Abstract
How neuroinflammatory activities affect signaling pathways leading to blood-brain barrier (BBB) injury during HIV/AIDS are currently unknown. Our previous work demonstrated that HIV-1 exposure activates pro-inflammatory genes in human brain microvascular endothelial cells (HBMEC) and showed that these genes are linked to the janus kinase (JAK)/signal transducers and activators of transcription (STAT) pathway. Here, we report that HIV-1 gp120 protein activated STAT1 and induced interleukin (IL)-6 and IL-8 secretion in HBMEC. IL-6, IL-8, and gp120 increased monocyte adhesion and migration across in vitro BBB models. The STAT1 inhibitor, fludarabine, prevented gp120-induced IL-6 and IL-8 secretion. Inhibitors of STAT1, mitogen activated protein kinase kinase (MEK) (PD98059), and phosphatidyl inositol 3 kinase (PI3K) (LY294002), blocked gp120-induced STAT1 activation and significantly diminished IL-8-, IL-6-, and gp120-induced monocyte adhesion and migration across in vitro BBB models. These data support the notion that STAT1 plays an important role in gp120-induced inflammation and BBB dysfunction associated with viral infection. Results also suggest crosstalk between STAT1, MEK, and PI3K pathways in gp120-induced BBB dysfunction. Inhibition of STAT1 activation could provide a unique therapeutic strategy to decrease neuroinflammation and BBB dysfunction in HIV/AIDS.
Keywords: HIV-1-gp120, STAT1, IL-6, IL-8, inflammation, blood-brain barrier injury
INTRODUCTION
Human immunodeficiency virus-1 (HIV-1) invades the brain in early stages of infection. Viral infection of the central nervous system (CNS) results in various clinical and pathological abnormalities, ranging from sub-clinical and mild cognitive motor deficits to dementia. Although the pathogenesis of HIV-1-associated dementia (HAD) is not well understood, analyses of the CNS and autopsies of brains from AIDS patients reveal brain atrophy, white matter gliosis, and neuronal cell loss (Ketzler et al., 1990; Price et al., 1988). These pathological abnormalities are caused by HIV-1 proteins, inflammatory and toxic factors resulting from systemic infection that cross the blood-brain barrier (BBB), or by toxic factors secreted in the brain by virus-infected cells. Structural BBB compromise is common amongst HIV-infected patients [for recent reviews, see (Banks et al., 2006; Toborek et al., 2005)] and has been documented in cellular and animal models, human clinical, and autopsy studies (Burger et al., 1997; Kanmogne et al., 2002; Kanmogne et al., 2005; Kanmogne et al., 2007). While dysfunction of the BBB is one critical feature of HIV-1 neuropathogenesis, the underlying mechanisms of BBB dysfunction and how it affects ongoing disease are incompletely understood. Through damaged BBB, free virus, activated or HIV-infected mononuclear phagocytes from the bloodstream can infiltrate the brain and spread infection to microglia and brain macrophages (Banks et al., 2006; Toborek et al., 2005). Elucidation of the signaling pathways mediating BBB compromise is important for understanding disease mechanisms and development of new therapies.
Inflammation and virus-induced cytokines and chemokines drive HIV-1 disease progression. HIV-1 infection alters cytokine secretion both in vivo and in vitro (Kedzierska and Crowe, 2001). Higher interleukin (IL)-6 and IL-8 levels are observed in HIV-1-infected humans and correlate with accelerated progression to AIDS (Breen et al., 1990; Matsumoto et al., 1993). IL-6 and IL-8 can signal through the Janus kinase (JAK)/signal transducers and activators of transcription (STAT) pathway, and exposure of human brain microvascular endothelial cells (HBMEC) to HIV-1 virions induce upregulation and activation of IL-6, IL-8, and STAT1 genes (Chaudhuri et al., 2008a; Chaudhuri et al., 2008b).
The HIV-1 envelope glycoprotein (gp) is initially synthesized as a polyprotein precursor of 160 kDa (gp160), which is subsequently cleaved into a surface exposed amino terminus subunit, gp120, and a carboxyl transmembrane subunit, gp41. To initiate viral infection, the gp120 subunit of the HIV envelope protein must first bind to the CD4 receptor or co-receptors on the cell surface; thus, gp120 protein plays an important role in viral mediated immunological response and cell injury. Cytotoxic gp120 protein is often released into the surrounding environment following cytopathic events during infection. In fact, cells infected with HIV-1 in vitro shed gp120 protein into the culture medium (Schneider et al., 1986), and gp120 has been detected in the sera and brains of HIV-1-infected patients (Jones et al., 2000; Oh et al., 1992).
In the present study, we demonstrated that HIV-1 gp120 proteins activated STAT1 and up-regulated IL-6 and IL-8 expression in primary HBMEC. This gp120-induced inflammation has functional consequences, as we demonstrated that gp120, IL-6, and IL-8 enhance adhesion and migration of monocyte across in vitro BBB models. A specific STAT1 inhibitor, fludarabine (FLUD), prevented gp120-induced IL-6 and IL-8 expression, diminished gp120-induced STAT1 activation, and diminished gp120-, IL-6- and IL-8-induced monocyte adhesion and migration across in vitro BBB models. Furthermore, specific inhibitors of mitogen activated protein kinase kinase (MEK), PD98059, and phosphatidyl inositol 3 kinase (PI3K), LY294002, blocked gp120-induced STAT1 activation, as well as gp120-, IL-6-, and IL-8-induced monocyte adhesion and transendothelial migration. These data support the notion that secreted gp120 proteins induce BBB inflammation through STAT1 and suggest a cross-talk between STAT1, MEK and PI3K pathways in gp120-induced BBB dysfunction.
MATERIALS AND METHODS
Endothelial cell culture
Primary HBMEC were isolated from the temporal cortex of brain tissue obtained during surgical removal of epileptogenic cerebral cortex in adult patients as described previously (Miller, 1992) and provided by Drs. Marlys Witte and Michael Bernas (University of Arizona, Tucson, AZ). Routine evaluation for von Willebrand factor, Ulex europeus lectin, and CD31 demonstrated that cells were > 99% pure. Freshly isolated HBMEC were seeded in the upper chamber of collagen I coated 6- or 96-well plates or 24-well tissue culture inserts (with 0.4 µm pore size) and cultured to confluence in EGM™-2 BulletKit® media (Cambrex, Walkersville, MD) supplemented with 5% fetal bovine serum. Cells at passage 1 to 4 were used in this study. All reagents were prescreened for endotoxin (<10 pg/ml, Associates of Cape Cod, Woods Hole, MA) and mycoplasma contamination (Gen-probe II, Gen-probe, San Diego, CA).
Detection of IL-6 and IL-8
HBMEC were exposed to 0.1, 1, 10, and 100 ng/ml gp120 proteins (gp120MN, ImmunoDiagnostics, Woburn, MA) for 2 to 24 hours (h). IL-6 and IL-8 levels in culture supernatant were quantified using the human IL-6 (eBioscience, San Diego, CA) and IL-8 (BD Biosciences, Franklin Lakes, NJ) ELISA kits according to the manufacturer’s instructions. IL-6 and IL-8 levels in untreated cells and cells exposed to 100 ng/ml heat-inactivated gp120 served as controls. For experiments testing the effect of STAT1 inhibition on gp120-induced cytokine expression, HBMEC were exposed to FLUD (20 µM) for 30 min before gp120 treatment.
Monocyte isolation, adhesion, and migration through an in vitro BBB model
Monocytes were obtained from HIV-1, HIV-2 and hepatitis B seronegative donor leukopaks, and separated by countercurrent centrifugal elutriation as previously described (Gendelman et al., 1988). These cells were identified as >98% pure monocytes by Wright staining and CD68 immunostaining (at 1:50 dilution, Dako, Carpentaria, CA). For monocyte adhesion, HBMEC were seeded in 96-well collagen-coated plates and cultured to confluency, and confluent cells were treated with gp120 (100 ng/ml), IL-6 (100 ng/ml), or IL-8 (100 ng/ml) for 2 to 24h. HBMEC were then rinsed to remove gp120 and cytokines and exposed for 15 min to 2.5 × 105 freshly elutriated monocytes labeled with calcein-AM (Invitrogen, Carlsbad, CA) at 5µM/1×106 cells for 45 min. After 15 min adhesion, endothelial cell monolayers were washed and the number of adherent monocytes quantified using a fluorescence plate reader (absorbance 494nm; emission, 517nm), with a standard curve derived from a serial dilution of a known number of calcein-labeled cells.
For monocyte migration, 2 × 104 HBMEC were seeded on collagen-coated FluoroBlok tinted tissue culture inserts (3 µm pore size, BD Biosciences). Because monolayers are not visible on these inserts, manual readings of transendothelial electric resistance (TEER) were taken with a voltmeter (EVOM, World Precision Instruments, Sarasota, FL) to confirm monolayer formation and confluence. Freshly elutriated monocytes were labeled with calcein-AM as detailed above and washed. Labeled-monocytes (2.5 × 105) were then placed on gp120-treated (2h or 24h treatment) and control HBMEC (upper chamber of the FluoroBlok insert) and allowed to migrate for 2h (37°C, 5% CO2). Migrated monocytes were quantified using a fluorescence plate reader (absorbance 494nm; emission, 517nm), with a standard curve derived from a serial dilution of a known number of calcein-labeled monocytes. For migration in response to cytokines, IL-8 (100 ng/ml) or IL-6 (100 ng/ml) was added to the bottom chamber of the insert immediately before migration in order to create a chemotactic gradient. For migration experiments testing the effects of STAT1, PI3K, or MEK inhibitors, HBMEC were exposed to FLUD (20 µM), LY294002 (50 µM) (both from Sigma, St Louis, MO), or PD98059 (50 µM) (Biomol, Plymouth, PA) for 30 min before migration.
Immunofluorescence microscopy
Confluent HBMEC monolayers on glass coverslips were exposed for 30 min to 2h to 100 ng/ml gp120, washed with phosphate-buffered saline (PBS) and fixed in methanol/acetone (1:1) for 20 min at –20°C. HBMEC were permeabilized with 0.1% triton X-100 and blocked for non-specific binding with 3% bovine serum albumin in PBS (10 min at 4°C). Cells were incubated with primary antibodies (1:50 dilution) for 1h at room temperature, followed by staining (1h in the dark at room temperature) with secondary antibodies coupled with Alexa-488 (1:500 dilution, Molecular Probes, Eugene, OR). Antibodies to CXCR2 and IL-6 receptor (IL6-R alpha, and gp130) were purchased respectively from R&D system (Minneapolis, MN) and Cell Signaling Technology (Danvers, MA). Stained cell monolayers were mounted in Prolong Gold (Molecular Probes) and examined using a fluorescent microscope (E800 Nikon, Melville, NY) connected to a color MagnaFire digital camera (Optronics, Goleta, CA). Microscopic images were processed with 20 iterations of 2-dimensional deconvolution at low noise level using Autoquant X software package (Media Cybernetics, Bethesda, MD).
Protein extraction and Western blot analysis
HBMEC samples were lysed in a mammalian total protein extraction reagent (M-PER®, Pierce, Rockford, IL) containing 1x Protease Inhibitor Cocktail (Sigma). Lysates were kept on ice for 20 min and then clarified by centrifugation for 30 min at 9000xg. Total protein concentration in the resulting supernatant was measured using the bicinchoninic acid assay (Pierce). Twenty-five µg of protein were fractionated in a 4–15% gradient gel and electrophoretically transferred to nitrocellulose membranes. Membranes were blocked for 1h with SuperBlock® T-20 (Pierce), blotted 2h or overnight with the primary antibodies, 1h with the secondary antibody, washed, and visualized using SuperSignal® West Pico Substrate (Pierce). For Western blot with antibodies to phospho-STAT1, phospho-STAT3, phospho-STAT5, and phospho-STAT6, membranes were stripped using Restore™ Western Blot Stripping Buffer (Pierce) and re-blotted with STAT1, STAT3, STAT5, and STAT6 antibodies, then stripped again and re-blotted with β-actin antibody. Results were expressed as ratio of relative intensity of the target protein (phospho-STAT) to that of the internal standard, β-actin, or to that of STAT. All STAT and phospho-STAT antibodies were from Cell Signaling Technology.
Statistical analysis
Statistical analysis was performed by one- or two-way analysis of variance (ANOVA) followed by Tukey’s Multiple Comparison Test, using GraphPad Prism 4. This statistical procedure tests differences among several means for significance without increasing Type I error rate (Klockars AJ, 1986). Threshold of significance level was 0.05.
RESULTS
HIV-1 gp120 proteins induce up-regulation of pro-inflammatory cytokines and chemokines in HBMEC
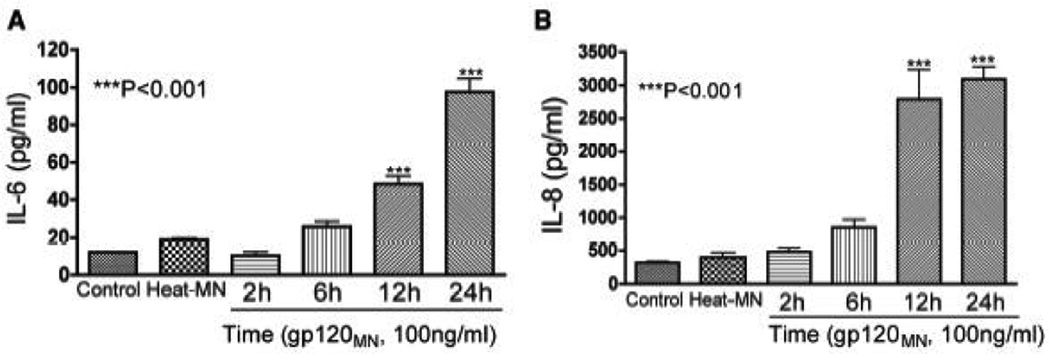
Inflammation enhanced leukocyte entry into the CNS implicates the involvement of inflammatory cytokines in neuroAIDS pathogenesis (Perrella et al., 1992). Therefore, we tested the effects of HIV-1 gp120 proteins on chemokine and cytokine expression in primary HBMEC. Exposure of HBMEC to HIV-1 gp120 protein induced increased expression of IL-6 and IL-8. Exposure of HBMEC to gp120 (100 ng/ml) induced secretion of 10.23 ± 2 pg/ml, 25.73 ± 2.86 pg/ml, 48.54 ± 4.3 pg/ml, and 97.55 ± 7.2 pg/ml IL-6 respectively at 2, 6, 12, and 24 h (P<0.001, Fig. 1A). IL-6 levels in control untreated cells and cells exposed for 24h to 100 ng/ml heat-inactivated gp120 were respectively 11.92 ± 0.1 pg/ml and 19 ± 0.76 pg/ml.
Figure 1. HIV-1 gp120 proteins increased IL-6 and IL-8 expression in HBMEC.
HBMEC were exposed for 2 to 24h to 100 ng/ml of HIV-1 gp120 (gp120MN), and IL-6 and IL-8 levels in culture supernatant were quantified by ELISA. Four replicates were used for each treatment condition. HIV-1 gp120 induced a time-dependent increase in IL-6 (A) and IL-8 (B) secretion in HBMEC (***P<0.001 compared to control). Control represents untreated cells; Heat-MN represents cells treated with heat-inactivated gp120MN.
Similarly, HBMEC exposure to gp120 (100 ng/ml) stimulated a time-dependent IL-8 production: 481.4 ± 57.78 pg/ml, 853 ± 120 pg/ml, 2787 ± 452 pg/ml, and 3095 ± 185 pg/ml at 2, 6, 12, and 24 h (P<0.001, Fig. 1B) respectively. IL-8 levels in control untreated cells and cells exposed to 100 ng/ml heat-inactivated gp120 were 318.6 ± 19.2 pg/ml and 396 ± 75.16 pg/ml, respectively.
STAT1 inhibitors prevented gp120-induced up-regulation of IL-6 and IL-8 expression
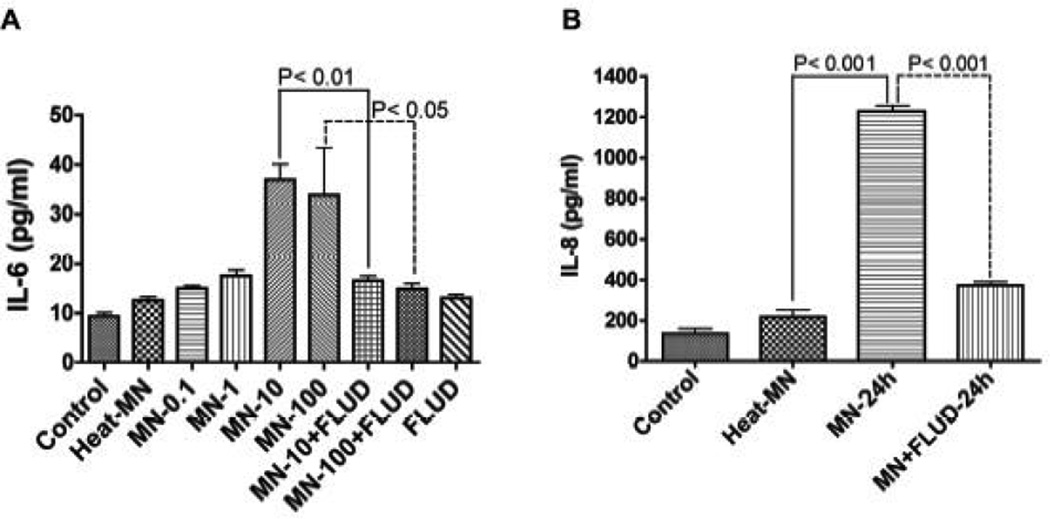
The JAK/STAT pathway plays a prominent role in cytokine-mediated inflammatory responses, and both IL-6 and IL-8 can signal through this pathway. Therefore, we tested the effect of STAT1 inhibitor FLUD on gp120-mediated IL-6 and IL-8 production. Our previous works determined that 20 μM was the optimal FLUD dose for preventing HIV-induced BBB inflammation without causing toxicity or functional changes in HBMEC (Chaudhuri et al., 2008b). HBMEC were exposed for 24 h to 0.1, 1, 10, and 100 ng/ml of gp120 or treated with gp120 and FLUD (20 μM). IL-6 levels in culture supernatant were determined by ELISA. HIV-1 gp120 induced a dose-dependent increase in IL-6 levels (Fig. 2A). IL-6 levels in HBMEC exposed to 0.1, 1, 10, and 100 ng/ml gp120 were respectively 15 ± 55pg/ml, 17.55 ± 1.15 pg/ml, 37 ± 3.17 pg/ml, and 34 ± 9.47 pg/ml (Fig. 2A). FLUD significantly diminished gp120-induced IL-6 expression (P<0.05 Fig. 2A). IL-6 levels in control untreated cells were 9.33 ± 0.75 pg/ml while levels in cells exposed to 100 ng/ml heat-inactivated gp120 were 12.6 ± 0.7 pg/ml.
Figure 2. FLUD inhibits gp120-induced IL-6 and IL-8 secretion.
(A) HBMEC were exposed for 24 h to 0.1 ng/ml (MN-0.1), 1 ng/ml (MN-1), 10 ng/ml (MN-10), and 100 ng/ml (MN-100) of HIV-1 gp120 (gp120MN) or exposed to gp120 + 20 μM FLUD, and IL-6 levels in culture supernatant were determined by ELISA. HIV-1 gp120 induced a dose-dependent increase in IL-6 levels and FLUD significantly diminished gp120-induced IL-6 secretion (P<0.01). (B) HBMEC were exposed for 24h to 100 ng/ml gp120 or 100 ng/ml gp120 + 20 μM FLUD and IL-8 levels in culture supernatant were determined by ELISA. Exposure of HBMEC to gp120 increased IL-8 secretion and FLUD significantly diminished gp120-induced IL-8 secretion (P<0.001). Controls are untreated cells, heat-MN are endothelial cells exposed to 100 ng/ml of heat-inactivated gp120. For each experimental condition, n=6.
Similarly, FLUD decreased gp120-induced IL-8 secretion by 3.3-fold. While IL-8 levels in cells exposed to gp120 (100 ng/ml for 24h) were 1228 ± 29.5 pg/ml, IL-8 levels in cells exposed to gp120 and FLUD (20 µM) were 373 ± 17 pg/ml (P<0.001, Fig. 2B). IL-8 levels in control untreated cells were 134.7 ± 25.5 pg/ml, while levels in cells exposed to 100 ng/ml heatinactivated gp120 were 216.8 ± 34.5 pg/ml (Fig. 2B).
HBMEC express IL-6 and IL-8 receptors and gp120 up-regulated IL-8 receptor expression
Since exposure of HBMEC to gp120 increased secretion of IL-6 and IL-8, we determined whether HBMEC express receptors for IL-6 and IL-8. Immunofluorescence analyses demonstrated that HBMEC express IL-8 receptor type II (CXCR2), IL-6 receptor-α, and gp130; the latter two comprising the α and β chains of the IL-6 receptor (Fig. 3). Next, we assessed whether expression of these receptors could be altered by gp120 exposure. HIV-1 gp120 increased CXCR2 immunostaining in HBMEC (Fig. 3A). We then performed semi-quantitative analyses of CXCR2 expression (percentage of area occupied per immunostaining) using computer-assisted image analysis (Image-Pro®Plus, Media Cybernetics, Silver Spring, MD). Image-Pro®Plus software measures the fluorescence intensity and densitometry of immunostained cells and normalize each measurement to individual cell size. Data from two independent experiments, using HBMEC from two different donors, showed that 30min, 1h, and 2h gp120 exposure increased CXCR2 expression for the two donors by 2.8- and 5.3-fold, 4.3- and 4.9-fold, and 3.6- and 8.5-fold, respectively at each time point (P<0.001, Fig. 3B and 3C). For each HBMEC donor, each experimental condition was performed in triplicate (Fig 3A and 3B). Heat-inactivated gp120 had no effect on CXCR2 expression. Interestingly, exposure of HBMEC to gp120 did not alter the expression of IL-6 receptor (Fig. 3D).
Figure 3. HBMEC express the IL-6 and IL-8 receptors.
(A) HBMEC from two different donors expressed CXCR2 (IL-8 receptor type II), and exposure of cells to gp120 (gp120MN) increase CXCR2 expression. (B, C) Semi-quantitative analysis (percentage of area occupied by immunostaining) carried out by computer-assisted image analysis (Image-Pro®Plus, Media Cybernetics, Silver Spring, MD) confirmed increase in CXCR2 expression in HBMEC exposed to gp120. (D) HBMEC express the alpha (IL6-R-α) and beta (gp130) subunits of the IL-6 receptor. Exposure of endothelial cells to gp120 did not change expression of the IL-6 receptor. Control represents cells exposed to heat-inactivated gp120.
HIV-1 gp120 proteins induce activation of STAT1 at serine 727 in HBMEC
STAT1 is activated by pro-inflammatory and regulatory factors. IL-6 signals principally through the JAK/STAT pathway and IL-8 can also use this pathway. Therefore, we tested whether HIV-1 gp120 activates STAT1 in HBMEC. Endothelial cells were treated with gp120 (100 ng/ml) for 5 min to 4h, followed by protein extraction and Western blot analysis for STAT1 and phospho-STAT1 (active form of STAT1). Exposure of HBMEC to gp120 induced phosphorylation of STAT1 at serine-727 (S727), with maximum levels of activation at 1h (Fig. 4A). To determine whether gp120 proteins activate other members of the STAT family in HBMEC, we tested the levels of phosphorylated STAT3, STAT5, and STAT6 in gp120-treated cells. HIV gp120 induced phosphorylation of STAT1 and STAT3 at S727 (Fig. 4A and 4B). HIV-1 gp120-induced STAT1 and STAT3 activation was seen only on serine residues, and no activation at tyrosine residues was detected. Gp120 exposure did not phosphorylate STAT2 (data not shown) and did not phosphorylate STAT5 or STAT6 (Fig. 4B). Figure 4 shows representative data from 3 independent experiments using HBMEC from 3 different donors.
Figure 4. HIV-1 gp120 proteins activate STAT1 at serine-727 in HBMEC; FLUD, PD, and LY inhibit gp120-induced STAT1 activation in HBMEC.
Exposure of HBMEC to gp120 (100 ng/ml) activates STAT1 at S727 (A, C, D) with maximal activation at 1h (A). No activation of STAT1 at tyrosine residues was detected. (B) HIV-1 gp120 activates STAT3 at S727 in HBMEC; no activation of STAT5 or STAT6 was detected. (C, D) Specific inhibitors of STAT1 (FLUD, 20 μM), MEK (PD, 50 μM), and PI3K (LY, 50 μM), significantly diminished gp120-induced STAT1 phosphorylation (***P<0.001).
Inhibitors of STAT1, MEK, and PI3K diminished gp120-induced STAT1 activation
In the classical JAK/STAT pathway, STATs are phosphorylated by JAK (Kovarik et al., 2001). To determine which effector upstream of STAT1 may be involved in gp120-induced BBB dysfunction, we analyzed JAK and tyrosine kinase 2 (TYK2) activation in gp120-exposed HBMEC. Phosphorylation of JAK1, JAK2, JAK3, or TYK2 was not detected in gp120-treated cells (data not shown). There is evidence that STAT1 and STAT3 can also be phosphorylated by serine threonine kinase, including mitogen activated protein kinase (MAPK) and MEK (Kovarik et al., 2001; Zhang et al., 2004). To investigate this possibility, we determined the effect of FLUD (a specific STAT1 inhibitor), PD98059 (a specific MEK inhibitor), SB202190 (a specific p38 MAPK inhibitor), and LY294002 (a specific PI3K inhibitor) on gp120-induced STAT1 activation. The STAT1 inhibitor, FLUD, diminished gp120-induced STAT1 activation by 2-fold (Fig. 4B and 4C). Similarly, using HBMEC from two different donors, the MEK and PI3K inhibitors diminished gp120-induced STAT1 phosphorylation by 2-fold to 4-fold (Fig 4B, donor #1), and 12-fold and 40-fold respectively (Fig. 4D, donor #2). The p38 MAPK inhibitor had no effect (data not shown).
STAT1 modulates cytokine-induced monocyte adhesion and migration across in vitro BBB models
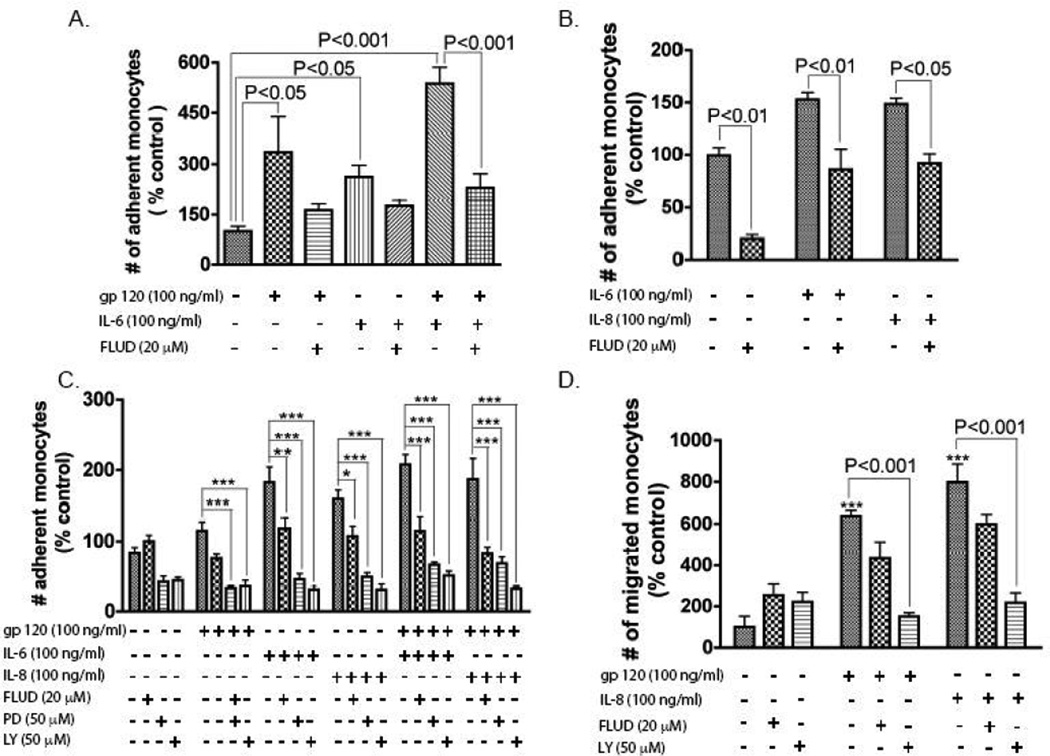
HIV-1 and secreted viral factors induce chemotaxis and migration of infected leukocytes across the BBB (Chaudhuri et al., 2008b; Kanmogne et al., 2007). To determine the functional significance of gp120-induced up-regulation of IL-6 and IL-8 expression, we performed adhesion and migration experiments in response to gp120, IL-6, and IL-8. Individually, gp120, IL-6, and IL-8 significantly increased monocyte adhesion to HBMEC (Fig. 5A–C), and gp120 + IL-6 further increased monocyte adhesion (Fig. 5A). Similarly, gp120 and IL-8 significantly increased monocyte migration across in vitro BBB models (Fig. 5D). STAT1 inhibitor diminished gp120-induced IL-8 and IL-6 expression, and inhibitors of STAT1, MEK, and PI3K prevented gp120-induced STAT1 activation; thus, we tested the effects of FLUD (a specific STAT1 inhibitor), PD98059 (PD, a specific MEK inhibitor), and LY294002 (LY, a specific PI3K inhibitor) on gp120-, IL-6- and IL-8-induced monocyte adhesion and migration. FLUD, PD, and LY significantly diminished gp120-induced monocyte adhesion, as well as IL-6- and IL-8-induced monocyte adhesion (Fig. 5A–C). Similarly, FLUD and LY diminished gp120- and IL-8-induced monocyte migration across in vitro BBB models (P<0.001, Fig. 5D).
Figure 5. Functional consequence of gp120- and cytokine-induced effects on HBMEC.
HIV-1 gp120, IL-6, and IL-8 increased monocyte adhesion (A – C) and migration (D) across in in vitro BBB models. Inhibitors of STAT1 (FLUD, 20 μM), MEK (PD, 50 μM), and PI3K (LY, 50 μM), significantly diminished IL-6-, IL-8-, and gp120-induced monocytes adhesion (A – C, *P<0.05, **P<0.01, ***P<0.001). FLUD and LY also diminished monocyte migration across in vitro BBB models (D, ***P<0.001, compared to untreated control). Adhesion and migration data are shown as percent of control, with control representing the number of monocytes that adhered to or migrated through untreated HBMEC.
DISCUSSION
BBB dysfunction is common in HIV-1 infected patients and plays an important role in the pathogenesis of HAD (Banks et al., 2006; Toborek et al., 2005). Impairment of the BBB could facilitate infiltration of viral particles, viral factors, and infected mononuclear phagocytes into the CNS where they accumulate, spread infection to resident macrophages and glial cells, and induce neuronal cell death. The mechanisms of BBB breakdown during viral infection are poorly understood. HBMEC, a major component of the BBB, constitute a physiological and functional barrier for entry of cells, fluids, and macromolecules into the brain. Therefore, cultures of HBMEC and in vitro BBB models are appropriate for studies of HIV-1-mediated effects on the brain endothelium.
We previously demonstrated that HIV-1 virions induce transcriptional up-regulation and expression of pro-inflammatory genes and the transcription factor STAT1 in HBMEC (Chaudhuri et al., 2008a; Chaudhuri et al., 2008b). Here, we report that exposure of primary HBMEC to the HIV-1 envelope protein, gp120, increased secretion of IL-6 and IL-8 via the STAT1 pathway. Our data are in agreement with previous studies showing that the JAK/STAT pathway is involved in inflammation and cytokine-mediated cell injury. IL-6 contain a STAT binding element in its promoter region, and inflammation and oxidative stress induce IL-6 expression in macrophages (Lee et al., 2006). Human clinical and autopsy studies suggest that viral-induced inflammation and dysregulation of cytokine expression play a major role in the pathogenesis of HIV-1 infection and disease progression (Breen et al., 1990; Kedzierska and Crowe, 2001; Matsumoto et al., 1993). HIV-1 gp120-induced dysregulation of cytokine expression was also demonstrated in laboratory and animal models studies, and gp120 upregulated IL-6 and IL-8 expression in mononuclear phagocytes and glial cells (Capobianchi et al., 1997; Oyaizu et al., 1991; Yeung et al., 1995). Intrathecal and intracerebroventricular injection of gp120 in mice and rats increases IL-6 expression in the hippocampus, meninges, and spinal cord; and the IL-6 released mediated inflammatory pain in these animals (Schoeniger-Skinner et al., 2007).
In vivo and in vitro studies also demonstrated that HIV-1 and its gp120 envelope protein cross the brain endothelium and can induce BBB injury (Kanmogne et al., 2002; Kanmogne et al., 2005; Toneatto et al., 1999). Our current data demonstrate that exposure of HBMEC to gp120 increased secretion of IL-6 and IL-8, gp120 and secreted cytokines enhanced leukocyte adhesion and transendothelial migration via STAT1 pathways. Our previous studies also demonstrated that HIV-1 virions increased IL-6 expression in HBMEC, and STAT1 inhibitor diminished HIV-induced IL-6 expression and blocked IL-6-induced monocyte migration across in vitro BBB models (Chaudhuri et al., 2008b). Our present results are congruent with previous studies showing that gp120 stimulated IL-6 secretion in children’s brain endothelial cells (Stins et al., 2001). High levels of IL-6 were found in the cerebrospinal fluids of HAD patients compared to HIV-1 seropositive patients without CNS disease (Perrella et al., 1992; Torre et al., 1992). The JAK/STAT pathway plays a prominent role in cytokine-mediated inflammatory responses, and STAT1 was implicated in the pathogenesis of HIV-1 infection and disease progression (Bovolenta et al., 1999; Bovolenta et al., 2002). There is enhanced activation of STAT1α in T-cells from HIV-1-infected individuals (Bovolenta et al., 1999), as well as in HIV-1- and human T-lymphotropic virus (HTLV)-2- infected T-lymphocytes (Bovolenta et al., 2002).
Activation of STAT1 is generally associated with phosphorylation of tyrosine at residue 701 (Y701) or serine at residue 727 (S727), and this activation correlates with pro-inflammatory and anti-proliferative responses (Bromberg et al., 1996). The STAT1 transactivating domain resides in the C-terminus, and phosphorylation of S727 increases the transcriptional activity of STAT1 (Kovarik et al., 2001). Taken together, our study points to STAT1 as an important modulator of gp120-induced BBB dysfunction. In the JAK/STAT pathway, phosphorylated STATs form dimers and associate with the interferon stimulated gene factor 3 gamma (ISGF3G) to form a complex transcription factor that translocates to the nucleus, binds to the interferon stimulated response element to activate the transcription of interferon or cytokine stimulated genes (Schindler and Brutsaert, 1999). HIV-1 also induces transcriptional upregulation of ISGF3G, a downstream effector of STAT1 (Chaudhuri et al., 2008a). We demonstrated that STAT1 and STAT3 are the STAT family members activated by gp120 proteins, which suggests that in gp120-induced BBB dysfunction, the STAT dimers that translocate to the nucleus consist of STAT1-STAT3 heterodimers. Upstream effectors of STATs include JAK and serine threonine kinase such as MEK and PI3K (Kovarik et al., 2001; Zhang et al., 2004). Other studies showed crosstalk between MAPK, PI3K, and protein kinase C (PKC) signaling pathways in gp120-induced toxicity of human umbilical vein endothelial cells (Langford et al., 2005). Our data show possible crosstalk between STAT1, MEK, and PI3K pathways, and suggest that following gp120 exposure, MEK and/or PI3K could be upstream effectors that phosphorylate STAT1. A subsequent study will further investigate this possibility, as well as the relationship between gp120-induced activation of STAT1 and PKC in BBB compromise. In fact, gp120 also activates PKC in HBMEC (Kanmogne et al., 2007), and STAT1 activation at S727 requires PKC (Xuan et al., 2005). The STAT1 inhibitor, FLUD, is an anti-inflammatory compound that is clinically approved and currently used in humans for the treatment of hematologic malignancies (Montillo et al., 2006), and also shows anti-viral effects in simian immunodeficiency virus -infected monkeys (Cervasi et al., 2006). During the course of HIV-1 infection, viral proteins are constantly shed in the blood and tissues causing BBB injury and enhancing infiltration of virus-infected leukocytes into the CNS (Jones et al., 2000; Oh et al., 1992; Schneider et al., 1986). Our current study suggests that inhibiting STAT1 activation could provide a unique therapeutic approach for preventing HIV-1 gp120-induced pro-inflammatory phenotype of the brain endothelium and BBB dysfunction.
Acknowledgements
We thank Drs. Marlys Witte, Michael Bernas, and Yuri Persidsky for providing HBMEC; Meg Marquardt and Michael Jacobsen from the Department Confocal Microscopy Facility for assistance with microscopy, Dr Lee Mosley for critical reading of the manuscript, and Robin Taylor for excellent editorial support.
This work was supported in part by NIH grants KO1 MH068214 and RO1 MH081780 to GDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banks WA, Ercal N, Price TO. The blood-brain barrier in neuroAIDS. Curr HIV Res. 2006;4(3):259–266. doi: 10.2174/157016206777709447. [DOI] [PubMed] [Google Scholar]
- Bovolenta C, Camorali L, Lorini AL, Ghezzi S, Vicenzi E, Lazzarin A, Poli G. Constitutive activation of STATs upon in vivo human immunodeficiency virus infection. Blood. 1999;94(12):4202–4209. [PubMed] [Google Scholar]
- Bovolenta C, Pilotti E, Mauri M, Panzeri B, Sassi M, Dall'Aglio P, Bertazzoni U, Poli G, Casoli C. Retroviral interference on STAT activation in individuals coinfected with human T cell leukemia virus type 2 and HIV-1. J Immunol. 2002;169(8):4443–4449. doi: 10.4049/jimmunol.169.8.4443. [DOI] [PubMed] [Google Scholar]
- Breen EC, Rezai AR, Nakajima K, Beall GN, Mitsuyasu RT, Hirano T, Kishimoto T, Martinez-Maza O. Infection with HIV is associated with elevated IL-6 levels and production. J Immunol. 1990;144(2):480–484. [PubMed] [Google Scholar]
- Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE., Jr Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci U S A. 1996;93(15):7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger DM, Boucher CA, Meenhorst PL, Kraayeveld CL, Portegies P, Mulder JW, Hoetelmans RM, Beijnen JH. HIV-1 RNA levels in the cerebrospinal fluid may increase owing to damage to the blood-brain barrier. Antivir Ther. 1997;2(2):113–117. [PubMed] [Google Scholar]
- Capobianchi MR, Barresi C, Borghi P, Gessani S, Fantuzzi L, Ameglio F, Belardelli F, Papadia S, Dianzani F. Human immunodeficiency virus type 1 gp120 stimulates cytomegalovirus replication in monocytes: possible role of endogenous interleukin-8. J Virol. 1997;71(2):1591–1597. doi: 10.1128/jvi.71.2.1591-1597.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervasi B, Paiardini M, Serafini S, Fraternale A, Menotta M, Engram J, Lawson B, Staprans SI, Piedimonte G, Perno CF, Silvestri G, Magnani M. Administration of fludarabine-loaded autologous red blood cells in simian immunodeficiency virus-infected sooty mangabeys depletes pSTAT-1-expressing macrophages and delays the rebound of viremia after suspension of antiretroviral therapy. J Virol. 2006;80(21):10335–10345. doi: 10.1128/JVI.00472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri A, Duan F, Morsey B, Persidsky Y, Kanmogne GD. HIV-1 activates proinflammatory and interferon-inducible genes in human brain microvascular endothelial cells: putative mechanisms of blood-brain barrier dysfunction. J Cereb Blood Flow Metab. 2008a;28(4):697–711. doi: 10.1038/sj.jcbfm.9600567. [DOI] [PubMed] [Google Scholar]
- Chaudhuri A, Yang B, Gendelman HE, Persidsky Y, Kanmogne GD. STAT1 signaling modulates HIV-1-induced inflammatory responses and leukocyte transmigration across the blood-brain barrier. Blood. 2008b;111(4):2062–2072. doi: 10.1182/blood-2007-05-091207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendelman HE, Orenstein JM, Martin MA, Ferrua C, Mitra R, Phipps T, Wahl LA, Lane HC, Fauci AS, Burke DS, et al. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J Exp Med. 1988;167(4):1428–1441. doi: 10.1084/jem.167.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MV, Bell JE, Nath A. Immunolocalization of HIV envelope gp120 in HIV encephalitis with dementia. Aids. 2000;14(17):2709–2713. doi: 10.1097/00002030-200012010-00010. [DOI] [PubMed] [Google Scholar]
- Kanmogne GD, Kennedy RC, Grammas P. HIV-1 gp120 proteins and gp160 peptides are toxic to brain endothelial cells and neurons: possible pathway for HIV entry into the brain and HIV-associated dementia. J Neuropathol Exp Neurol. 2002;61(11):992–1000. doi: 10.1093/jnen/61.11.992. [DOI] [PubMed] [Google Scholar]
- Kanmogne GD, Primeaux C, Grammas P. HIV-1 gp120 proteins alter tight junction protein expression and brain endothelial cell permeability: implications for the pathogenesis of HIV-associated dementia. J Neuropathol Exp Neurol. 2005;64(6):498–505. doi: 10.1093/jnen/64.6.498. [DOI] [PubMed] [Google Scholar]
- Kanmogne GD, Schall K, Leibhart J, Knipe B, Gendelman HE, Persidsky Y. HIV-1 gp120 compromises blood-brain barrier integrity and enhances monocyte migration across blood-brain barrier: implication for viral neuropathogenesis. J Cereb Blood Flow Metab. 2007;27(1):123–134. doi: 10.1038/sj.jcbfm.9600330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierska K, Crowe SM. Cytokines and HIV-1: interactions and clinical implications. Antivir Chem Chemother. 2001;12(3):133–150. doi: 10.1177/095632020101200301. [DOI] [PubMed] [Google Scholar]
- Ketzler S, Weis S, Haug H, Budka H. Loss of neurons in the frontal cortex in AIDS brains. Acta Neuropathol (Berl) 1990;80(1):92–94. doi: 10.1007/BF00294228. [DOI] [PubMed] [Google Scholar]
- Klockars AJGS. Multiple comparisons. Quantitative applications in the social sciences series #61. Thousand Oaks, CA: Sage Publications; 1986. [Google Scholar]
- Kovarik P, Mangold M, Ramsauer K, Heidari H, Steinborn R, Zotter A, Levy DE, Muller M, Decker T. Specificity of signaling by STAT1 depends on SH2 and C-terminal domains that regulate Ser727 phosphorylation, differentially affecting specific target gene expression. Embo J. 2001;20(1–2):91–100. doi: 10.1093/emboj/20.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford D, Hurford R, Hashimoto M, Digicaylioglu M, Masliah E. Signalling crosstalk in FGF2-mediated protection of endothelial cells from HIV-gp120. BMC Neurosci. 2005;6(1):8. doi: 10.1186/1471-2202-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Lim HK, Sakong J, Lee YS, Kim JR, Baek SH. Janus kinase-signal transducer and activator of transcription mediates phosphatidic acid-induced interleukin (IL)-1beta and IL-6 production. Mol Pharmacol. 2006;69(3):1041–1047. doi: 10.1124/mol.105.018481. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Miike T, Nelson RP, Trudeau WL, Lockey RF, Yodoi J. Elevated serum levels of IL-8 in patients with HIV infection. Clin Exp Immunol. 1993;93(2):149–151. doi: 10.1111/j.1365-2249.1993.tb07957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DW, Audus KL, Borchardt RT. Application of cultured endothelial cells of the brain microvasculature in the study of the blood-brain barrier. Journal of Tissue Culture Methods. 1992;14:217–224. [Google Scholar]
- Montillo M, Ricci F, Tedeschi A. Role of fludarabine in hematological malignancies. Expert Rev Anticancer Ther. 2006;6(9):1141–1161. doi: 10.1586/14737140.6.9.1141. [DOI] [PubMed] [Google Scholar]
- Oh SK, Cruikshank WW, Raina J, Blanchard GC, Adler WH, Walker J, Kornfeld H. Identification of HIV-1 envelope glycoprotein in the serum of AIDS and ARC patients. J Acquir Immune Defic Syndr. 1992;5(3):251–256. [PubMed] [Google Scholar]
- Oyaizu N, Chirmule N, Ohnishi Y, Kalyanaraman VS, Pahwa S. Human immunodeficiency virus type 1 envelope glycoproteins gp120 and gp160 induce interleukin-6 production in CD4+ T-cell clones. J Virol. 1991;65(11):6277–6282. doi: 10.1128/jvi.65.11.6277-6282.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrella O, Carrieri PB, Guarnaccia D, Soscia M. Cerebrospinal fluid cytokines in AIDS dementia complex. J Neurol. 1992;239(7):387–388. doi: 10.1007/BF00812156. [DOI] [PubMed] [Google Scholar]
- Price RW, Brew B, Sidtis J, Rosenblum M, Scheck AC, Cleary P. The brain in AIDS: central nervous system HIV-1 infection and AIDS dementia complex. Science. 1988;239(4840):586–592. doi: 10.1126/science.3277272. [DOI] [PubMed] [Google Scholar]
- Schindler C, Brutsaert S. Interferons as a paradigm for cytokine signal transduction. Cell Mol Life Sci. 1999;55(12):1509–1522. doi: 10.1007/s000180050391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Kaaden O, Copeland TD, Oroszlan S, Hunsmann G. Shedding and interspecies type sero-reactivity of the envelope glycopolypeptide gp120 of the human immunodeficiency virus. J Gen Virol. 1986;67(Pt 11):2533–2538. doi: 10.1099/0022-1317-67-11-2533. [DOI] [PubMed] [Google Scholar]
- Schoeniger-Skinner DK, Ledeboer A, Frank MG, Milligan ED, Poole S, Martin D, Maier SF, Watkins LR. Interleukin-6 mediates low-threshold mechanical allodynia induced by intrathecal HIV-1 envelope glycoprotein gp120. Brain Behav Immun. 2007 doi: 10.1016/j.bbi.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stins MF, Shen Y, Huang SH, Gilles F, Kalra VK, Kim KS. Gp120 activates children's brain endothelial cells via CD4. J Neurovirol. 2001;7(2):125–134. doi: 10.1080/13550280152058780. [DOI] [PubMed] [Google Scholar]
- Toborek M, Lee YW, Flora G, Pu H, Andras IE, Wylegala E, Hennig B, Nath A. Mechanisms of the blood-brain barrier disruption in HIV-1 infection. Cell Mol Neurobiol. 2005;25(1):181–199. doi: 10.1007/s10571-004-1383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toneatto S, Finco O, van der Putten H, Abrignani S, Annunziata P. Evidence of bloodbrain barrier alteration and activation in HIV-1 gp120 transgenic mice. Aids. 1999;13(17):2343–2348. doi: 10.1097/00002030-199912030-00005. [DOI] [PubMed] [Google Scholar]
- Torre D, Zeroli C, Ferraro G, Speranza F, Tambini R, Martegani R, Fiori GP. Cerebrospinal fluid levels of IL-6 in patients with acute infections of the central nervous system. Scand J Infect Dis. 1992;24(6):787–791. doi: 10.3109/00365549209062465. [DOI] [PubMed] [Google Scholar]
- Xuan YT, Guo Y, Zhu Y, Wang OL, Rokosh G, Messing RO, Bolli R. Role of the protein kinase C-epsilon-Raf-1-MEK-1/2-p44/42 MAPK signaling cascade in the activation of signal transducers and activators of transcription 1 and 3 and induction of cyclooxygenase-2 after ischemic preconditioning. Circulation. 2005;112(13):1971–1978. doi: 10.1161/CIRCULATIONAHA.105.561522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung MC, Pulliam L, Lau AS. The HIV envelope protein gp120 is toxic to human brain-cell cultures through the induction of interleukin-6 and tumor necrosis factor-alpha. Aids. 1995;9(2):137–143. [PubMed] [Google Scholar]
- Zhang Y, Cho YY, Petersen BL, Zhu F, Dong Z. Evidence of STAT1 phosphorylation modulated by MAPKs, MEK1 and MSK1. Carcinogenesis. 2004;25(7):1165–1175. doi: 10.1093/carcin/bgh115. [DOI] [PubMed] [Google Scholar]