Abstract
Introduction
Aspartyl-asparaginyl-β-hydroxylase (AAH) regulates cell motility and invasiveness by enhancing Notch signaling. Invasive trophoblastic cells, which mediate placentation, normally express high levels of AAH. Previously, we showed that ethanol-impaired placentation is associated with reduced AAH expression. The present study determines the degree to which inhibition of AAH expression is sufficient to impair functions required for placentation.
Methods
Immortalized, first trimester-derived, human trophoblastic cells (HTR-8/SVneo) were transfected with siRNA targeting AAH (siRNA-AAH) or no specific sequences (siRNA-Scr) using the Amaxa electroporation system. Directional motility was measured using an ATP Luminescence-based assay. For in vivo studies, we microinjected siRNA-AAH or siRNA-Scr directly into the implantation sites (mesometrial triangle) of gestation-day-17, Long Evans pregnant rats, and harvested placentas 24 h later for histologic and molecular studies.
Results
siRNA-AAH transfection reduced AAH expression and directional motility in HTR-8/SVneo cells. In vivo delivery of siRNA-AAH reduced AAH expression and mean number of invasive trophoblastic cells at the implantation site. These adverse effects of siRNA-AAH were associated with impaired fetal growth and significantly reduced expression of Notch-signaling network genes.
Conclusions
AAH is an important, positive regulator of trophoblastic cell motility, and inhibition of AAH in vivo leads to impaired implantation and fetal growth, and alters Notch-signaling mechanisms, similar to the effects of chronic ethanol exposure.
Keywords: Intra-placental gene delivery, Aspartyl-asparaginyl-β-hydroxylase, Notch, Intrauterine growth restriction, Ethanol, Small interfering RNA, In vivo transfection, Trophoblast migration
INTRODUCTION
Aspartyl-(asparaginyl)-β-hydroxylase (AAH) is a type 2 transmembrane protein that catalyzes the hydroxylation of aspartyl and asparaginyl residues in epidermal growth factor (EGF)-like domains [29] of proteins, such as Notch and Jagged [15,16]. AAH can physically interact with both Notch and Jagged [6], which have known roles in cell motility [8,25]. Overexpression of AAH results in increased nuclear translocation and accumulation of Notch, and activation of Notch’s downstream target genes, including Hairy and Enhancer of Split 1 (HES-1) [6]. AAH is over-expressed in highly invasive, malignant neoplasms [13,23,26]. Placenta is one of the few non-transformed organs that expresses high levels of AAH and extravillous cytotrophoblasts, which are motile and invasive, expressing higher levels of AAH than the non-motile villous counterparts [20]. The invasive properties of extravillous cytotrophoblasts are critical for implantation and placentation. In humans, placentas from pregnancies with intrauterine growth restriction (IUGR) or early spontaneous abortion have reduced levels of AAH expression, especially in extravillous cytotrophoblasts [20]. Correspondingly, both conditions are associated with impairments in implantation and placentation, and experimental depletion of the AAH gene in mice reduces litter size [15].
Notch signaling is evolutionarily conserved and regulates cell fate decisions in various tissues during embryogenesis and postnatal development [3,33,4,2]. Notch-receptor family is composed of membrane-tethered transcription factors whose N-terminal fragments translocate to the nucleus upon activation by Delta-like (DII 1, 3, and 4) or Serrate-like (Jagged 1 and 2) ligands [18]. Notch ligands themselves are transmembrane proteins that have extracellular EGF-like repeat arrays [17]. Notch pathway signaling is activated by ligand binding to Notch receptors, which triggers proteolytic cleavage and attendant release of Notch’s N-terminal intracellular domain (ICD). The ICD translocates to the nucleus where it gets recruited by the transcription factor of the CBF-1/suppressor of hairless/Lag-1 (CSL) family to regulate target genes, such as HES and HEY [4].
Implantation, trophoblast cell differentiation, and establishment of the maternal and fetal circulations within the placenta are major processes required for placentation, and experimental evidence suggests that intact Notch signaling is required for normal placental development [18]. AAH mediates cell motility and invasiveness, and functions by increasing Notch signaling. AAH and Notch are both expressed in extravillous cytotrophoblasts [11,20], and reduced expression of either occurs in human diseases that cause intrauterine growth restriction, e.g. preeclampsia [9,20]. Similarly, placental AAH expression is reduced by gestational ethanol exposure, which induces IUGR in rat pups [21]. However, the effects of AAH inhibition with regard to placentation have been largely indirect. This study examines the functional consequences of transiently inhibiting AAH expression with small interfering RNA (siRNA) molecules that target the 5′ end of the AAH gene [13]. In vitro studies, conducted with human trophoblastic cells, were used to measure directional motility and Notch expression following siRNA-AAH transfection. In addition, in pregnant Long Evans rats, siRNA-AAH was microinjected into the implantation sites on gestation-day 17, and the effects on placentation, Notch signaling, and fetal growth were examined twenty-four hours later.
MATERIALS AND METHODS
Trophoblast Cell Culture
HTR-8/SVneo cells are immortalized, first trimester, human trophoblast cells that have properties of invasive extravillous cytotrophoblasts. Cultures were maintained in Roswell Park Memorial Institute (RPMI) 1640 media (Lonza Walkersville, Inc., Walkersville, MD) supplemented with 10% fetal bovine serum (FBS) and 2.1 mM L-glutamine [19] at 37°C in a 5% CO2 incubator. Cells (1.6 x 106) were transfected with commercially generated and validated (Dharmacon, Inc., Chicago, IL) ON-TARGETplus SMARTpool siRNA duplexes (0.16 nM) that targeted either AAH [ASPH NM_004318] (siRNA-AAH) or no specific sequences (siRNA-Scr), together with green fluorescent protein (GFP) expressing plasmid (1 μg) [13,27] as a reporter gene. Transfections were performed using Amaxa “T” nucleofector cell-line reagents and the Amaxa nucleofector apparatus (Amaxa, Inc. Gaithersburg, MD) set at the HEK293-A023 program. We consistently achieved transfection efficiencies between 75% and 90% based on the percentages of GFP-positive cells. Twenty-four hours after transfection, cells were harvested to measure motility, immunoreactivity, and gene expression.
Directional Motility Assay
Directional motility was measured using the ATP Luminescence-Based Motility-Invasion assay [12]. Blind well-chambers partitioned by polycarbonate filters with 12 μm pore diameter were used for the experiments. 105 viable (Trypan blue excluded) siRNA-SCR or siRNA-AAH transfected cells suspended in 100 μL serum-free medium, were seeded into the upper chamber, and medium containing 1% FBS and 0.1 μg/ml IGF2 as the trophic factor was supplied in the lower chamber. Cell migration was allowed to proceed for 30 minutes at 37°C in a CO2 incubator. Cells harvested from the upper chambers (sessile), undersurfaces of the membranes (motile, adherent), and lower chambers (motile non-adherent) were quantified using ATPlite reagent, and luminescence was measured in a TopCount machine (Perkin-Elmer, Waltham, MA). The percentages of sessile (non-motile), motile adherent, motile non-adherent, and total motile (adherent + non-adherent) cells were calculated, and statistical analyses were performed using results from eight replicate assays.
In vivo AAH silencing
Timed pregnant Long Evans rats were purchased from Harlan Sprague Dawley, Inc. (Indianapolis, IN). On gestational day (GD) 17, under isofluorane inhalation anesthesia and using sterile surgical technique, a midline vertical laparotomy incision was made to externalize the uterine horns and gestational sacs. Using an insulin syringe (28 gauge, 0.5 mL), we injected the mesometrial triangle, i.e. the implantation site, of each gestational sac with a 10 μL solution containing 0.2 nM siRNA targeting rat AAH [Asphd2, NM_001009716] (N=4 dams) or siRNA-Scr (N=2 dams), together with 100 ng GFP, 1 μL Lipofectamine 2000, and siRNA buffer. Targeting and non-targeting ON-TARGETplus SMARTpool siRNA complexes were commercially prepared and validated (Dharmacon, Inc., Chicago, IL). GD 17 was selected for siRNA delivery because GD18 is the peak period of trophoblast cell invasion of the mesometrial triangle [32], and our goal was to maximally inhibit trophoblast cell activity. Once any minor bleeding was stopped, the uterine horns and gestational sacs were returned to the peritoneal cavity, and the incision was sutured. The entire surgical procedure was completed within twenty minutes. We observed no post-operative complications or deaths. Twenty-four hours after gene delivery, fetuses and placentas were harvested for study. Fetal body weights and crown-rump lengths were measured. GD was confirmed based on fetal developmental stage [1]. The implantation sites were dissected. Placental tissue with underlying mesometrial triangle was weighed, immersion fixed in Histochoice, and embedded in paraffin for histological sectioning [21]. Alternatively, micro-dissected placental and mesometrial triangle tissues [21] were snap-frozen in a dry ice-methanol bath and stored at −80°C for later mRNA and protein studies. Successful gene delivery was confirmed by fluorescence microscopic detection of GFP in cryostat sections of placenta and mesometrium in representative specimens from each dam. The Institutional Animal Welfare Committee at Rhode Island Hospital approved this study.
Immunohistochemistry
Histological sections of placenta with underlying mesometrial triangle (five μm thick) were immunostained with monoclonal antibody to cytokeratin (CK) to detect invasive trophoblastic cells at the implantation site [32]. Briefly, deparaffinized and rehydrated tissue slides were incubated in preheated EnVision™ FLEX Target Retrieval Solution (high pH=9.0) for 20 minutes at 95–100 °C. After rinsing the slides in EnVision™ FLEX wash buffer, they were treated with 3% hydrogen peroxide for 5 minutes to quench the endogenous peroxidase activity. After rinsing in wash buffer, the slides were incubated for 20 minutes at room temperature with a 1:50 dilution of monoclonal cytokeratin antibody (DAKO # M0821, clone MNF-116). Antibody binding was detected with EnVision™ FLEX/HRP detection reaction, which consists of a dextran backbone to which a large number of peroxidase (HRP) molecules and secondary antibody molecules have been coupled. Diaminobenzidine tetrahydrochloride (DAB) was used as the chromogen. The sections were then counterstained lightly with hematoxylin. All immunohistochemical-staining reactions were performed using the Dako Autostainer (Dako, Carpenteria, CA).
Stereology was utilized to quantify the CK-positive invasive trophoblastic cells within the mesometrial triangle. Numerical density calculation was performed with the aid of an Olympus BX60 light microscope (Olympus America Inc., Center Valley, PA) with attached MS-2000 XYZ Inverted Stage (Applied Scientific Instrumentation, Eugene, OR), and Stereologer software (Stereology Resource Center, Inc., Chester, MD). Briefly, entire mesometrial triangle tissue including the decidua was selected as the anatomical area of interest at low power magnification (4X). Trophospongial-decidual junction was used as the upper boundary of the mesometrial triangle. This junction was easily identifiable with CK immunostaining because trophospongium is CK-positive, whereas decidua is CK-negative except for the invasive trophoblastic cells. To maintain uniformity, sections showing linear maternal spiral artery with maternal channel continuation were selected for the evaluation. Unbiased counting frames were applied under software control. CK-positive cells were counted at high-power magnification (40X) using optical dissector method and normalized to the volume, which was based on Cavalieri point grid method.
Enzyme-linked immunosorbent assay (ELISA)
Tissues and cells were homogenized in radioimmunoprecipitation assay (RIPA) buffer containing protease and phosphatase inhibitors [10]. Protein concentrations were determined using the bicinchoninic acid (BCA) assay (Pierce, Rockford, IL). We performed direct binding ELISAs to measure AAH, Notch-1, and Jagged-1, immunoreactivity as previously described [10]. Also, we measured the levels of p85 subunit of PI3 kinase to rule out the off-target effects of transfection. The results were normalized to protein content in each well, which was measured using the NanoOrange® Protein Quantitation Kit [22]. Negative control reactions included omission of sample protein, primary antibody, secondary antibody, or both antibodies. Immunoreactivity was measured in triplicate assay reactions.
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) analysis
We used qRT-PCR analysis to measure mRNA levels of genes encoding AAH, Notch-1, Jagged-1 and HES-1. Briefly, cells or tissues were homogenized in Qiazol reagent, and total RNA was isolated using EZ1 RNA universal tissue kit and the BIO Robot EZ1 (Qiagen Inc., Valencia, CA). RNA was reverse transcribed using random oligodeoxynucleotide primers and the AMV First Strand cDNA synthesis kit. PCR amplifications were performed in 20 μl reactions containing cDNA generated from 2.5 ng of original RNA template, 300 nM each of gene-specific forward and reverse primer pairs (Table 1), and 10 μl of 2x QuantiTect SYBR Green PCR Mix (Qiagen Inc, Valencia, CA). Primers were designed using MacVector 10 software (MacVector, Inc., Cary, NC), and their target specificity was verified using the NCBI-BLAST program (National Center for Biotechnology Information-Basic Local Alignment Search Tool). Amplified signals were detected and analyzed using the Mastercycler ep realplex instrument and software (Eppendorf North America, Westbury, NY). Relative mRNA abundance was calculated from the ng ratios of specific mRNA to 18S rRNA measured in the same samples. Inter-group statistical comparisons were made using the calculated mRNA/18S ratios.
Table 1.
Primer Pair Sequences for Real-Time Quantitative RT-PCR
| Gene-Specific Primer | Forward/ Reverse | Sequence 5′ - 3′ | Position (mRNA) | Amplicon (bp) |
|---|---|---|---|---|
| 18s rRNA | Forward | GGA CAC GGA CAG GAT TGA GCA | 1278 | 50 |
| 18S rRNA | Reverse | ACC CAC GGA ATC GAG AAA GA | 1327 | |
| AAH (human) | Forward | GGG AGA TTT TAT TTC CAC CTG GG | 1842 | 257 |
| AAH (human) | Reverse | CCT TTG GCT TTA TCC ATC ACT GC | 2098 | |
| AAH (rat) | Forward | TGC CTG CTC GTC TTG TTT GTG | 666 | 118 |
| AAH (rat) | Reverse | ATC CGT TCT GTA ACC CGT TGG | 783 | |
| Notch-1 (rat) | Forward | GGT GGA CAT TGA CGA GTG TG | 1918 | 204 |
| Notch-1 (rat) | Reverse | CCC TTG AGG CAT AAG CAG AG | 2121 | |
| Jagged-1 (rat) | Forward | CTG AGG ACT ACG AGG GCA AG | 2041 | 191 |
| Jagged-1 (rat) | Reverse | ACA GGT GAA TTT GCC TCC TG | 2231 | |
| HES-1 (rat) | Forward | AGC GCT ACC GAT CAC AAA GT | 31 | 182 |
| HES-1 (rat) | Reverse | TCA GCT GGC ATT TTC CTT TT | 212 |
RT-PCR: reverse transcriptase polymerase chain reaction; Position: initial nucleotide for primer binding; bp: base pair size of amplicon; AAH: aspartyl-asparaginyl β-hydroxylase; HES: hairy and enhancer of split 1.
Sources of reagents
Polyclonal antibodies to Notch-1 and Jagged-1 were purchased from Santa Cruz (Santa Cruz, CA). Monoclonal antibody to cytokeratin (clone MNF-116) was purchased from Dako (Carpenteria, CA). Polyclonal antibody to p85 was purchased from Millipore (Temecula, CA). The 85G6 AAH monoclonal antibody was generated to human recombinant protein and purified over Protein G columns (Healthcare, Piscataway, NJ) [14]. QuantiTect SYBR Green PCR Mix and Qiazol reagent were purchased from Qiagen Inc. (Valencia, CA). AMV First Strand cDNA synthesis kits were obtained from Roche Diagnostics Corp. (Indianapolis, IN). Purified ON-TARGETplus SMARTpool duplex siRNA molecules were purchased from Dharmacon, Inc., (Chicago, IL). NanoOrange, Lipofectamine 2000, and oligodeoxynucleotide primers were obtained from Invitrogen (Carlsbad, CA). Histochoice fixative was purchased from Amresco Corp. (Solon, OH). BCA reagents were purchased from the Pierce Chemical Company (Rockford, IL). ATPLite substrate was obtained from Perkin-Elmer Corp. (Waltham, MA). Polycarbonate filters used in directional motility assays were purchased from GE Osmonics Labstore (Minnetonka, MN). All other fine chemicals and reagents were purchased from Calbiochem-EMD Biosciences (La Jolla, CA), Pierce Biotechnology Inc. (Rockford, IL), or Sigma-Aldrich (St. Louis, MO).
Statistical analyses
Data are depicted as means ± S.E.M. in the graphs. Inter-group comparisons were made using Student t-tests or two-way analysis of variance (ANOVA) with the Bonferroni posttests. Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA). Significant P-values are indicated over the graphs.
RESULTS
Inhibition of AAH impairs trophoblast cell motility
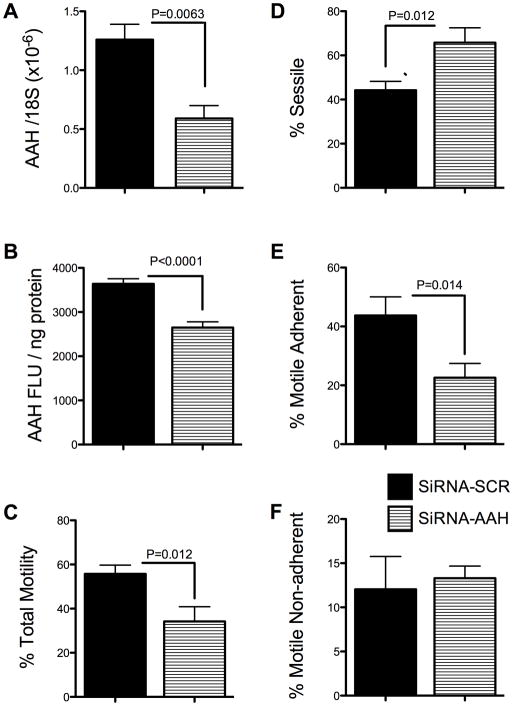
To demonstrate siRNA inhibition of AAH expression in HTR-8/SVneo cells, we measured AAH mRNA and protein levels by qRT-PCR and ELISA. Those studies demonstrated significantly reduced levels of AAH mRNA (P=0.0063; Fig. 1A) resulting in 55% inhibition of protein expression (Fig. 1B) in siRNA-AAH compared with siRNA-Scr (negative control) transfected cells (P<0.0001). Correspondingly, directional motility was significantly impaired (P=0.012; Fig. 1C), and the mean percentage of sessile (non-motile) cells was significantly increased (P=0.012) in siRNA-AAH transfected relative to control cells (Fig. 1D). The major effect of siRNA-AAH was to reduce the mean percentages of motile adherent cells (P=0.014; Fig. 1E). The mean percentages of motile non-adherent cells were similar in siRNA-AAH and siRNA-Scr transfected cultures (Fig. 1F). These results suggest that AAH has an important role in regulating adhesiveness of motile trophoblastic cells, similar to the findings with respect to other cell type [13,28,30].
Fig. 1.
siRNA-AAH inhibits AAH expression and directional motility in HTR-8/SVneo cells. HTR-8/SVneo cells were transiently transfected with siRNA-Scr or siRNA-AAH, and AAH (A) mRNA and (B) protein were measured by qRT-PCR and ELISA, and (C–F) directional motility was measured using the ATP luminescence-based motility-invasion assay. AAH mRNA levels were normalized to 18S rRNA, and immunoreactivity was normalized to protein content (NanoOrange assay). For the directional motility assays, we calculated percentages of (C) motile (adherent + non-adherent), (D) sessile, (E) motile adherent, and (F) motile non-adherent cells in eight replicate assay chambers. Graphs depict the mean ± S.E.M. levels. Inter-group comparisons were made using Student t-tests.
In vivo silencing of AAH causes intrauterine growth restriction and reduces invasive trophoblastic cells in the mesometrial triangle
In the two control group dams, siRNA-Scr was injected into 11 or 14 implantation sites; and in the 4 experimental group dams, siRNA-AAH was injected into 9, 10, 11, or 12 implantation sites. Pups harvested from siRNA-AAH injected gestational sacs had significantly lower mean body weights (P<0.0001) and shorter crown-rump lengths (P=0.0001) relative to pups harvested from siRNA-Scr injected gestational sacs (Fig. 2). In contrast, the mean placental weights were similar for the two groups (Fig. 2B).
Fig. 2.
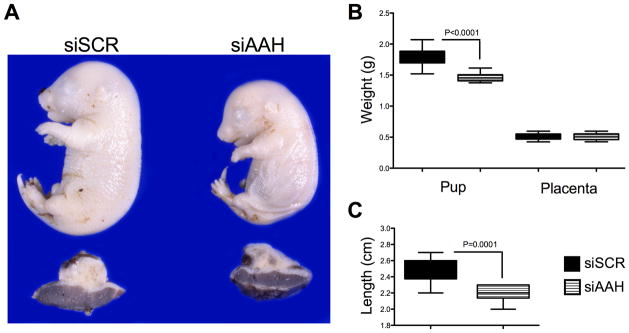
In vivo siRNA-AAH microinjection into the implantation site causes intrauterine growth restriction. The implantation sites (mesometrial triangles) of GD17 pregnant Long Evans rats were microinjected with siRNA-AAH or siRNA-Scr (See Methods), and 24 hours later, the (A) pups and placentas were harvested to examine effects on body growth. The graphs depict the mean ± S.E.M. corresponding to (B) pup weight, placental weight or (C) pup length. Inter-group comparisons were made using Student t-tests.
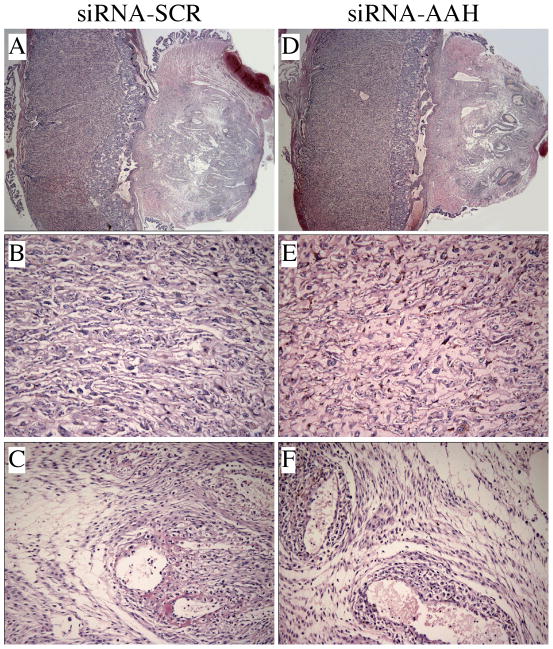
Hematoxylin and eosin-stained sections of full-thickness placenta with underlying mesometrial triangle (Fig. 3A,D) revealed no detectable structural differences in the labyrinthine region of placenta, i.e. the main region of maternal-fetal exchange, between the siRNA-Scr (Fig. 3B) and siRNA-AAH (Fig. 3E) injected groups. The only evidence of injection trauma observed in the mesometrial triangle was mild interstitial edema, which was similar in the siRNA-Scr (Fig. 3C) and siRNA-AAH (Fig. 3F) injected groups. There was no evidence of hemorrhage or necrosis in any of the placenta or implantation sites, and both groups exhibited normal degrees of decidual artery transformation into flaccid vessels, which is needed for maternal-fetal nutrient and gas exchange.
Fig. 3.
Placental morphology is not altered by in vivo siRNA microinjection into the implantation site. Hematoxylin and eosin-stained, full thickness, placental sections with underlying mesometrial triangles from (A) siRNA-SCR and (D) siRNA-AAH are demonstrated in the upper panels. Middle panels represent the labyrinthine region of (B) siRNA-SCR and (E) siRNA-AAH injected placentas. Interstitial edema was noted in the mesometrial triangles of both (C) siRNA-SCR and (F) siRNA-AAH injected dams. Physiologic transformation of spiral arteries was comparable in both groups. [Original magnification X20 (A, D), X400 (B–F)]
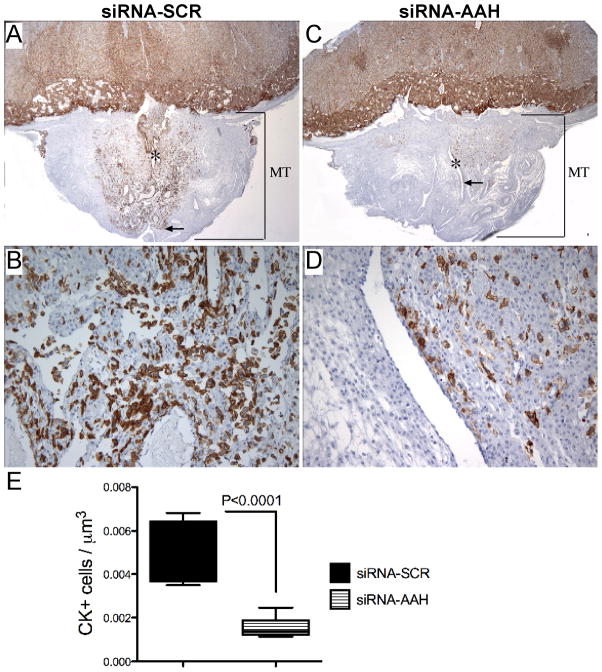
Cytokeratin (CK) immunostaining highlighted the trophoblastic cells within the placenta and mesometrial triangle. Our studies focused on the invasive throphoblastic cells in the mesometrial triangle (Fig. 4). Invasive trophoblastic cells were located at the center of the mesometrial triangle, predominantly around the decidual spiral artery. In the siRNA-SCR injected dams, the mesometrial triangles exhibited dense and widely distributed invasive trophoblastic cells (Fig. 4A), the majority of which were interstitial with foci of endovascular invasion into the spiral artery and arterioles (Fig. 4B). In siRNA-AAH injected dams, the invasive trophoblastic cells were significantly reduced in number and density, with a patchy rather than diffuse distribution in the mesometrial triangles (Fig. 4C–D). Moreover, the depth of trophoblastic cell invasion was shallower in the siRNA-AAH (Fig. 4C) relative to control group (Fig. 4A). Image analysis demonstrated significantly reduced mean densities of CK-positive cells in siRNA-AAH compared to siRNA-SCR injected mesometrial triangles (P<0.0001; Fig. 4E).
Fig. 4.
In vivo siRNA-AAH microinjection reduces trophoblastic cell invasion at the implantation site. Cytokeratin (CK) immunostained placental sections with underlying mesometrial triangle (MT) from (A–B) siRNA-SCR and (C–D) siRNA-AAH injected dams demonstrate the invasive trophoblastic cell distribution around the linear maternal spiral artery (*) as well as depth of invasion (arrow). Quantitative evaluation of the invasive trophoblastic cells was carried by stereology. Graph (E) depicts the mean ± S.E.M. corresponding to the numerical density of CK-positive cells in 8–10 samples per group. Inter-group comparison was made using Student t-test. Significant P-value is shown in the panel. [Original magnification x20 (A, C), X200 (B, D)].
Notch-signaling pathway is impaired by AAH inhibition
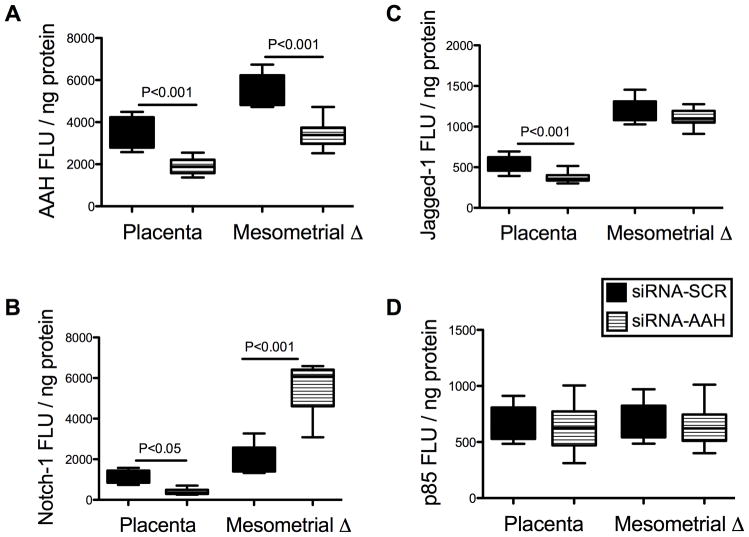
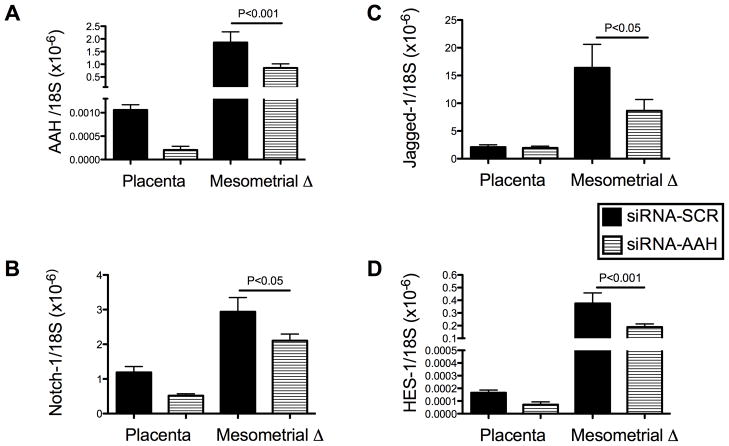
Previous studies demonstrated that AAH protein can physically interact with Notch and Jagged, and positively regulate Notch signaling [6]. To determine the role of AAH inhibition on Notch signaling, we measured Notch-1 expression by ELISA in siRNA-AAH and siRNA-SCR transfected cells. Notch-1 protein expression was significantly reduced in siRNA-AAH (1127 ± 35.65 FLU/ng protein) relative to siRNA-Scr (1597 ± 33.72 FLU/ng protein) transfected HTR-8/SVneo cells (P<0.0001). These analyses were extended to our in vivo model by measuring mesometrial triangle and placental expression of AAH, Notch-1, Jagged-1, and HES-1 by ELISA and qRT-PCR analysis. Corresponding with previous observations [21], AAH expression in siRNA-Scr injected control samples was significantly higher in mesometrial triangle tissue relative to corresponding placenta (P<0.001; Fig. 5A). In addition, Notch-1 and Jagged-1 immunoreactivities, which co-localize with AAH, were also significantly higher in mesometrial triangle compared with placental tissue (P=0.007 and P<0.0001 respectively; Fig. 5B–C). The siRNA-AAH injection significantly reduced the mean levels of AAH immunoreactivity in both mesometrial triangle and placenta relative to corresponding control tissues (F=93.04, P<0.0001). Nonetheless, the mean levels of AAH immunoreactivity were higher in mesometrial triangle relative to placenta in the siRNA-AAH injected group (F=88.02, P<0.0001; Fig. 5A). The siRNA-AAH injections also significantly altered Notch-1 protein expression (F=77.59, P<0.0001), resulting in decreased levels of immunoreactivity in placenta (P<0.05) and increased levels in the mesometrial triangle (P<0.001) (Fig. 5B). While siRNA-AAH injection reduced Jagged-1 expression (F=20.22, P<0.0001) in both tissue compartments, the inter-group difference was only significant in placental tissue (P<0.001) relative to control (Fig. 5C). In addition, we confirmed the on-target effects of siRNA duplexes by demonstrating comparable expression levels of p85 subunit of PI3 kinase in both groups by ELISA (Fig. 5D).
Fig. 5.
In vivo siRNA-AAH inhibition of Notch-Signaling Network – ELISA Studies. The implantation sites (mesometrial triangles) of GD17 pregnant Long Evans rats were microinjected with siRNA-AAH or siRNA-Scr, and 24 hours later, placental and mesometrial tissues were micro-dissected and homogenized in RIPA buffer. ELISA was used to measure levels of (A) AAH, (B) Notch-1, (C) Jagged-1, and (D) p85 (control) immunoreactivity. Results were normalized to protein content in each well. Graphs depict the mean ± S.E.M. corresponding to the levels of immunoreactivity in eight samples per group (all assays were performed in triplicate). Inter-group comparisons were made using two-way ANOVA with Bonferroni posttests. Significant P-values are shown within each panel.
Corresponding with the ELISA analysis results, qRT-PCR analysis demonstrated mRNA expression of AAH, Notch-1, Jagged-1 and HES-1 in both placental and mesometrial triangle tissues. Independent of treatment, the mean levels of AAH (F=129.0, P<0.0001), Notch-1 (F=59.25, P<0.0001), Jagged-1 (F=25.46, P<0.0001) and HES-1 (F=314.7, P<0.0001) mRNA were significantly higher in the mesometrial triangle than in placenta (Fig. 6A–D). Microinjection of siRNA-AAH significantly reduced AAH mRNA expression (F=17.74, P<0.0004) in both tissue compartments. The degree of reduction was significant at the mesometrial triangle (46%; P<0.001) relative to placenta (20%). More importantly, siRNA-AAH microinjection significantly reduced Notch-1 (F=12.0, P<0.002) and its downstream effector HES-1 (F=34.17, P<0.0001) mRNA levels in both the mesometrial triangle and placenta (Fig. 6), whereas, Jagged-1 expression was only significantly reduced in mesometrial triangle by siRNA-AAH microinjection (Fig. 6C).
Fig. 6.
In vivo siRNA-AAH inhibition of Notch-Signaling Network – qRT-PCR Studies. The implantation sites (mesometrial triangles) of GD17 pregnant Long Evans rats were microinjected with siRNA-AAH or siRNA-Scr, and 24 hours later, placental and mesometrial tissues were micro-dissected and homogenized in Qiazol buffer to extract RNA and cDNA was generated using random hexamers and the AMV 1st Strand cDNA synthesis kit. (A) AAH, (B) Notch-1, (C) Jagged-1, and (D) HES-1 gene expression was measured by qRT-PCR with results normalized to 18S rRNA. Graphs depict the mean ± S.E.M. corresponding to levels of immunoreactivity in eight samples per group (all assays were performed in triplicate). Inter-group comparisons were made using two-way ANOVA with Bonferroni posttests. Significant P-values are shown within each panel.
DISCUSSION
The findings herein demonstrated that inhibition of AAH expression by transfection of HTR-8/SVneo human trophoblastic cells impairs directional motility, which is needed for placentation. Furthermore, in vivo experiments demonstrated that microinjection of siRNA-AAH with attendant inhibition of AAH expression results in intrauterine fetal-growth restriction, reduction in trophoblastic cell invasion at the implantation site, and decreased expression of Notch signaling molecules including Notch-1, Jagged-1, and HES-1 in placenta and/or the implantation site. The significance of these studies are two-fold: 1) they demonstrate that microinjection of siRNA, and probably also recombinant plasmid DNA, can be used to interrogate short-term consequences of gene inhibition or over-expression without the need to generate transgenic lines; and 2) the findings demonstrate the importance of AAH inhibition as a mediator of intrauterine-growth restriction, placentation and constitutive impairment of Notch-signaling networks in placenta and at the site of implantation. The in vivo delivery of siRNA was easily mastered, caused minimal trauma and virtually no fetal loss, and was reproducible. Therefore it serves as a convenient model for testing hypotheses.
Trophoblastic cell motility is required for implantation and placentation [5], and previous studies showed that AAH, under the control of insulin and insulin-like growth factors (IGFs), mediates cell motility and invasion in various transformed cell types, as well as during development [6,7,13]. The fact that siRNA-AAH impaired directional motility and adhesion of HTR-8/SVneo first-trimester, human trophoblast cells, which have properties of invasive extravillous cytotrophoblasts [19], suggests that AAH has an important role in regulating placentation and probably implantation in humans. While insulin and IGFs support multiple functions, including growth, viability, metabolism, and motility, its regulation of AAH expression mediates cell motility. Therefore, inhibition of signaling through IGFs with attendant impairment of placentation, as occurs with chronic ethanol exposure [21] could be attributed to reduced AAH expression.
One of the most interesting observations in this study was that siRNA-AAH injection into the implantation site with attendant inhibition of AAH and Notch-signaling mechanisms in the mesometrial triangle and placenta impaired fetal growth, as was manifested by the significantly reduced weights and lengths of the fetuses relative to control. In accordance with in vitro gene-silencing studies, in vivo siRNA-AAH injection resulted in marked reduction in invasive trophoblastic cells at the implantation site. However, contrary to our expectations and the abnormalities produced by chronic gestational exposure to ethanol [21], we did not observe any structural abnormalities in the placenta or implantation site, and vascular transformation appeared to be intact following siRNA-AAH injection. Reduction in trophoblastic cell invasion without impairment in vascular transformation might be explained by the short duration of the experiment. We also demonstrated targeted gene inhibition with siRNA in placenta and mesometrial triangle by qRT-PCR analysis. Reductions in Notch-1, its ligand Jagged-1, and its downstream effector HES-1 mRNA expression in siRNA-AAH injected placentas and mesometrial triangles, suggest that Notch transcriptional activity is regulated by AAH. Moreover, the finding of decreased HES-1 expression in the context of decreased Notch-1 mRNA expression suggests that Notch signaling is clearly suppressed by AAH inhibition because Notch functions by activating HES gene transcription [24]. Elevated Notch-1 protein levels in the mesometrial triangle might be a compensatory response to reductions in its ligand Jagged. Despite elevated Notch-1 protein expression levels, the overall interaction between AAH and Notch/Jagged would be impaired, because AAH functions by hydroxylating Jagged as well as Notch [6]. In addition to their role in trophoblast motility, the potential regulatory link between AAH and Notch would have further implications in placental development, especially in vasculogenesis, which could have an important role in disease processes such as preeclampsia. Moreover, interactions between AAH and Notch signaling might involve unknown mechanisms involving pathways affected by AAH.
In the absence of histologic evidence for impaired placentation, the effects of siRNA-AAH on fetal growth were likely functional and might be related to impairments in Notch signaling. Targeted deletion of Notch-1 causes embryonal growth arrest either at, or shortly before E9.5 [31]; however, its role in relation to fetal growth at later stages of development is largely unknown. Members of Notch-signaling pathway are critical for chorioallontoic attachment (E8.5) leading to angiogenic vascular remodeling in placenta [18]. In our study, placental development was beyond this stage and vascular morphogenesis of the placenta was not altered. Future studies will determine the role of endothelial cell function and/or maternal-fetal transport of nutrients in this setting.
In conclusion, AAH is an important regulator of trophoblastic cell motility, and it mediates intrauterine fetal growth. Intrauterine microinjection of rat implantation sites with siRNA-AAH reduces Notch-signaling molecules and might have further implications for placental development and/or fetal growth.
Acknowledgments
Supported by AA-016783, AA-11431, AA-12908, and AA-16126 from the National Institutes of Health
References
- 1.Altman PL, Dittmer DS. Biology Data Book. Federation of American Societies for Experimental Biology; Maryland: 1972. pp. 178–180. [Google Scholar]
- 2.Alva JA, Iruela-Arispe ML. Notch signaling in vascular morphogenesis. Curr Opin Hematol. 2004;11:278–83. doi: 10.1097/01.moh.0000130309.44976.ad. [DOI] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–32. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 4.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 5.Bischof P, Irminger-Finger I. The human cytotrophoblastic cell, a mononuclear chameleon. Int J Biochem Cell Biol. 2005;37:1–16. doi: 10.1016/j.biocel.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Cantarini MC, de la Monte SM, Pang M, Tong M, D’Errico A, Trevisani F, Wands JR. Aspartyl-asparagyl beta hydroxylase over-expression in human hepatoma is linked to activation of insulin-like growth factor and notch signaling mechanisms. Hepatology. 2006;44:446–57. doi: 10.1002/hep.21272. [DOI] [PubMed] [Google Scholar]
- 7.Carter JJ, Tong M, Silbermann E, Lahousse SA, Ding FF, Longato L, Roper N, Wands JR, de la Monte SM. Ethanol impaired neuronal migration is associated with reduced aspartyl-asparaginyl-beta-hydroxylase expression. Acta Neuropathol. 2008;116:303–15. doi: 10.1007/s00401-008-0377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christiansen JH, Coles EG, Wilkinson DG. Molecular control of neural crest formation, migration and differentiation. Curr Opin Cell Biol. 2000;12:719–24. doi: 10.1016/s0955-0674(00)00158-7. [DOI] [PubMed] [Google Scholar]
- 9.Cobellis L, Mastrogiacomo A, Federico E, Schettino MT, De Falco M, Manente L, Coppola G, Torella M, Colacurci N, De Luca A. Distribution of Notch protein members in normal and preeclampsia-complicated placentas. Cell Tissue Res. 2007;330:527–34. doi: 10.1007/s00441-007-0511-6. [DOI] [PubMed] [Google Scholar]
- 10.Cohen AC, Tong M, Wands JR, de la Monte SM. Insulin and insulin-like growth factor resistance with neurodegeneration in an adult chronic ethanol exposure model. Alcohol Clin Exp Res. 2007;31:1558–73. doi: 10.1111/j.1530-0277.2007.00450.x. [DOI] [PubMed] [Google Scholar]
- 11.De Falco M, Cobellis L, Giraldi D, Mastrogiacomo A, Perna A, Colacurci N, Miele L, De Luca A. Expression and distribution of notch protein members in human placenta throughout pregnancy. Placenta. 2007;28:118–26. doi: 10.1016/j.placenta.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 12.de la Monte SM, Lahousse SA, Carter J, Wands JR. ATP luminescence-based motility-invasion assay. Biotechniques. 2002;33:98–100. 102. doi: 10.2144/02331rr01. 104 passim. [DOI] [PubMed] [Google Scholar]
- 13.de la Monte SM, Tamaki S, Cantarini MC, Ince N, Wiedmann M, Carter JJ, Lahousse SA, Califano S, Maeda T, Ueno T, D’Errico A, Trevisani F, Wands JR. Aspartyl-(asparaginyl)-beta-hydroxylase regulates hepatocellular carcinoma invasiveness. J Hepatol. 2006;44:971–83. doi: 10.1016/j.jhep.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 14.de la Monte SM, Tong M, Carlson RI, Carter JJ, Longato L, Silbermann E, Wands JR. Ethanol inhibition of aspartyl-asparaginyl-beta-hydroxylase in fetal alcohol spectrum disorder: potential link to the impairments in central nervous system neuronal migration. Alcohol. 2009;43:225–40. doi: 10.1016/j.alcohol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinchuk JE, Focht RJ, Kelley JA, Henderson NL, Zolotarjova NI, Wynn R, Neff NT, Link J, Huber RM, Burn TC, Rupar MJ, Cunningham MR, Selling BH, Ma J, Stern AA, Hollis GF, Stein RB, Friedman PA. Absence of post-translational aspartyl beta-hydroxylation of epidermal growth factor domains in mice leads to developmental defects and an increased incidence of intestinal neoplasia. J Biol Chem. 2002;277:12970–7. doi: 10.1074/jbc.M110389200. [DOI] [PubMed] [Google Scholar]
- 16.Dinchuk JE, Henderson NL, Burn TC, Huber R, Ho SP, Link J, O’Neil KT, Focht RJ, Scully MS, Hollis JM, Hollis GF, Friedman PA. Aspartyl beta -hydroxylase (Asph) and an evolutionarily conserved isoform of Asph missing the catalytic domain share exons with junctin. J Biol Chem. 2000;275:39543–54. doi: 10.1074/jbc.M006753200. [DOI] [PubMed] [Google Scholar]
- 17.Ehebauer M, Hayward P, Arias AM. Notch, a universal arbiter of cell fate decisions. Science. 2006;314:1414–5. doi: 10.1126/science.1134042. [DOI] [PubMed] [Google Scholar]
- 18.Gasperowicz M, Otto F. The notch signalling pathway in the development of the mouse placenta. Placenta. 2008;29:651–9. doi: 10.1016/j.placenta.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Graham CH, Hawley TS, Hawley RG, MacDougall JR, Kerbel RS, Khoo N, Lala PK. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp Cell Res. 1993;206:204–11. doi: 10.1006/excr.1993.1139. [DOI] [PubMed] [Google Scholar]
- 20.Gundogan F, Elwood G, Greco D, Rubin LP, Pinar H, Carlson RI, Wands JR, de la Monte SM. Role of aspartyl-(asparaginyl) beta-hydroxylase in placental implantation: relevance to early pregnancy loss. Hum Pathol. 2007;38:50–9. doi: 10.1016/j.humpath.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Gundogan F, Elwood G, Longato L, Tong M, Feijoo A, Carlson RI, Wands JR, de la Monte SM. Impaired placentation in fetal alcohol syndrome. Placenta. 2008;29:148–57. doi: 10.1016/j.placenta.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gundogan F, Elwood G, Mark P, Feijoo A, Longato L, Tong M, de la Monte SM. Ethanol-Induced Oxidative Stress and Mitochondrial Dysfunction in Rat Placenta: Relevance to Pregnancy Loss. Alcohol Clin Exp Res. 2010;34:415–423. doi: 10.1111/j.1530-0277.2009.01106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ince N, de la Monte SM, Wands JR. Overexpression of human aspartyl (asparaginyl) beta-hydroxylase is associated with malignant transformation. Cancer Res. 2000;60:1261–6. [PubMed] [Google Scholar]
- 24.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–55. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 25.Jia S, VanDusen WJ, Diehl RE, Kohl NE, Dixon RA, Elliston KO, Stern AM, Friedman PA. cDNA cloning and expression of bovine aspartyl (asparaginyl) beta-hydroxylase. J Biol Chem. 1992;267:14322–7. [PubMed] [Google Scholar]
- 26.Lavaissiere L, Jia S, Nishiyama M, de la Monte S, Stern AM, Wands JR, Friedman PA. Overexpression of human aspartyl(asparaginyl)beta-hydroxylase in hepatocellular carcinoma and cholangiocarcinoma. J Clin Invest. 1996;98:1313–23. doi: 10.1172/JCI118918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawton M, Tong M, Gundogan F, Wands JR, Monte SM. Aspartyl-(asparaginyl) beta-hydroxylase, hypoxia-inducible factor-alpha and notch cross-talk in regulating neuronal motility. Oxid Med Cell Longev. 2010;3:347–56. doi: 10.4161/oxim.3.5.13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda T, Sepe P, Lahousse S, Tamaki S, Enjoji M, Wands JR, de la Monte SM. Antisense oligodeoxynucleotides directed against aspartyl (asparaginyl) beta-hydroxylase suppress migration of cholangiocarcinoma cells. J Hepatol. 2003;38:615–22. doi: 10.1016/s0168-8278(03)00052-7. [DOI] [PubMed] [Google Scholar]
- 29.Monkovic DD, VanDusen WJ, Petroski CJ, Garsky VM, Sardana MK, Zavodszky P, Stern AM, Friedman PA. Invertebrate aspartyl/asparaginyl beta-hydroxylase: potential modification of endogenous epidermal growth factor-like modules. Biochem Biophys Res Commun. 1992;189:233–41. doi: 10.1016/0006-291x(92)91549-6. [DOI] [PubMed] [Google Scholar]
- 30.Sepe PS, Lahousse SA, Gemelli B, Chang H, Maeda T, Wands JR, de la Monte SM. Role of the aspartyl-asparaginyl-beta-hydroxylase gene in neuroblastoma cell motility. Lab Invest. 2002;82:881–91. doi: 10.1097/01.lab.0000020406.91689.7f. [DOI] [PubMed] [Google Scholar]
- 31.Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–19. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 32.Vercruysse L, Caluwaerts S, Luyten C, Pijnenborg R. Interstitial trophoblast invasion in the decidua and mesometrial triangle during the last third of pregnancy in the rat. Placenta. 2006;27:22–33. doi: 10.1016/j.placenta.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Weinmaster G. The ins and outs of notch signaling. Mol Cell Neurosci. 1997;9:91–102. doi: 10.1006/mcne.1997.0612. [DOI] [PubMed] [Google Scholar]