Eukaryotic Gene Expression. 2000;10(2):179–212. [PubMed] [Google Scholar]
- 25.Croft JA, et al. Differences in the localization and morphology of chromosomes in the human nucleus. Journal of Cell Biology. 1999;145(6):1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bystricky K, et al. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(47):16495–16500. doi: 10.1073/pnas.0402766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosak ST, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296(5565):158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 28.Bertolino E, et al. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nature Immunology. 2005;6(8):836–843. doi: 10.1038/ni1226. [DOI] [PubMed] [Google Scholar]
- 29.Prado GN, et al. Mechanisms regulating the expression: self-maintenance, and signaling-function of the bradykinin B2 and B1 receptors. Journal of Cellular Physiology. 2002;193(3):275–286. doi: 10.1002/jcp.10175. [DOI] [PubMed] [Google Scholar]
- 30.Chen D, et al. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. Journal of Cell Biology. 2005;168(1):41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szczerbal I, Foster HA, Bridger JM. The spatial repositioning of adipogenesis genes is correlated with their expression status in a porcine mesenchymal stem cell adipogenesis model system. Chromosoma. 2009;118(5):647–663. doi: 10.1007/s00412-009-0225-5. [DOI] [PubMed] [Google Scholar]
- 32.Johnson AJ, et al. Characterization of the TCL-1 transgenic mouse as a preclinical drug development tool for human chronic lymphocytic leukemia. Blood. 2006;108(4):1334–1338. doi: 10.1182/blood-2005-12-011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zink D, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. Journal of Cell Biology. 2004;166(6):815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meister P, et al. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes and Development. 2010;24(8):766–782. doi: 10.1101/gad.559610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy KL, et al. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452(7184):243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 36.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Molecular Cell. 2010;38(4):603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Degner SC, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(23):9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin YC, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nature Immunology. 2012;13(12):1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osborne CS, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biology. 2007;5(8):e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlimgen RJ, et al. Initiation of allelic exclusion by stochastic interaction of Tcrb alleles with repressive nuclear compartments. Nature Immunology. 2008;9(7):802–809. doi: 10.1038/ni.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finlan LE, Bickmore WA. Porin new light onto chromatin and nuclear organization. Genome Biology. 2008;9(5):222. doi: 10.1186/gb-2008-9-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453(7197):948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 43.Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein–DNA interactions using DamID in mammalian cells. Nature Protocols. 2007;2(6):1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 44.Zullo JM, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149(7):1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 45.Meuleman W, et al. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Research. 2012 doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naumova N, Dekker J. Integrating one-dimensional and three-dimensional maps of genomes. Journal of Cell Science. 2010;123(12):1979–1988. doi: 10.1242/jcs.051631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Misteli T. Higher order genome organization in human disease. Cold Spring Harbor Symposia on Perspective Biology. 2010;2(8):a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pegoraro G, et al. Ageing-related chromatin defects through loss of the NURD complex. Nature Cell Biology. 2009;11(10):1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boban M, et al. Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors. Journal of Cell Biology. 2006;173(5):695–707. doi: 10.1083/jcb.200601011. http://dx.doi.org/10.1083/jcb.200601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldman RD, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(24):8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maraldi NM, et al. Nuclear envelope proteins and chromatin arrangement: a pathogenic mechanism for laminopathies. European Journal of Histochemistry. 2006;50(1):1–8. [PubMed] [Google Scholar]
- 53.Redwood AB, Gonzalez-Suarez I, Gonzalo S. Regulating the levels of key factors in cell cycle and DNA repair: new pathways revealed by lamins. Cell Cycle. 2011;10(21):3652–3657. doi: 10.4161/cc.10.21.18201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gonzalez-Suarez I, Gonzalo S. Nurturing the genome: A-type lamins preserve genomic stability. Nucleus. 2010;1(2):129–135. doi: 10.4161/nucl.1.2.10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilbert SF. Ageing and cancer as diseases of epigenesis. Journal of Bioscience. 2009;34(4):601–604. doi: 10.1007/s12038-009-0077-4. [DOI] [PubMed] [Google Scholar]
- 56.Espada J, et al. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Research. 2007;35(7):2191–2198. doi: 10.1093/nar/gkm118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dahl KN, et al. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson–Gilford progeria syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(27):10271–10276. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Agrelo R, et al. Inactivation of the lamin A/C gene by CpG island promoter hypermethylation in hematologic malignancies: and its association with poor survival in nodal diffuse large B-cell lymphoma. Journal of Clinical Oncology. 2005;23(17):3940–3947. doi: 10.1200/JCO.2005.11.650. [DOI] [PubMed] [Google Scholar]
- 59.Stadelmann B, et al. Repression of nuclear lamin A and C gene expression in human acute lymphoblastic leukemia and non-Hodgkin’s lymphoma cells. Leukemia Research. 1990;14(9):815–821. doi: 10.1016/0145-2126(90)90076-l. [DOI] [PubMed] [Google Scholar]
- 60.Belt EJT, et al. Loss of lamin A/C expression in stage II and III colon cancer is associated with disease recurrence. European Journal of Cancer. 2011;47(12):1837–1845. doi: 10.1016/j.ejca.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 61.Rickman DS, et al. Oncogene-mediated alterations in chromatin conformation. Proceedings of the National Academy of Sciences. 2012;109(23):9083–9088. doi: 10.1073/pnas.1112570109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Filion GJ, van Steensel B. Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells. Nature Genetics. 2010;42(1):4. doi: 10.1038/ng0110-4. [author reply 5–6]. [DOI] [PubMed] [Google Scholar]
- 63.Wen B, et al. Euchromatin islands in large heterochromatin domains are enriched for CTCF binding and differentially DNA-methylated regions. BMC Genomics. 2012;13:566. doi: 10.1186/1471-2164-13-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hawkins RD, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6(5):479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lienert F, et al. Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS Genetics. 2011;7(6):e1002090. doi: 10.1371/journal.pgen.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nature Genetics. 2011;43(8):768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berman BP, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nature Genetics. 2012;44(1):40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McDonald OG, et al. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nature Structural & Molecular Biology. 2011;18(8):867–874. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Towbin Benjamin D, et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150(5):934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]