Abstract
We analyzed the frequency of four mitochondrial DNA haplogroups in 424 individuals from 21 Colombian Amerindian tribes. Our results showed a high degree of mtDNA diversity and genetic heterogeneity. Frequencies of mtDNA haplogroups A and C were high in the majority of populations studied. The distribution of these four mtDNA haplogroups from Amerindian populations was different in the northern region of the country compared to those in the south. Haplogroup A was more frequently found among Amerindian tribes in northern Colombia, while haplogroup D was more frequent among tribes in the south. Haplogroups A, C and D have clinal tendencies in Colombia and South America in general. Populations belonging to the Chibcha linguistic family of Colombia and other countries nearby showed a strong genetic differentiation from the other populations tested, thus corroborating previous findings. Genetically, the Ingano, Paez and Guambiano populations are more closely related to other groups of south eastern Colombia, as also inferred from other genetic markers and from archeological data. Strong evidence for a correspondence between geographical and linguistic classification was found, and this is consistent with evidence that gene flow and the exchange of customs and knowledge and language elements between groups is facilitated by close proximity.
Keywords: mitochondrial DNA, Amerindian, Colombia, Chibcha, genetic relationships
Introduction
Studies about genetic variation among human populations are of great value for understanding genetic structure, migration routes and possible genetic relationships among different continental populations, and mitochondrial DNA (mtDNA) analysis has frequently put to such use in American populations (Schurr et al., 1990; Torroni et al., 1992, 1993a,b, 1994; Horai et al., 1993; Bailliet et al., 1994; Merriwether et al., 1994; Santos et al., 1994a,b; Bianchi et al., 1995; Lorenz and Smith, 1996; Merriwether and Ferrell, 1996; Bonatto and Salzano, 1997; Bisso-Machado et al., 2012). Despite its maternal inheritance (Giles et al., 1980), the mitochondrial genome is extremely useful for determining genetic histories because of its rapid rate of mutation (Brown et al., 1979) and lack of recombination and repair mechanisms. Most mtDNA polymorphisms are single nucleotide substitutions, but insertions and deletions have also been described (Brown et al., 1980; Cann and Wilson 1983; Cann et al., 1984; Wallace et al., 1985; Horai et al., 1993; Torroni et al., 1992, 1993a,b, 1994; Howell and Smejkal, 2000;). By revealing specific geographic locations for mitochondrial haplogroups, such studies helped to clarify migration patterns of human populations throughout history and over all continents (Fernandez-Dominguez, 2005).
Previous studies based on mtDNA analysis in Native American populations revealed the presence of four distinct haplogroups called A, B, C and D. Haplogroup A is characterized by the gain of a HaeIII restriction site at position 663, haplogroup B by the 9 bp COII/tARNlys intergenic deletion, and haplogroup C by the loss of a HincII site at 13259 bp. Haplogroup D is characterized by the loss of an AluI restriction site at position 5176 and a gain of a HincII site at 13259 bp (Torroni et al., 1992, 1993a,b. A fifth haplogroup, X, has been predominantly characterized in some primarily North American populations (Eshleman et al., 2003), but is absent in South America (Dornelles et al., 2005).
Colombia has great cultural and genetic diversity. Its indigenous population is distributed in 89 different ethnic groups which are estimated to represent 1.83% of the total population (Arango and Sánchez, 2006). Based on the theory that peopling of the Americas occurred by migration from northeast Asia across the Bering Strait and subsequent migration through Central America to South America (Turner, 1984; Greenberg et al., 1986; Dillehay and Meltzer, 1991), the nowadays Colombian territory at the northern tip of South America became an obligatory passage for people migrating to the southern cone.
In Colombia, several mtDNA studies of indigenous communities have been carried out (Mesa et al., 2000; Keyeux et al., 2002; Rodas et al., 2002; Torres et al., 2006; Melton et al., 2007; Rondon et al., 2007). In this study, we analyzed 424 individuals from 21 Amerindian populations to determine genetic structure and relationships among them based on geographical and historical information, as well as linguistic and genetic relationships with other tribes of the Americas.
Subjects and Methods
Samples
We analyzed 424 blood samples from individuals unrelated by maternal lineage from 21 Amerindian tribes of Colombia (Table 1). Blood samples were collected between 1989 and 1992 after proper informed consent had been obtained. Informed consent included approval of each tribal Chief or Governor. The linguistic affiliation of each tribe is shown in Table 1. No Ge-Pano Carib speaking tribes were included in this study (Table 1).
Table 1.
Geographic location, sample size and linguistic classification for 21 Colombian Amerindian tribes analyzed.
| mtDNA haplogroups (%)
|
Linguistic classification a | Geographic location (Department / region) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Population | n | A | B | C | D | E | h | ||
| Chimila | 21 | 95.2 | 4.8 | 0.0 | 0.0 | 0.0 | 0.0952 | Chibcha | Magdalena/ Caribbean region |
| Arsario | 8 | 0.0 | 12.5 | 75.0 | 0.0 | 12.5 | 0.4643 | Chibcha | Magdalena / Caribbean region |
| Kogui | 32 | 71.9 | 0.0 | 28.1 | 0.0 | 0.0 | 0.4173 | Chibcha | Cesar / Caribbean region |
| Arhuaco | 21 | 90.4 | 4.8 | 4.8 | 0.0 | 0.0 | 0.1857 | Chibcha | Cesar / Caribbean region |
| Embera | 21 | 9.5 | 52.4 | 28.6 | 9.5 | 0.0 | 0.6571 | Choco-Chibcha | Chocó / Pacific region |
| Ingano | 48 | 39.6 | 35.4 | 22.9 | 2.1 | 0.0 | 0.5326 | Andean | Putumayo / Amazonian region |
| Guambiano | 24 | 4.2 | 12.5 | 66.6 | 16.7 | 0.0 | 0.6791 | Without Classification | Cauca /Pacific region |
| Páez | 36 | 27.8 | 8.3 | 27.8 | 33.3 | 2.8 | 0.7476 | Without Classification | Cauca /Pacific region |
| Wayuu | 17 | 29.4 | 17.6 | 47.1 | 0.0 | 5.9 | 0.6985 | Tucano-ecuatorial | Guajira / Caribbean region |
| Tucano | 14 | 7.1 | 21.4 | 7.1 | 57.1 | 7.1 | 0.6593 | Tucano-ecuatorial | Vaupés-Guainía/ Orinoquian region |
| Guanana | 10 | 20.0 | 0.0 | 20.0 | 50.0 | 10.0 | 0.7333 | Tucano-ecuatorial | Vaupés-Guainía/ Orinoquian region |
| Cubeo | 24 | 33.3 | 8.3 | 29.2 | 25.0 | 4.2 | 0.7645 | Tucano-ecuatorial | Vaupés-Guainía/ Orinoquian region |
| Curripaco | 22 | 4.5 | 40.9 | 36.4 | 13.7 | 4.5 | 0.7100 | Tucano-ecuatorial | Vaupés-Guainía/ Orinoquian region |
| Desano | 20 | 15.0 | 15.0 | 45.0 | 25.0 | 0.0 | 0.7263 | Tucano-ecuatorial | Vaupés-Guainía/ Orinoquian region |
| Barasano | 5 | 80.0 | 0.0 | 0.0 | 20.0 | 0.0 | 0.4000 | Tucano-ecuatorial | Vaupés / Orinoquian region |
| Tatuyo | 10 | 40.0 | 0.0 | 50.0 | 10.0 | 0.0 | 0.6444 | Tucano-ecuatorial | Vaupés / Orinoquian region |
| Piratapuyo | 8 | 12.5 | 12.5 | 50.0 | 12.5 | 12.5 | 0.7857 | Tucano-ecuatorial | Vaupés / Orinoquian region |
| Tuyuca | 6 | 17.0 | 50.0 | 0.0 | 17.0 | 16.0 | 0.8000 | Tucano-ecuatorial | Vaupés / Orinoquian Region |
| Puinave | 61 | 8.2 | 50.8 | 32.8 | 6.6 | 1.6 | 0.6333 | Tucano-ecuatorial | Guainía / Orinoquian region |
| Yeral | 8 | 12.5 | 12.5 | 50.0 | 25.0 | 0.0 | 0.7500 | Tucano-ecuatorial | Guainía / Orinoquian region |
| Piapoco | 8 | 12.5 | 25.0 | 25.0 | 12.5 | 25.0 | 0.8929 | Tucano-ecuatorial | Guainía / Orinoquian region |
| Frequency | 31.0 | 22.4 | 30.4 | 13.4 | 2.8 | 0.7447 | |||
| Total | 424 | 131 | 95 | 129 | 57 | 12 | |||
Linguistic affiliations (Ruhlen, 1987).
DNA extraction and mtDNA haplogroup analysis
DNA was extracted using the salting out method (Gustincich et al., 1991) with the DNA Wizard Genomic DNA Extraction Kit (Promega Corporation, Madison WI), following manufacturer’s recommendations.
Four regions of the human mtDNA representing mtDNA haplogroups A, B, C and D were PCR amplified with the use of primers that were described elsewhere (Parra et al., 1998).
Each amplification reaction consisted of 2.5 μL of DNA, 1.25 μL of each set of primers (10 nmol/μL), 2.0 μL of dNTPs (10 mM), and 0.125 μL of DNA Taq polymerase (Promega Corporation, Madison WI). The reaction mixture also contained 1.5 μL of MgCl2 (25 mM) for haplogroup A, and 2.0 μL of MgCl2 (25 mM) for the other haplogroups, respectively, in a final volume of 25 μL.
Amplification conditions consisted of a first denaturing cycle at 94 °C for 5 min; followed by 34 cycles of denaturing at 94 °C for 30 s, annealing at 50 °C for 30 s (Haplogroups B and D) or at 55 °C for 30 s (haplogroups A and C), extension at 72 °C for 30 s, and a final extension step at 72 °C for 5 min. The amplification products were evaluated by electrophoresis in a 2% agarose Nusieve/Seakem gel that was stained with ethidium bromide and photographed under UV light. 15 μL aliquots of the amplified products for groups A, C and D were digested with restriction enzymes for 3 h at 37 °C, while haplogroup B was only analyzed by electrophoresis. The digestion products were separated by electrophoresis in a 3% Nusieve/Seakem gel and processed as described above.
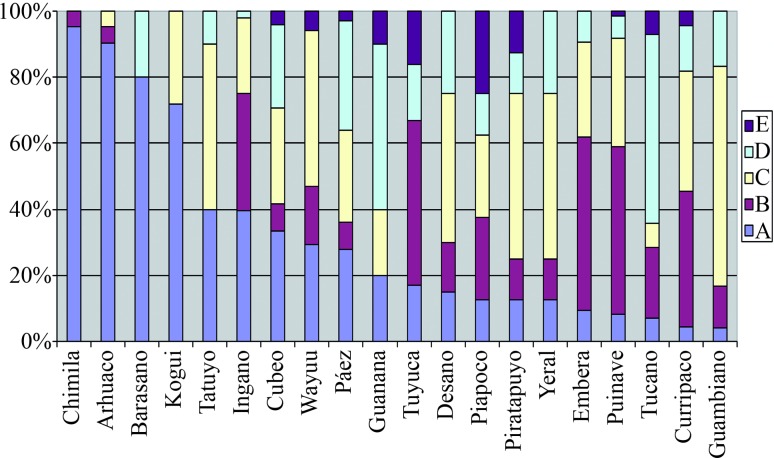
The haplogroup frequency of each population was estimated by direct counting (Table 1, Figure 1). Genetic diversity was estimated as (n/(n−1))(1−∑pi2), where n is the sample size and pi the haplogroup frequency estimate for haplogroup i (Nei, 1978). Genetic distance estimates were based on mtDNA haplogroup frequencies calculated from FST pairs with the aid of Arlequin software (Excoffier et al., 2005). Frequency data for mtDNA haplogroups belonging to Amerindian populations of South and Central America used in the analysis were obtained from the literature, see Table S1 (Ginther et al., 1993; Horai et al., 1993; Torroni et al., 1993a, 1994; Bailliet et al., 1994; Santos et al., 1994a; Bianchi et al., 1995; Kolman et al., 1995; Merriwether et al., 1995, 1997; Easton et al., 1996; Lalueza-Fox, 1996; Ward et al., 1996; Bonatto and Salzano, 1997; Kolman and Bermingham, 1997; Lalueza et al., 1997; Dipierri et al., 1998; Rickards et al., 1999; Mesa et al., 2000; Moraga et al., 2000; Bert et al., 2001; Demarchi et al., 2001; Lobato-da-Silva et al., 2001; Rothhammer et al., 2001; Keyeux et al., 2002; Williams et al., 2002; Briceño et al., 2003; Fuselli et al., 2003; Garcia-Bour et al., 2004; Lewis et al., 2004; Sandoval et al., 2004; Dornelles et al., 2005; Cabana et al., 2006; Torres et al., 2006; Marrero et al., 2007; Melton et al., 2007; Barreto et al., 2008). The results are presented in Figures 2 and 3.
Figure 1.
Mitochondrial DNA (mtDNA) haplogroup frequency for 21 Amerindian populations of Colombia.
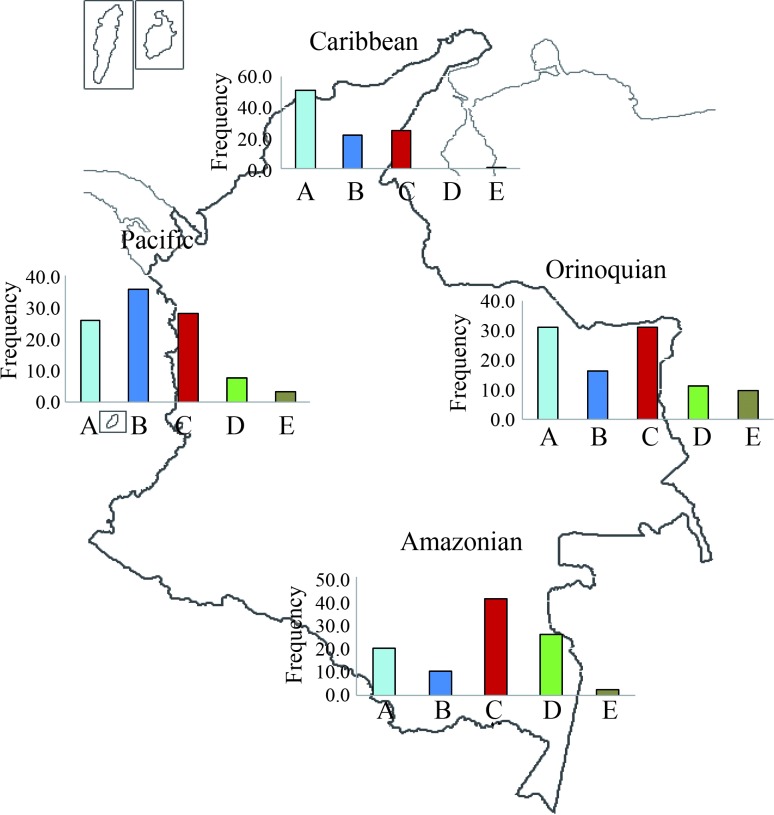
Figure 2.
mtDNA Haplogroup distribution of Amerindian tribes in Colombia according to their geographic location. Data reported by Torroni et al. (1994), Kolman and Bermingham (1997), Merriwether et al. (1997), Mesa et al. (2000), Keyeux et al. (2002), Briceño et al. (2003), Barreto et al. (2006), Torres et al. (2006) and Melton et al. (2007) was also included.
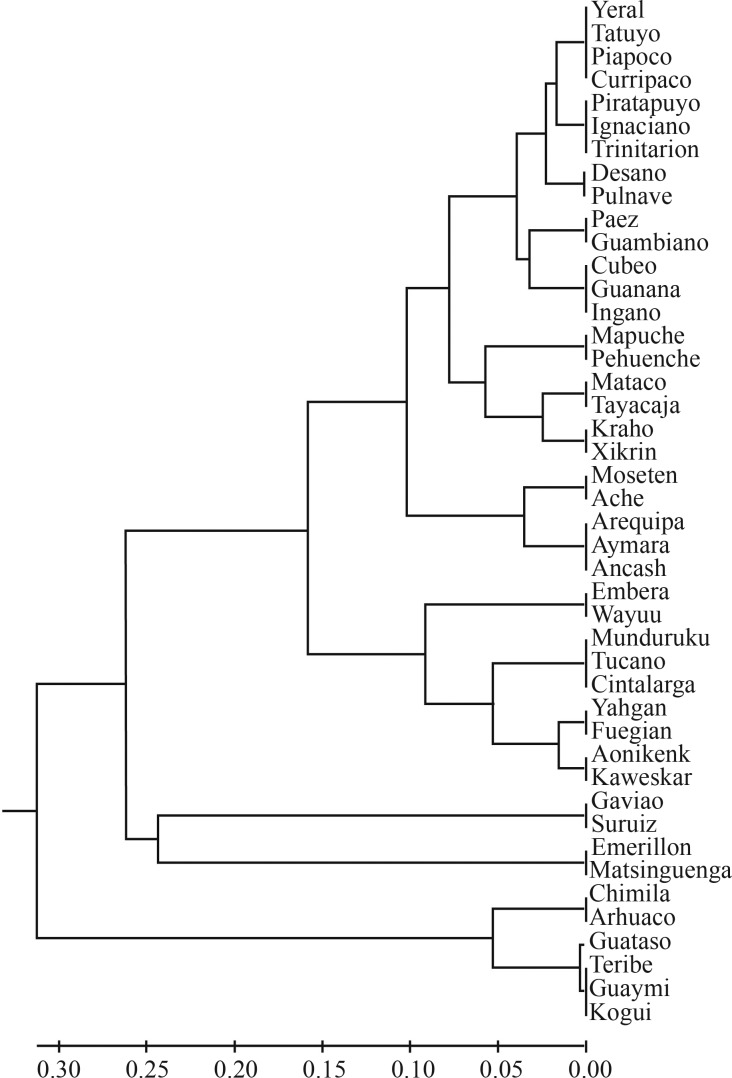
Figure 3.
UPGMA tree based on FST distances for Colombian and Central and South American Amerindian tribes analyzed for mtDNA haplogroups.
In addition we calculated the degree of genetic differentiation among subpopulations (GST) based on the genetic diversity of the total population. An AMOVA analysis using Arlequin (Excoffier et al., 2005) was carried out using linguistic classification or geographical location as testing parameters. In the first analysis, we evaluated the linguistic classification of each tribe, and whether differences could be attributed to belonging or not to the Chibcha speaking family. In the second analysis, we tested groups by geographic location (Tribes located in the north; tribes located in the east-Orinoquian/Amazonian region, and tribes located in the Pacific region-west). We also conducted a comparison to determine if the Andes mountain range was a factor in genetic differentiation (Table 2).
Table 2.
Analysis of Molecular Variance (AMOVA) for mtDNA haplogroups. Grouping based on linguistics or geographical criteria for Colombian Amerindian tribes.
| Groups | AMOVA values (%) | Fixation index | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| a | b | c | FSC | FST | FCT | |
| 1. Chibchas and Non-Chibcha | 69.06 | 9.87 | 21.07 | 0.12505 * |
0.30939 * |
0.21069 (p = 0.001) |
| 2. All linguistic families | 77.26 | 15.83 | 6.91 | 0.17003 * |
0.22739 * |
0.06911 (p = 0.082) |
| 3. North East and Pacific | 72.54 | 11.26 | 16.2 | 0.13435 * |
0.27458 * |
0.162 (p = 0.013) |
| 4. Andes barrier | 79.2 | 17.66 | 3.14 | 0.18229 * |
0.20796 * |
0.03139 (p = 0.150) |
AMOVA analysis based on linguistic classification (groups 1 and 2) and geographical location (3 and 4). Group 1:Chibcha (Arsario, Kogui, Arhuaco, Chimila); Non-Chibcha (Wayuu, Curripaco, Piapoco, Yeral, Cubeo, Desano, Tatuyo, Piratapuyo, Puinave, Guanana, Ingano, Tucano). Group 2: Chibcha (Arsario, Arhuaco, Kogui, Chimila, Embera); Ecuatorial -Tucano: (Wayuu, Curripaco, Piapoco, Yeral, Cubeo, Desano, Tatuyo, Piratapuyo, Puinave, Guanana, Tucano); Andean: (Ingano). Group 3: North (Kogui, Chimila, Arsario, Arhuaco, Wayuu); Pacific (Paéz, Guambiano, Embera); East (Ingano, Cubeo, Curripaco, Desano, Yeral, Tucano, Piapoco, Puinave, Guanana, Tatuyo, Piratapuyo). Group 4: Andean mountains: (Kogui, Arhuaco, Arsario, Chimila, Wayuu, Embera, Guambiano, Paéz); Amazonian/Orinoquian (Ingano, Guanana, Curripaco, Desano, Cubeo, Tatuyo, Piratapuyo, Yeral, Piapoco, Puinave, Tucano).
Finally, we compared the genetic (FST values), geographical (distance in km using the AMIGLOBE program) (Collard, 2006) and linguistic distance based on Ruhlen’s classification (Ruhlen, 1987) matrices to calculate a possible relationship between these three variables. This was done with the aid of Arlequin, V3.1 software (Excoffier et al., 2005) by using the Mantel test with 100,000 permutations (Figure 3).
Results
Mitochondrial DNA haplogroup frequencies from 424 individuals belonging to 21 Amerindian tribes of Colombia are shown in Table 1 and Figure 1. Haplogroup A was found most frequently; its average frequency was 31% (131/424 individuals), followed by haplogroup C with 30.4% (129/424), haplogroup B with 22.4% (95/424) and haplogroup D with 13.4% (57/424). The 12 out of 424 individuals who did not show any of the four mtDNA founder haplogroups (2.8%) were listed as haplogroup E. At least two of four mitochondrial haplogroups were present in the 21 populations studied. The frequency distribution of these haplogroups ranged from 2.1% to 95.2%.
Genetic diversity index values are shown in Table 1. The least genetic diversity was found among the Chimila tribe (h = 0.0952) while the greatest one was found among the Piapoco (h = 0.8929). The average diversity index for all populations studied was h = 0.7447 (n = 424).
Figure 2 shows the mtDNA haplogroup frequency distribution based on four geographical location groups: Caribbean (northern region), Amazonian (southern region), Pacific (western region) and Orinoquian (eastern region). We also included data from other studies (Torroni et al., 1994; Kolman and Bermingham, 1997; Merriwether et al., 1997; Mesa et al., 2000; Keyeux et al., 2002; Briceño et al., 2003; Torres et al., 2006; Melton et al., 2007; Barreto et al., 2008) in this analysis. There was a marked clinal pattern for mtDNA haplogroup distribution among Amerindian tribes of Colombia. Haplogroup A frequency was higher in the northern region of Colombia (50% frequency) decreasing to 20% in the southern region of the country while haplogroup C frequency was lower in the north and highest in the south. The pattern for haplogroup D was similar, being almost absent in the northern part of Colombia, and showing the highest value in the southern part of the country (25%). Haplogroup B was more frequent in the west, declining towards the east and south.
We constructed a UPGMA tree based on FST genetic distances which includes other Amerindian populations from Central and South America (Figure 3). One cluster included the Kogui, Arhuaco and Chimila tribes of Colombia and the Teribe, Guaymi and Guataso Chibcha-speaking tribes of Central America, which are all characterized by high frequencies of haplogroup A. An exception was found for the Arsario tribe, where none of the individuals tested in this Chibcha speaking tribe carried haplogroup A. The remaining Colombian tribes clustered together with other Amerindian tribes of South America that do not belong to the Chibcha linguistic family. The Guambianos, Paez and Ingano tribes were grouped within this cluster, reflecting their relationships to these non-Chibcha Amerindian populations.
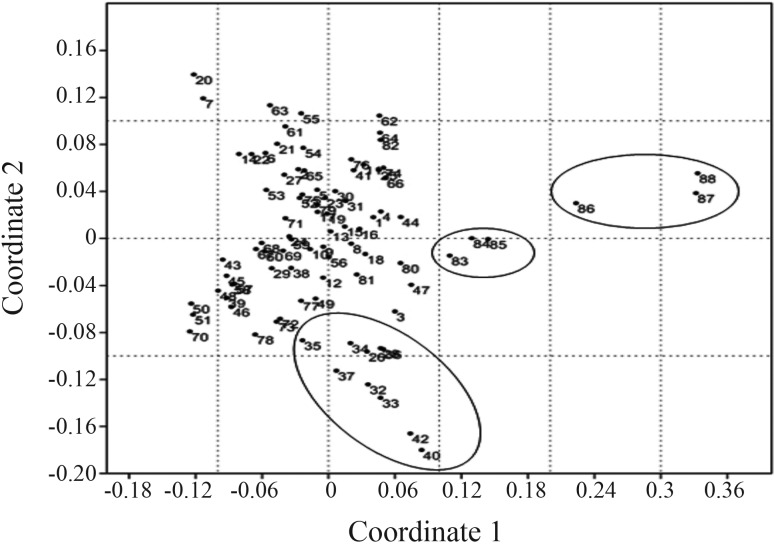
We performed a non-metric multidimensional scaling analysis based on the mtDNA haplogroups identified (Figure 4,). Herein we included the results of other Amerindian populations (Table S1) as well as populations of African descent of Colombia (Nuqui, Guangui and Providencia) described by Rodas et al. (2002), and African populations as an outgroup (Chen et al., 2000) (Figure 4). Most of the Amerindian tribes are clustered together due to the heterogeneous presence of the four mtDNA haplogroups among them. However, the Chibcha speaking tribes have a tendency to cluster much closer together due to the high frequency of haplogroup A and low frequencies for haplogroups C and D. The African descent populations from Colombia are located intermediately between the Amerindian populations and the African population used as outgroup. This is due to the admixture process that resulted in the presence of some of the four mtDNA haplogroups among the Colombian African-descent populations.
Figure 4.
NM-MDS plot of Amerindian populations from Central and South America, Colombian African descendant and African populations. The lower circle includes most of the Chibcha Speaking groups. The circle in the middle includes the populations of African descent in Colombia, and the circle to the left includes African populations used as outgroup. For haplogroup data see Table S1.
The AMOVA analysis based on linguistic affiliation was used to test for differences based on belonging or not to the Chibcha linguistic family (Table 2). The Guambiano and Paez tribes were not included since their languages have not been classified yet. The results showed that 69% variations were due to variations within populations and 21% was due to whether or not a tribe belonged to the Chibcha linguistic family (p < 0.001). Another AMOVA analysis based on the geographical location of Colombian Amerindian tribes detected no significant differences when the tribes were grouped according to the side of the Andes mountain range they were located. Significant differences were found among tribes residing in the northern part of Colombia (most of the Chibcha speaking tribes analyzed here), compared to the Pacific region and the Orinoquian/Amazonian region (p = 0.013), but not so for the Andes as a separating barrier (p = 0.150) (Table 2).
Finally, the Mantel test was used to evaluate the possible relationship between genetic, linguistic and geographical distance. There was a strong correlation between linguistic and geographic distances, and a less strong correlation between genetic and geographic distances. There was no correlation between genetic and linguistic distance (Table 3).
Table 3.
Mantel test correlating genetic, linguistic and geographical distances for 21 Colombian Amerindian populations studied.
| Matrix 1 | Matrix 2 | r | p-value |
|---|---|---|---|
| Genetic | Linguistic | 0.156272 | 0.06825 |
| Genetic | Geographic | 0.287512 | 0.00939 |
| Linguistic | Geographic | 0.648335 | 0.00001 |
Discussion
This study provides additional information on mtDNA haplogroup distribution in several Colombian Amerindian populations to previous studies (Keyeux et al., 2002). Haplogroup A, with an average frequency of 31% (131/424 individuals) was found most frequently. It was followed by haplogroup C with 30.4% (129/424), haplogroup B with 22.4% (95/424) and haplogroup D with 13.4% (57/424).
Previous studies of Colombian Amerindian populations have shown high frequencies of haplogroups A and C and lower frequencies for haplogroup D (Keyeux et al., 2002; Torres et al., 2006; Melton et al., 2007; Rondon et al., 2007). Our results are in agreement with those reports. However, the 13.4% average haplogroup D frequency we found was higher than that previously published for Colombian Amerindian populations of 6.6% by Keyeux et al. (2002) and 9.95% by Torres et al. (2006). These differences could be attributed to the fact that these three studies chose different populations to study, or may even be due to differences within groups of the same population. For example, Keyeux et al. (2002) found no haplogroup D in the Paez tribe, whereas we found this haplogroup in 33% of the Paez individuals. Similar situations occurred in the cases of the other haplogroups. For instance, in our study, the Arsario tribe did not carry haplogroup A (but only 8 individuals were tested), while 68% of the individuals of the Arsario tribe tested by Melton et al. (2007) were reported to carry haplogroup A. These results indicate an even greater genetic heterogeneity within the same populations than has been described before.
Only 12 out of 424 individuals showed none of the four founder mtDNA haplogroups (2.8%). These individuals may either have unrecognized founder lineages (Bailliet et al., 1994), recent racial admixture (Torroni et al., 1993a) or reversal of a mutation. The second possibility could be the case for the Wayuu, Arsario and Paez tribes in which admixture has been documented by blood groups and HLA class II genes (Yunis et al., 1994, 2001). The third possibility, which is termed haplogroup C revertant, is common in populations found in the Colombian Orinoquian and Amazonian basin (Torres et al., 2006). This may be the case for the Piapoco tribe of our study that showed a 25% frequency of non A–D haplogroups. A high frequency (59%) for the revertant C haplogroup had previously been found by Torres et al. (2006) for this tribe. The same scenario is possible for the Piartapuyo (12.5%), Tuyuca (16%) and Guanana (10%) Amerindian tribes that live geographically close together in the Northern Amazonian region of Colombia. They present low genetic admixture based on Y STR haplotypes (Campo, D and, JJY, unpublished data) and HLA Class II genes (unpublished data).
The Amerindian tribes considered in this study showed a high degree of genetic heterogeneity (Table 2) and diversity (similar to or greater than populations found throughout South America) as has been described before (Santos et al., 1994a; Batista et al., 1995; Kolman et al., 1995; Ward et al., 1996; Bonatto and Salzano, 1997; Mesa et al., 2000; Keyeux et al., 2002).
Genetic diversity values were higher among the Tucano-Equatorial speaking tribes (0.60 to 0.80) while the Chibchan-speaking groups showed lower values (0.09 to 0.50). These results are consistent with those reported for Chibcha speaking tribes from Central and South America, including Colombia (Torroni et al., 1994; Kolman et al., 1995; Keyeux et al., 2002). The higher diversity values found in Amazonian populations may be a result of gene flow between these populations, as has been shown for other genetic markers such as the Y-chromosome (Mesa et al., 2000). Alternately, it could be the result of fission, fragmentation and founder effects (Cavalli-Sforza et al., 1992). The population that showed the lowest genetic diversity value (and the highest frequency for haplogroup A) was the Chimila (h = 0.0952). The low diversity found in this population has been reported by others (Keyeux et al., 2002) looking at different genetic markers and is probably due to inbreeding (unpublished data).
The high genetic diversity found in our study and others indicates that it is unlikely that bottleneck events took place during the early Amerindian settlement of South America. However, it is evident that Amerindian populations located in northern Colombia that belong to the Chibcha linguistic family differ from non-Chibcha speaking tribes, as has been described before with nuclear genetic markers (Yunis et al., 1994, 2001). Previous studies have shown that Amerindian populations of northern Colombia are close to Central American tribes and North American Amerindian populations (Stone and Stoneking, 1993; Lorenz and Smith, 1996; O’Rourke et al., 2000; Keyeux et al., 2002; Melton et al., 2007). Our results provide further support indicating that Chibcha speaking tribes in Central and South America genetically differentiated from non-Chibcha speaking tribes prior to entering South America.
There were marked clinal patterns for mtDNA haplogroup distribution among Amerindian tribes of Colombia. When populations were grouped according to their geographical location (northern-Caribbean; southern-Amazonian, western-Pacific and eastern-Orinoquian) (see Figure 2), haplogroup A frequency was high in the northern part of Colombia (50% frequency) but decreased to 20% in the southern part of the country. Haplogroup C frequency was lower in the north and had its highest value in the south. Similarly, haplogroup D was almost absent in the north, but had the highest value in the southern part of the country (25%). Haplogroup B was more frequent in the west and had decreasing frequencies towards the east and south. These clinal patterns are similar to those described earlier (Torroni et al., 1994; Lalueza-Fox, 1996; Lorenz and Smith, 1996; Lalueza et al., 1997; Keyeux et al., 2002; Bisso-Machado et al., 2012).
A UPGMA tree constructed from data for the Amerindian tribes analyzed in this study plus data from several Amerindian populations from Central and South America described elsewhere showed a cluster of Chibcha speaking tribes (Chimila, Arhuaco, Kogui, Teribe, Guaymi tribes), which are genetically distant from other Amerindian tribes analyzed. The second cluster includes the remaining tribes including the Guambiano, and Paez tribes. The results for these two tribes, which currently have unclassified languages, provide further support of a genetic relationship to Tucano-Equatorial or Andean linguistic families rather than to the Chibcha linguistic family where they had been classified before. Similar results have also been obtained with HLA genes (Yunis et al., 2001). Some authors have postulated that the Paez originated from the Amazonian region and migrated northeast to their present location before the Spanish conquest (Arboleda, 1993). Recently, archeological findings in the west Amazonian region of Colombia have provided further support for an Amazonian ancestral origin of the Guambiano and Paez tribes.
The AMOVA analysis showed a significant association (p < 0.001) due to variation based on whether or not a tribe belonged to the Chibcha linguistic family (21%) (Table 2). On the other hand, our results do not support the hypothesis that the Andes mountain range served as a differentiation factor for the Amerindian tribes studied.
The strong genetic differentiation between the Chibcha and non-Chibcha speaking tribes is likely due to the high frequency of haplogroup A among these populations. Similar results were obtained in the past using the major histocompatibility complex and other genetic markers (Yunis et al., 1994, 2001).
The correlation analysis between the geographical, linguistic and genetic data (Table 3) showed the highest correlation value for the linguistic-geographical pair followed by the genetic-geographic comparison. These results are explained by the fact that many populations that belong to the same linguistic family are also geographically close, so it is difficult to infer whether there is a linguistic-genetic relationship based solely on mtDNA haplogroups. The Amerindian tribes that are closely related are also geographically close, which facilitates gene flow and exchange of customs, knowledge and languages. Both geographic and linguistic factors are associated with genetic differentiation in the Amerindian populations analyzed in Colombia. As has been found for other Amerindian tribes, these three parameters have evolved together in a historical and strongly correlated fashion.
Acknowledgments
We would like to thank all the Colombian Amerindian communities that kindly contributed by providing samples for this study. This research was financed in part by grants from Colciencias to EJY and by the Universidad Nacional de Colombia to JJY.
Footnotes
Associate Editor: Francisco Mauro Salzano
Supplementary Material
The following online material is available for this article:
Table S1 - mtDNA haplogroup frequencies of Colombian and South America Amerindian tribes.
This material is available as part of the online article from http://www.scielo.br/gmb.
References
- Arango R, Sánchez E. Los Pueblos Indígenas de Colombia en el Umbral del Nuevo Milenio. Tercer Mundo; Bogotá: 2006. p. 426. [Google Scholar]
- Arboleda JE. Inganos, Paeces y Coconucos: Notas para la Etnohistoría. Incora; Popayán: 1993. p. 218. [Google Scholar]
- Bailliet G, Rothhammer F, Carnese F, Bravi C, Bianchi N. Founder mitochondrial haplotypes in American populations. Am J Hum Genet. 1994;54:27–33. [PMC free article] [PubMed] [Google Scholar]
- Barreto G, Osorio JC, Peña AV, Garcés HA, Rondón F. Diversidad genética en poblaciones humanas de dos regiones colombianas. Colombia Médica. 2008;39:52–60. [Google Scholar]
- Batista O, Kolman CJ, Bermingham E. Mitochondrial DNA diversity in the Kuna Amerinds of Panama. Hum Mol Genet. 1995;4:921–929. doi: 10.1093/hmg/4.5.921. [DOI] [PubMed] [Google Scholar]
- Bert F, Corella A, Gene M, Perez-Perez A, Turbon D. Major mitochondrial DNA haplotype heterogeneity in highland and lowland Amerindian populations from Bolivia. Hum Biol. 2001;73:1–16. doi: 10.1353/hub.2001.0001. [DOI] [PubMed] [Google Scholar]
- Bianchi N, Baillet G, Bravi C. Peopling of the Americas as inferred through the analysis of mtDNA. Braz J Genet. 1995;18:661–668. [Google Scholar]
- Bisso-Machado R, Bortolini MC, Salzano FM. Uniparental genetic markers in South Amerindians. Genet Mol Biol. 2012;35:365–387. doi: 10.1590/S1415-47572012005000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonatto SL, Salzano FM. Diversity and age of the four major mtDNA haplogroups, and their implications for the peopling of the New World. Am J Hum Genet. 1997;61:1413–1423. doi: 10.1086/301629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briceño I, Gómez A, Lozano PAU, Mitchell RJ, Papiha S. Mitochondrial variation in Colombia: Study of matrilineal lineages among amerindian tribes; XIX International Congress of Genetics Proceedings; Melbourne. 2003. [Google Scholar]
- Brown WM, George M, Jr, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W, George M, Wilson A. Polymorphism in mitocondrial DNA of humans as revealed by restriction endonuclease analysis. Proc Natl Acad Sci USA. 1980;77:3605–3609. doi: 10.1073/pnas.77.6.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabana GS, Merriwether DA, Hunley K, Demarchi DA. Is the genetic structure of Gran Chaco populations unique? Interregional perspectives on Native South American mitochondrial DNA variation. Am J Phys Anthropol. 2006;131:108–119. doi: 10.1002/ajpa.20410. [DOI] [PubMed] [Google Scholar]
- Cann RL, Wilson AC. Length mutations in human mitochondrial DNA. Genetics. 1983;104:699–711. doi: 10.1093/genetics/104.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann RL, Brown WM, Wilson AC. Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics. 1984;106:479–499. doi: 10.1093/genetics/106.3.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Minch E, Mountain JL. Coevolution of genes and languages revisited. Proc Natl Acad Sci USA. 1992;89:5620–5624. doi: 10.1073/pnas.89.12.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, Olckers A, Schurr TG, Kogelnik AM, Huoponen K, Wallace DC. mtDNA variation in the South African Kung and Khwe-and their genetic relationships to other African populations. Am J Hum Genet. 2000;66:1362–1383. doi: 10.1086/302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarchi DA, Panzetta-Dutari GM, Motran CC, Lopez de Basualdo MA, Marcellino AJ. Mitochondrial DNA haplogroups in Amerindian populations from the Gran Chaco. Am J Phys Anthropol. 2001;115:199–203. doi: 10.1002/ajpa.1074. [DOI] [PubMed] [Google Scholar]
- Dillehay TD, Meltzer DJ. The First Americans. CRC Press; Boca Raton: 1991. p. 310. [Google Scholar]
- Dipierri JE, Alfaro E, Martinez-Marignac VL, Bailliet G, Bravi CM, Cejas S, Bianchi NO. Paternal directional mating in two Amerindian subpopulations located at different altitudes in northwestern Argentina. Hum Biol. 1998;70:1001–1010. [PubMed] [Google Scholar]
- Dornelles CL, Bonatto SL, De Freitas LB, Salzano FM. Is haplogroup X present in extant South American Indians? Am J Phys Anthropol. 2005;127:439–448. doi: 10.1002/ajpa.20103. [DOI] [PubMed] [Google Scholar]
- Easton RD, Merriwether DA, Crews DE, Ferrell RE. mtDNA variation in the Yanomami: Evidence for additional New World founding lineages. Am J Hum Genet. 1996;59:213–225. [PMC free article] [PubMed] [Google Scholar]
- Eshleman JA, Malhi RS, Smith DG. Mitochondrial DNA Studies of Native Americans: Conceptions and misconceptions of the population prehistory of the Americas. Evol Anthropol. 2003;12:7–18. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.1: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Dominguez E. Polimorfismos de DNA Mitocondrial en Poblaciones Antiguas de la Cuenca Mediterránea. Universidad de Barcelona; Barcelona: 2005. p. 670. [Google Scholar]
- Fuselli S, Tarazona-Santos E, Dupanloup I, Soto A, Luiselli D, Pettener D. Mitochondrial DNA diversity in South America and the genetic history of Andean highlanders. Mol Biol Evol. 2003;20:1682–1691. doi: 10.1093/molbev/msg188. [DOI] [PubMed] [Google Scholar]
- Garcia-Bour J, Perez-Perez A, Alvarez S, Fernandez E, Lopez-Parra AM, Arroyo-Pardo E, Turbon D. Early population differentiation in extinct aborigines from Tierra del Fuego-Patagonia: Ancient mtDNA sequences and Y-chromosome STR characterization. Am J Phys Anthropol. 2004;123:361–370. doi: 10.1002/ajpa.10337. [DOI] [PubMed] [Google Scholar]
- Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci USA. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginther C, Corach D, Penacino GA, Rey JA, Carnese FR, Hutz MH, Anderson A, Just J, Salzano FM, King MC. Genetic variation among the Mapuche Indians from the Patagonian region of Argentina: Mitochondrial DNA sequence variation and allele frequencies of several nuclear genes. EXS. 1993;67:211–219. doi: 10.1007/978-3-0348-8583-6_17. [DOI] [PubMed] [Google Scholar]
- Greenberg J, Turner CG, Zegura SL. The settlement of the Americas: A comparison of the linguistic, dental and genetic evidence. Curr Anthropol. 1986;4:477–497. [Google Scholar]
- Gustincich S, Manfioletti G, Del Sal G, Schneider C, Carninci P. A fast method for high-quality genomic DNA extraction from whole human blood. Biotechniques. 1991;11:298–300. 302. [PubMed] [Google Scholar]
- Horai S, Kondo R, Nakagawa-Hattori Y, Hayashi S, Sonoda S, Tajima K. Peopling of the Americas, founded by four major lineages of mitochondrial DNA. Mol Biol Evol. 1993;10:23–47. doi: 10.1093/oxfordjournals.molbev.a039987. [DOI] [PubMed] [Google Scholar]
- Howell N, Smejkal CB. Persistent heteroplasmy of a mutation in the human mtDNA control region: Hyper-mutation as an apparent consequence of simple-repeat expansion/contraction. Am J Hum Genet. 2000;66:1589–1598. doi: 10.1086/302910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyeux G, Rodas C, Gelvez N, Carter D. Possible migration routes into South America deduced from mitochondrial DNA studies in Colombian Amerindian populations. Am J Hum Genet. 2002;74:211–233. doi: 10.1353/hub.2002.0022. [DOI] [PubMed] [Google Scholar]
- Kolman CJ, Bermingham E, Cooke R, Ward RH, Arias TD, Guionneau-Sinclair F. Reduced mtDNA diversity in the Ngobe Amerinds of Panama. Genetics. 1995;140:275–283. doi: 10.1093/genetics/140.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolman CJ, Bermingham E. Mitochondrial and nuclear DNA diversity in the Choco and Chibcha Amerinds of Panama. Genetics. 1997;147:1289–1302. doi: 10.1093/genetics/147.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalueza-Fox C. Mitochondrial DNA haplogroups in four tribes from Tierra del Fuego-Patagonia: Inferences about the peopling of the Americas. Hum Biol. 1996;68:855–871. [PubMed] [Google Scholar]
- Lalueza C, Perez-Perez A, Prats E, Cornudella L, Turbon D. Lack of founding Amerindian mitochondrial DNA lineages in extinct aborigines from Tierra del Fuego-Patagonia. Hum Mol Genet. 1997;6:41–46. doi: 10.1093/hmg/6.1.41. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Tito RY, Lizarraga B, Stone AC. Land, language, and loci: mtDNA in Native Americans and the genetic history of Peru. Am J Phys Anthropol. 2004;127:351–360. doi: 10.1002/ajpa.20102. [DOI] [PubMed] [Google Scholar]
- Lobato-da-Silva DF, Ribeiro-dos-Santos AKC, Santos SEB. Diversidade genética de populações humanas na Amazônia. In: Guimarães Vieira IC, Cardoso da Silva JM, Oren DC, D’Ineao MA, editors. Diversidade Humana e Cultural na Amazônia. Museu Paraense Emilio Goeldi; Belém: 2001. pp. 167–193. [Google Scholar]
- Lorenz JG, Smith DG. Distribution of four founding mtDNA haplogroups among native North Americans. Am J Phys Anthropol. 1996;101:307–323. doi: 10.1002/(SICI)1096-8644(199611)101:3<307::AID-AJPA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Marrero AR, Silva-Junior WA, Bravi CM, Hutz MH, Petzl-Erler ML, Ruiz-Linares A, Salzano FM, Bortolini MC. Demographic and evolutionary trajectories of the Guarani and Kaingang natives of Brazil. Am J Phys Anthropol. 2007;132:301–310. doi: 10.1002/ajpa.20515. [DOI] [PubMed] [Google Scholar]
- Melton PE, Briceño I, Gómez A, Devor EJ, Bernal JE, Crawford MH. Biological relationship between Central and South American Chibchan speaking populations: Evidence from mtDNA. Am J Phys Anthropol. 2007;133:753–770. doi: 10.1002/ajpa.20581. [DOI] [PubMed] [Google Scholar]
- Merriwether DA, Ferrell RE. The four founding lineages hypothesis for the New World. A critical reevaluation. Mol Phylogen Evol. 1996;5:241–246. doi: 10.1006/mpev.1996.0017. [DOI] [PubMed] [Google Scholar]
- Merriwether DA, Rothhammer F, Ferrell RE. Genetic variation in the New World: Ancient teeth, bone and tissue as sources of DNA. Experientia. 1994;50:592–601. doi: 10.1007/BF01921730. [DOI] [PubMed] [Google Scholar]
- Merriwether DA, Rothhammer F, Ferrell RE. Distribution of the four founding lineage haplotypes in Native Americans suggests a single wave of migration for the New World. Am J Phys Anthropol. 1995;98:411–430. doi: 10.1002/ajpa.1330980404. [DOI] [PubMed] [Google Scholar]
- Merriwether DA, Reed DM, Ferrell RE. Ancient and contemporary mitochondrial DNA variation in the Maya. In: Whittington SL, Reed DM, editors. Bones of the Maya: Studies of Ancient Skeletons. Smithsonian Institution Press; Washington, DC: 1997. pp. 208–217. [Google Scholar]
- Mesa NR, Mondragon MC, Soto ID, Parra MV, Duque C, Ortiz-Barrientos D, Garcia LF, Velez ID, Bravo ML, Munera JG, et al. Autosomal, mtDNA, and Y-chromosome diversity in Amerinds: Pre- and post-Columbian patterns of gene flow in South America. Am J Hum Genet. 2000;67:1277–1286. doi: 10.1016/s0002-9297(07)62955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga ML, Rocco P, Miquel JF, Nervi F, Llop E, Chakraborty R, Rothhammer F, Carvallo P. Mitochondrial DNA polymorphisms in Chilean aboriginal populations: Implications for the peopling of the southern cone of the continent. Am J Phys Anthropol. 2000;113:19–29. doi: 10.1002/1096-8644(200009)113:1<19::AID-AJPA3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke DH, Hayes MG, Carlyle SW. Spatial and temporal stability of mtDNA haplogroup frequencies in native North America. Hum Biol. 2000;72:15–34. [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickards O, Martinez-Labarga C, Lum JK, De Stefano GF, Cann RL. mtDNA history of the Cayapa Amerinds of Ecuador: Detection of additional founding lineages for the Native American populations. Am J Hum Genet. 1999;65:519–530. doi: 10.1086/302513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodas C, Gelvez N, Keyeux G. Mitochondrial DNA Studies show asymmetrical Amerindian admixture in Afro-Colombian and Mestizo populations. Hum Biol. 2002;75:13–30. doi: 10.1353/hub.2003.0026. [DOI] [PubMed] [Google Scholar]
- Rondon F, Braga Y, Cardenas H, Barreto G. Análisis de la diversidad y el grado de estructura genética presente en poblaciones humanas colombianas a partir del uso de marcadores RFLPs de mtDNA. Rev Asoc Colomb Cienc Biol. 2007;19:94–103. [Google Scholar]
- Rothhammer F, Llop E, Carvallo P, Moraga M. Origin and evolutionary relationships of native Andean populations. High Alt Med Biol. 2001;2:227–233. doi: 10.1089/152702901750265323. [DOI] [PubMed] [Google Scholar]
- Ruhlen M. A Guide to the World’s Languages. Stanford University Press; Stanford: 1987. p. 469. [Google Scholar]
- Sandoval J, Fujita R, Delgado B, Rivas L, Bonilla B, Nugent D. Variants of mtDNA among islanders of the lake Titicaca: Highest frequency of haplotype B1 and evidence of founder effect. Rev Peru Biol. 2004;11:161–168. [Google Scholar]
- Santos M, Ward RH, Barrantes R. mtDNA variation in the Chibcha Amerindian Huetar from Costa Rica. Hum Biol. 1994a;66:963–977. [PubMed] [Google Scholar]
- Santos M, Ward RH, Barrantes R. D-Loop mtDNA deletion as a unique marker of Chibchan Amerindians. Am J Hum Genet. 1994b;55:413–414. [PMC free article] [PubMed] [Google Scholar]
- Schurr TG, Ballinger SW, Gan YY, Hodge JA, Merriwether DA, Lawrence DN, Knowler WC, Weiss KM, Wallace DC. Amerindian mitochondrial DNAs have rare Asian mutations at high frequencies, suggesting they derived from four primary maternal lineages. Am J Hum Genet. 1990;46:613–623. [PMC free article] [PubMed] [Google Scholar]
- Stone A, Stoneking M. Ancient DNA from a pre-Columbian Amerindian population. Am J Phys Anthropol. 1993;92:463–471. doi: 10.1002/ajpa.1330920405. [DOI] [PubMed] [Google Scholar]
- Torres MM, Bravi CM, Bortolini MC, Duque C, Callegari-Jacques S, Ortiz D, Bedoya G, Groot de Restrepo H, Ruiz-Linares A. A revertant of the major founder Native American haplogroup C common in populations from northern South America. Am J Hum Biol. 2006;18:59–65. doi: 10.1002/ajhb.20461. [DOI] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Yang CC, Szathmary EJ, Williams RC, Schanfield MS, Troup GA, Knowler WC, Lawrence DN, Weiss KM, et al. Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics. 1992;1:153–162. doi: 10.1093/genetics/130.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo CM, Wallace DC. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet. 1993a;53:563–590. [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Sukernik RI, Schurr TG, Starikorskaya YB, Cabell MF, Crawford MH, Comuzzie AG, Wallace DC. mtDNA variation of aboriginal Siberians reveals distinct genetic affinities with Native Americans. Am J Hum Genet. 1993b;53:591–608. [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Chen YS, Semino O, Santachiara-Beneceretti AS, Scott CR, Lott MT, Winter M, Wallace DC. mtDNA and Y-chromosome polymorphisms in four Native American populations from southern Mexico. Am J Hum Genet. 1994;54:303–318. [PMC free article] [PubMed] [Google Scholar]
- Turner C. Advances in the dental search for Native American origins. Acta Anthopogenetica. 1984;8:23–78. [PubMed] [Google Scholar]
- Wallace DC, Garrison K, Knowler WC. Dramatic founder effects in Amerindian mitochondrial DNAs. Am J Phys Anthropol. 1985;68:149–155. doi: 10.1002/ajpa.1330680202. [DOI] [PubMed] [Google Scholar]
- Ward RH, Salzano FM, Bonatto SL, Hutz MH, Coimbra CE, Santos RV. Mitochondrial DNA polymorphism in three Brazilian Indian tribes. Am J Hum Biol. 1996;8:317–323. doi: 10.1002/(SICI)1520-6300(1996)8:3<317::AID-AJHB2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Williams SR, Chagnon NA, Spielman RS. Nuclear and mitochondrial genetic variation in the Yanomamo: A test case for ancient DNA studies of prehistoric populations. Am J Phys Anthropol. 2002;117:246–259. doi: 10.1002/ajpa.10035. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Ossa H, Salazar M, Delgado MB, Deulofeut R, de la Hoz A, Bing DH, Ramos O, Yunis EJ. Major histocompatibility complex class II alleles and haplotypes and blood groups of four Amerindian tribes of northern Colombia. Hum Immunol. 1994;41:248–258. doi: 10.1016/0198-8859(94)90043-4. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Yunis EJ, Yunis E. Genetic relationship of the Guambino, Paez, and Ingano Amerindians of southwest Colombia using major histocompatibility complex class II haplotypes and blood groups. Hum Immunol. 2001;62:970–978. doi: 10.1016/s0198-8859(01)00295-6. [DOI] [PubMed] [Google Scholar]
Internet Resources
- Collard O. AMIGLOBE. 2006 p Amiglobe is a world atlas and database with information about every country in the world, http://www.downloadatoz.com/home-education_directory/amiglobe-2006/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 - mtDNA haplogroup frequencies of Colombian and South America Amerindian tribes.