Abstract
Kaempfer’s Woodpecker (Celeus obrieni) is the only species of the genus Celeus endemic to Brazil. The description of this taxon as a subspecies of the Rufous-headed Woodpecker (Celeus spectabilis) was based on a single specimen. While C. obrieni and C. spectabilis are now considered separate species based on morphological and limited molecular evidence, no study has critically tested the reciprocal monophyly and degree of evolutionary independence between these taxa with several specimens. Herein, fragments of the mitochondrial and nuclear DNA of three recently-collected specimens of C. obrieni were analyzed to evaluate the degree of evolutionary differentiation of this taxon with respect to C. spectabilis. The results confirm the reciprocal monophyly between the specimens of C. obrieni and C. spectabilis. The genetic divergence values for the two taxa also support their classification as independent species, given that they are greater than the values recorded among other closely-related but separate species of the same genus. Estimates of the divergence time between C. obrieni and C. spectabilis indicate that cladogenesis occurred in the mid-Pleistocene, during a period of major climatic fluctuations and landscape change, consistent with the hypothesis of a corridor of open bamboo dominated forests and woodland stretching.
Keywords: Bamboo, biogeography, Celeus obrieni, Celeus spectabilis, taxonomy
Introduction
The woodpeckers of the genus Celeus are endemic to the Neotropics. At the present time, 10 species are recognized (Remsen et al., 2012), of which, nine are found in Brazil. The Kaempfer’s Woodpecker (Celeus obrieni Short 1973) is the only Celeus endemic to this country (Winkler and Christie, 2002; CBRO, 2011).
Celeus obrieni (Short 1973) was originally described as a subspecies of the Rufous-headed Woodpecker (Celeus spectabilis), based on a specimen collected in 1926 by Emil Kaempfer in the region of Uruçuí, in the Brazilian state of Piauí (Hidasi et al., 2008). More than thirty years later, the South American Classification Committee (SACC - Remsen et al., 2012) re-evaluated the status of Celeus s. obrieni based on a comparison of plumage characters with specimens of C. spectabilis from Ecuador, Peru, and Bolivia, and concluded that obrieni was a distinct species. The total lack of new records over almost a century led some ornithologists to believe that the taxon had become extinct (Tobias et al., 2006). The first record since the original specimen collected by Emil Kaempfer was obtained only in 2006, when the species was rediscovered in Goiatins, in the northeast of the Brazilian state of Tocantins, some 400 km from the type locality (Prado, 2006).
Subsequently, several new records were obtained from the state of Maranhão (Santos and Vasconcelos, 2007; Santos et al., 2010), in addition to the states of Tocantins (Pinheiro and Dornas, 2008), Goiás (Dornas et al., 2009; Pinheiro et al., 2012), and Mato Grosso (Dornas et al., 2011). All of these new records extended the known geographic range of C. obrieni quite considerably. Even though, the size of the species population has yet to be defined, since its rediscovery more than 50 individuals have been recorded within an area extending more than one thousand kilometers between extreme localities of distribution. Despite this expansion in the known range of the species, currently estimated at some 280,000 km2 (Benz and Robbins, 2011; BirdLife International, 2011), a cautious estimate of the total population is 50–250 individuals, which is consistent with the IUCN critically threatened (CR) category (IUCN, 2010).
Recently, Benz and Robbins (2011) published a phylogeny for the genus Celeus based on molecular and morphological data, including the genetic material from the holotype of C. obrieni, collected in 1926. This specimen was identified as the sister taxon of C. spectabilis, as predicted by the traditional classification. In their study, four mitochondrial (ND2, ND3, ATP6-8/COIII, and the Control Region) and two nuclear markers (Intron 7 of the β-fibrinogen gene and HMGN2) were analyzed, although only the ND2 and ND3 genes and part of the HMGN2 sequence were amplified successfully for C. obrieni (Benz and Robbins, 2011). The results indicated a genetic divergence of approximately 1% between C. obrieni and C. spectabilis for the mitochondrial marker ND2. The authors suggested that further sampling would be needed to confirm the reciprocal monophyly of these forms and their status as distinct evolutionary lineages (Benz and Robbins, 2011). Nonetheless, Benz and Robbins (2011) treated species treatment for C. obrieni.
In the present study, a multi-locus molecular approach was used to confirm the validity of the species status of C. obrieni, an essential initial step in the development of a conservation plan for this threatened taxon. Three recently collected specimens of C. obrieni were sequenced, together with individuals representing three other species of the genus (C. spectabilis, Celeus undatus Waved Woodpecked, and Celeus grammicus Scale-breasted Woodpecked) to estimate phylogenetic relationships and pairwise genetic distances within this group.
Materials and Methods
Sampling
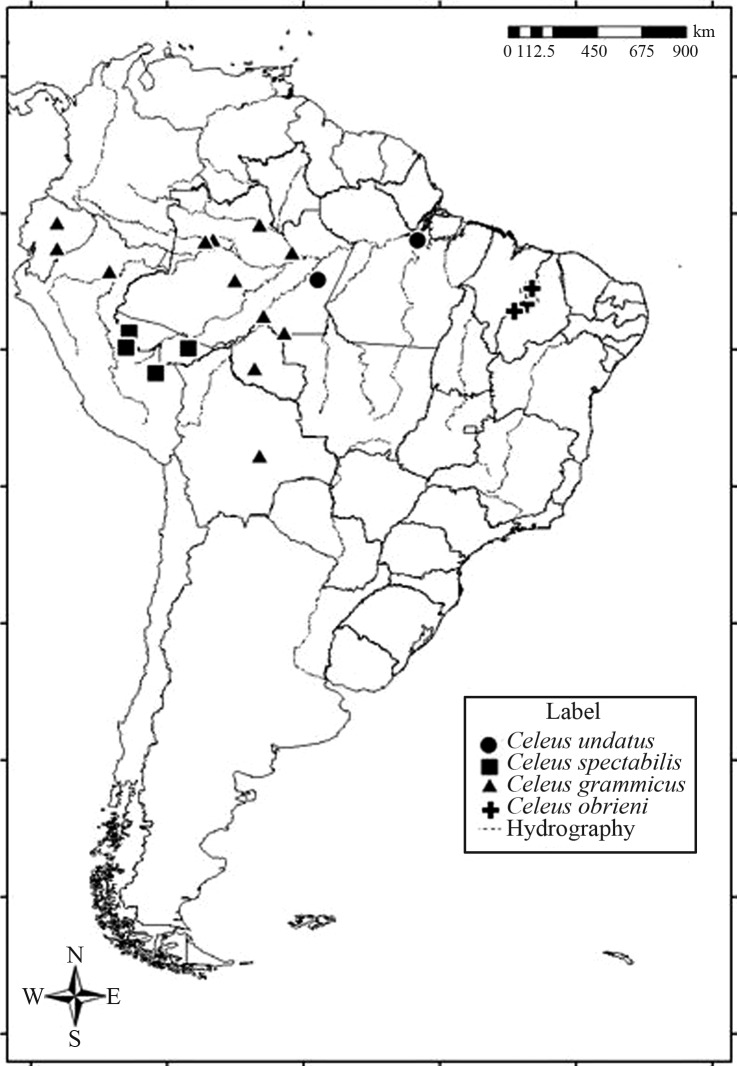
Three specimens of C. obrieni were collected by MPDS during surveys of bird populations at three sites in the Brazilian state of Maranhão (Figure 1, Table 1) - Serra da Raposa, (06°35′S, 43°37′W), in the municipality of São João dos Patos (specimen registered in the Museu Paraense Emílio Goeldi [MPEG] under the accession number 61549), Fazenda Castiça (05°28′ S, 43°13′W), in the municipality of Matões (MPEG 69978), and Fazenda Normasa (05°36′S, 43°28′W), in the municipality of Parnarama (MPEG 69979). Specimens were collected under special license 20902-1 issued to MPDS.
Figure 1.
Map showing the localities where Celeus specimens analyzed in the present study were collected.
Table 1.
Celeus specimens analyzed in the present study, showing the species name, number of specimens analyzed, identification code, collecting locality, and GenBank accession numbers for the sequences of the different molecular markers analyzed.
| Species (number of specimens) | Identification code (Voucher specimen number) | Collecting locality | Cyt-B | 16S | ND2 | I7BF |
|---|---|---|---|---|---|---|
| C. obrieni (n = 4) | ||||||
| Cob1109 (MPEG 61549) | Brazil: Maranhão | KC858911 | KC858892 | KC858943 | KC858930 | |
| Cob1399 (MPEG 69978) | Brazil: Maranhão | KC858912 | KC858893 | KC858944 | KC858931 | |
| Cob1414 (MPEG 69979) | Brazil: Maranhão | KC858913 | KC858894 | KC858945 | - | |
| AMNH242687* | Brazil: Piauí | - | - | JF433290 | - | |
| C. spectabilis (n = 7) | ||||||
| Csp70 (MPEG 58371) | Brazil: Acre | KC858914 | KC858895 | KC858946 | - | |
| Csp786 (MPEG 61256) | Brazil: Acre | KC858915 | KC858896 | KC858947 | KC858932 | |
| Csp789 (MPEG 61254) | Brazil: Acre | KC858916 | KC858897 | KC858948 | KC858933 | |
| Csp791 (MPEG 61255) | Brazil: Acre | KC858917 | KC858898 | KC858949 | KC858934 | |
| Csp846 (MPEG 61257) | Brazil: Acre | KC858918 | KC858899 | KC858950 | KC858935 | |
| LSUMNS 45460* | Peru: Madre de Díos | - | - | JF433281 | - | |
| LSUMNS 10664* | Peru: Ucayalí | - | - | JF433280 | JF433138 | |
| C. undatus (n = 4) | ||||||
| Cun228 (MPEG 61715) | Brazil: Pará | KC858919 | KC858900 | KC858951 | - | |
| Cun229 (MPEG 61716) | Brazil: Pará | KC858920 | KC858901 | KC858952 | KC858936 | |
| KUNHM 5765* | Guyana | - | - | JF433267 | JF433143 | |
| KUNHM 5829* | Guyana | - | - | JF433266 | JF433142 | |
| C. grammicus (n = 14) | ||||||
| Cgr74 (MPEG 57567) | Brazil: Amazonas | KC858921 | KC858902 | KC858953 | - | |
| Cgr89 (MPEG 59384) | Brazil: Amazonas | KC858922 | KC858903 | KC858954 | KC858937 | |
| Cgr102 (MPEG 57021) | Brazil: Amazonas | KC858923 | KC858904 | KC858955 | KC858938 | |
| Cgr103 (MPEG 57020) | Brazil: Amazonas | KC858924 | KC858905 | KC858956 | KC858939 | |
| Cgr168 (MPEG 62597) | Brazil: Amazonas | KC858925 | KC858906 | KC858957 | KC858940 | |
| Cgr180 (MPEG 62598) | Brazil: Amazonas | KC858926 | KC858907 | KC858958 | KC858941 | |
| Cgr376 (MPEG 59385) | Brazil: Amazonas | KC858927 | KC858908 | KC858959 | KC858942 | |
| Cgr380 (MPEG 62599) | Brazil: Amazonas | KC858928 | KC858909 | KC858960 | - | |
| Cgr613 (MPEG 58677) | Brazil: Amazonas | KC858929 | KC858910 | KC858961 | - | |
| ANSP 2477* | Ecuador: Morona-Santiago | - | - | JF433272 | - | |
| LSUMNS 6892* | Peru: Loreto | - | - | JF433271 | JF433140 | |
| FMNH 389782* | Brazil: Rondônia | - | - | JF433270 | JF433141 | |
| LSUMNS 105252* | Bolivia: Santa Cruz | - | - | JF433269 | - | |
| ANSP 3253* | Ecuador: Napo | - | - | JF433268 | - | |
| C. elegans (n = 3) | ||||||
| KUNHM 5764* | Guyana | - | - | JF433261 | - | |
| LSU 4364** | Peru: Loredo | AY940795 | - | - | - | |
| ANSP 4364* | Peru: Loredo | - | - | - | JF433131 |
sequences obtained from Benz and Robbins (2011).
sequences obtained from Webb and Moore (2005).
Samples of muscle tissue were obtained from each of the three specimens of C. obrieni, together with 16 samples of specimens representing three other Celeus species - C. spectabilis (n = 5), C. undatus (n = 2), and C. grammicus (n = 9) - all of which were provided by the ornithological collection of the Goeldi Museum (Table 1). Samples of C. undatus and C. grammicus, two sister lineages historically regarded as separate species by traditional taxonomy and close to C. obrieni / spectabilis (Benz and Robbins, 2011), were included in the analysis so that genetic distances between these species pairs could be contrasted. The samples were divided into aliquots and kept frozen at −20 °C until processing in the UFPA Genetics and Molecular Biology Laboratory.
Extraction, amplification and sequencing of the DNA
Once the samples were processed, the genetic material was extracted using the standard phenol-chloroform protocol, followed by precipitation in sodium acetate and isopropanol (Sambrook et al., 1989). The samples were then electrophoresed in 1% agarose gel, stained with ethidium bromide and viewed under an UV transilluminator to the confirm the successful extraction of genetic material, its integrity and concentration. Using specific primers, the polymerase chain reaction (PCR) technique was used to amplify three regions of the mitochondrial genome (rDNA 16S, Cytochrome b, subunit 2 of the NADH dehydrogenase region) and one nuclear marker, intron 7 of the β-fibrinogen gene. The primers described by Sorenson et al. (1999) - L-15298 and H-16064 - were used for the cytochrome b gene (Cyt b), and those presented by Palumbi et al. (1991) - L-1987 and H-2609 -for the rDNA 16S (16S) gene. For subunit 2 of the NADH dehydrogenase region (ND2), the primers described by Hackett (1996) were used - H-6313 and L-5215. Part of intron 7 of the β-fibrinogen gene (I7BF) was amplified using the primers (FIB-BI7U and FIB-BI7L) described by Prychitko and Moore (1997).
Each reaction was conducted in a final volume of 25 μL, containing 4 μL of the deoxynucleotides (1.25 mM), 2.5 μL of 10× Taq buffer, 1 μL of MgCl2 (25 mM), 0.5 μL of each primer (200 ng/μL), approximately 80 ng of the total DNA extracted from the samples, 0.25 μL of Taq polymerase (5 U/μL, Taq DNA Polymerase, Recombinant - Invitrogen) and sterile distilled water to complete the final reaction volume. The PCR for each of the genetic markers was run in a thermocycler (GeneAmp, PCR System 9700 - Applied Biosystems). For the mitochondrial markers (rDNA 16S, Cyt b, and ND2), the protocol began with 3 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, one minunte at 55 °C, two min at 72 °C, with a final extension of 7 min at 72 °C. For the nuclear fragment (I7BF), the protocol was 3 min at 94 °C, followed by 35 cycles of 20 s at 94 °C, 15 s at 50 °C, and 1 min at 72 °C, with a final extension of 7 min at 72 °C.
The amplified products were purified using an ExoSAP-IT kit (Amersham Pharmacia Biotech. Inc., UK). The purified samples were used for the sequencing reaction, using the dideoxy-terminal method (Sanger et al., 1977) with reagents of the Big Dye kit (ABI Prism™ Dye Terminator Cycle Sequencing Ready Reaction - Applied Biosystems, USA), following the manufacturer’s instructions. Reagents not incorporated during the reaction were eliminated through ethanol washes. The gene fragments resulting from this sequencing reaction had their nucleotide sequences determined by an ABI 3500 automatic sequencer (Applied Biosystems). Sequences obtained from GenBank for the Cyt b (Webb and Moore, 2005), ND2, and I7BF segments (Benz and Robbins, 2011) were also used in the present analysis (see Table 1 for access numbers).
Sequence alignment
The sequences obtained by electrophoresis were aligned automatically using the CLUSTAL-W application (Thompson et al., 1994), with the parameters suggested by Schneider (2007). The file generated by this procedure was converted into the FASTA format and transferred to the BioEdit sequence editor (Hall, 1999) for visual inspection of the alignment and possible correction of the codification of observed insertions or suppressions. The nucleotide composition, transition/transversion rates, polymorphic sites, and divergence rates (P distances) within and between species were calculated using the MEGA software, version 4.0. (Tamura et al., 2007). The DnaSP program, version 5.10 (Librado and Rozas, 2009) was used to determine the haplotypes obtained, and the Network program, version 4.6 (Bandelt et al., 1999) was used to produce a haplotype network.
Statistical analyses
The best fitting evolutionary model for each separate gene region and the concatenated data was determined by the Akaike Information Criterion (AIC), run in jModeltest 0.1.1 (Posada, 2008). The phylogenetic arrangements of the species were obtained using the PAUP* program, v. 4.0b 10 (Swofford, 2002), for the production of maximum parsimony (MP) and maximum likelihood (ML) trees, with the bootstrap support for nodes being based on 1000 pseudo-replicates (Felsenstein, 1985), which provides an estimate of the confidence limits for the arrangement of each tree. The Bayesian Inference analysis was run in MrBayes v3.0b4 (Huelsenbeck and Ronquist, 2001) with three heated chains and one “cold” chain, each with five million generations, sampled every 100 generations, with the application of the stopping rule command. The sequences of the species Celeus elegans - Chestnut Woodpecker were obtained from GenBank (Table 1) for use as the outgroup to root the phylogenetic arrangements.
The BEAST 1.7.2 program (Drummond and Rambaut, 2007) was used to visualize the phylogenetic relationships among the different taxa by generating species trees using the *BEAST procedure (Heled and Drummond, 2010), with 60 million generations. This approach is based on the prior selection of the most appropriate evolutionary model for each gene, which is adapted to the database for the generation of the species tree. This approach contrasts with the simple concatenation of the different models, with only a single model being selected for the concatenated data. The results of this analysis were visualized in Tracer v. 1.5 (Drummond and Rambaut, 2007) to determine the quality of the Markov chain search process. The trees were visualized and edited in FigTree v. 1.3.1 (Drummond and Rambaut, 2007). The divergence time among the Celeus species was estimated based on a relaxed molecular-clock analysis of the species tree, assuming a 2.1% nucleotide substitution rate per million years for Cyt b (Weir and Schluter, 2008), using the *BEAST methodology implemented in BEAST 1.7.2 software.
Results
A total of 3113 base pairs were sequenced, of which 2234 belonged to the mitochondrial markers (ND2, Cyt b, and 16S), while the other 879 represent the nuclear segment, I7BF. The analyses were complemented by sequences obtained from GenBank, belonging to the four species analyzed here, together with C. elegans (Webb and Moore, 2005; Benz and Robbins, 2011), which was used as the outgroup for the phylogenetic analyses. No evidence was found that the mitochondrial segments analyzed here may have been nuclear copies (NUMTs) of the mitochondrial genome, based on standard analytical criteria (Lacerda et al., 2007; Rêgo et al., 2010).
The nucleotide composition of the mitochondrial sequences was broadly similar among the four species, with an AT content of 51% for the mitochondrial sequences and 64% for the nuclear ones. Most changes in the mitochondrial sequences were transitions, at rates four to ten times higher than those recorded for transversions, indicating a lack of saturation in the sequences. This ratio was more balanced (ts/tv = 1.7) for the nuclear I7BF segment. A total of 28 amino-acid substitutions were recorded for the two coding regions (Cyt b and ND2). Of these, 10 were exclusive to C. obrieni and C. spectabilis.
A total of 66 variable sites were identified for Cyt b, 97 for ND2, 16 for 16S, and nine for I7BF in the 19 Celeus specimens examined. In all, 170 of these 188 sites were informative for phylogenetic analysis. There were 36 variable sites between C. obrieni and C. spectabilis, of which 26 were informative for parsimony analysis, with ND2 and Cytb, once again, being the most variable.
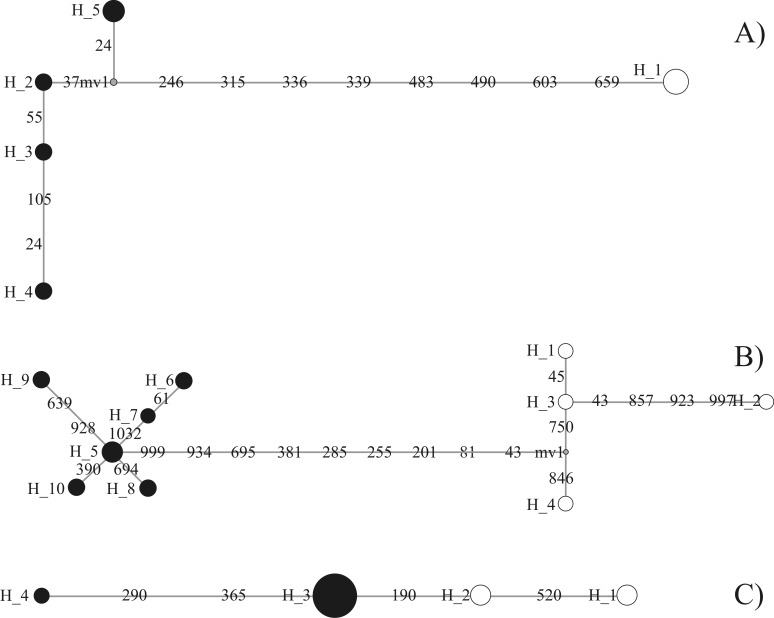
The genetic divergence values (p distance) recorded within and between species for the four gene segments are presented in Tables 2 and 3. Within taxa, the greatest divergence was observed in Cyt b and ND2, with lower levels being observed in 16S and I7BF. Divergence between C. obrieni and C. spectabilis was much higher for Cyt b (1.3%) and ND2 (1.2%) than that observed between C. undatus and C. grammicus - Cyt b (0.5%) and ND2 (0.7%). The haplotype network for Cyt b and ND2 indicated a clear differentiation between C. obrieni and C. spectabilis, with no shared haplotypes (Figure 2). As expected for a relatively well-conserved marker, the 16S fragment presented only a single haplotype for each species, separated by two mutations.
Table 2.
Mean pairwise distances (p distances) and standard deviations for Cyt b (752 bps; below the diagonal) and ND2 (1041 bps; above the diagonal) for the four Celeus species analyzed in the present study. Intraspecific distances in parentheses.
| C. obrieni (0.002 ± 0.001) | C. spectabilis (0.002 ± 0.001) | C. undatus (0.007 ± 0.002) | C. grammicus (0.002 ± 0.001) | |
|---|---|---|---|---|
| C. obrieni (0.000 ± 0.000) | - | 0.012 ± 0.003 | 0.073 ± 0.007 | 0.075 ± 0.007 |
| C. spectabilis (0.003 ± 0.001) | 0.013 ± 0.004 | - | 0.072 ± 0.007 | 0.075 ± 0.008 |
| C. undatus (0.000 ± 0.000) | 0.076 ± 0.009 | 0.068 ± 0.009 | - | 0.007 ± 0.002 |
| C. grammicus (0.005 ± 0.002) | 0.077 ± 0.009 | 0.069 ± 0.009 | 0.005 ± 0.002 | - |
Table 3.
Mean pairwise distances (p distances) and standard deviations for 16S (441 bps; below the diagonal) and I7BF (879 bps; above the diagonal) for the four Celeus species analyzed in the present study. Intraspecific distances in parentheses.
| C. obrieni (0.001 ± 0.001) | C. spectabilis (0.000 ± 0.000) | C. undatus (0.000 ± 0.000) | C. grammicus (0.001 ± 0.000) | |
|---|---|---|---|---|
| C. obrieni (0.000 ± 0.000) | - | 0.002 ± 0.001 | 0.004 ± 0.002 | 0.004 ± 0.002 |
| C. spectabilis (0.000 ± 0.000) | 0.005 ± 0.003 | - | 0.002 ± 0.002 | 0.002 ± 0.002 |
| C. undatus (0.000 ± 0.000) | 0.034 ± 0.008 | 0.029 ± 0.007 | - | 0.000 ± 0.000 |
| C. grammicus (0.000 ± 0.000) | 0.036 ± 0.008 | 0.032 ± 0.008 | 0.002 ± .002 | - |
Figure 2.
Haplotype networks for Celeus obrieni (black circles) and Celeus spectabilis (white circles) for three molecular markers - Cyt b (A), ND2 (B), and I7BF (C). Haplotypes are represented by circles, the sizes of which are proportional to their frequencies. Numbers on the lines that join the circles correspond to the positions of the divergent nucleotides in the region studied.
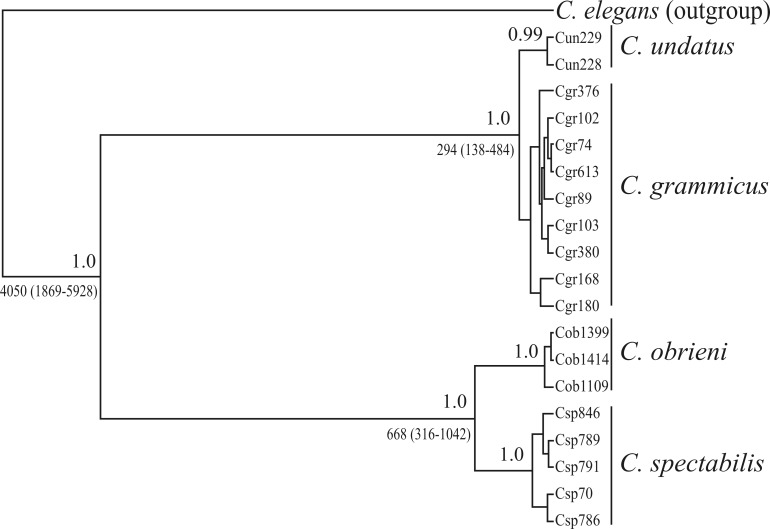
The most appropriate evolutionary models selected by the jModeltest program for the Bayesian Inference (BI) criterion of the four markers were HKY (rRNA 16S), TPM3uf+G (Cyt b), HKY+I (ND2), and F81 (I7BF). The topology obtained for the species tree indicated a high degree of statistical support for both the C. obrieni / C. spectabilis and the C. undatus / C. grammicus groupings (Figure 3). Similarly, the phylogenetic analyses of the concatenated data provided by the MP, ML, and BI approaches confirmed the results of the species tree, particularly the distinction between C. obrieni and C. spectabilis, which represent two reciprocally monophyletic groups, with branch support values of over 95% in all the analyses (data not shown). The estimated divergence time obtained based on the Cyt b marker indicated that the cladogenesis of C. obrieni and C. spectabilis was probably completed between 400,000 and 800,000 years ago, which coincides with the mid-Pleistocene (Figure 3).
Figure 3.
Bayesian Inference for the species tree based on the mitochondrial and nuclear data run in *BEAST. Posterior probability values are shown above branches, whereas divergence times from the common ancestor are shown below branches.
Discussion
The conservation of species is often hampered by the uncertain taxonomic status of different forms, and the reliable definition of such status is a primary preoccupation in conservation biology (Assis et al., 2006). The use of molecular techniques is fundamental to the analysis of the taxonomic status of most uncertain forms (Silveira and Olmos, 2007; Lacerda et al., 2007), although this approach is limited by the need for an adequate supply of biological material from which DNA can be isolated. Celeus obrieni presents a good example of the importance of this approach, given that the existence of a single specimen, deposited at the American Museum of Natural History in New York in 1926, was used for the confirmation of the species status of this form in relation to C. spectabilis (Benz and Robbins, 2011). However, the availability of only a single specimen from almost a century ago led these authors to emphasize the need for a larger sample in order to confirm the reciprocal monophyly of these forms, and the reliability of the evolutionary lineages identified. The present study provides additional support for the status of C. obrieni as a valid species, distinct from C. spectabilis, thus reconfirming the findings of Benz and Robbins (2011). The present findings were based on a larger number of C. obrieni specimens, with the results of all the analyses - intra- and interspecific divergence values, haplotype networks, and species trees - confirming reciprocal monophyly between C. obrieni and C. spectabilis, with a high degree of statistical support.
In addition, the comparative analysis indicated a greater degree of divergence between C. obrieni and C. spectabilis in comparison with C. undatus and C. grammicus, and the species tree (Figure 1) was consistent with the complete separation of these four taxa. These analyses, however, did not include the specimens used by Benz and Robbins (2011), given that sequences for the genetic markers used in both studies were not available for all the specimens.
One important detail in the results of Benz and Robbins (2011) is that the mtDNA divergence value within C. grammicus was larger than those recorded between the grammicus and undatus forms, resulting in a paraphyletic arrangement. Nonetheless, a number of substitutions observed in the nuclear loci indicated a different arrangement, in which the taxa were clearly separated, indicating the possibility that the mtDNA data were the result of introgression. Given this, Benz and Robbins (2011) indicated the need for a more definitive evaluation of the taxonomic arrangement of this group (grammicus-undatus) based on a larger sample from a wider range of populations and a larger number of independent genetic markers for the evaluation of possible gene flow, especially in the western Amazonian populations of grammicus, and in the eastern contact zone mentioned above, to provide a more conclusive assessment of the status of these taxa. The low distance values separating undatus and grammicus that we found also call into question the specific distinctiveness of these two taxa.
While C. obrieni had long been considered a subspecies of C. spectabilis (Short, 1982), based on the availability of a single specimen, subsequent morphological and ecological data indicated that C. obrieni and C. spectabilis are distinct, albeit closely-related species (Santos and Vasconcelos, 2007; Remsen et al., 2012). The geographic distribution of the two species is probably related to the availability of habitats with bamboo (Pinheiro et al., 2012). In the western Amazon basin, C. spectabilis is dependent on patches of humid bamboo forest, in which it forages and reproduces (Kratter, 1997, 1998; Whittaker and Oren, 1999; Guilherme and Santos, 2009). Tobias et al. (2006) also highlight that the known geographic range of C. obrieni (from the type specimen) coincided with open and forested Cerrado habitats, interspersed with narrow and discontinuous belts of gallery forest. New observations (Pinheiro and Dornas, 2008; Pinheiro et al., 2012) indicate that this species is specialized for occupation of patches of bamboo habitat within this landscape, especially those dominated by Guadua paniculata, in which approximately 98% of the records of C. obrieni foraging behavior were obtained at Cerrado sites. The marked dependence of the species on a bamboo habitat could be related to the dietary preferences of this bird species (Kratter, 1997; Winkler and Christie, 2002). The heterogeneous distribution of C. obrieni within the Cerrado may thus be related to this dependence on bamboo habitats, which are relatively widely dispersed within this biome.
The present-day distribution of C. obrieni includes the Brazilian states of Goiás, Maranhão, Piauí, and Tocantins (Pinheiro and Dornas, 2008; Dornas et al., 2011), and more recently, Mato Grosso (Pinheiro et al., 2012). This distribution encompasses wooded environments - gallery and semideciduous forests, both with bamboo habitats -within the Cerrado biome, and reduces the distance (to approximately 1000 km) between the known range of C. obrieni and the nearest recorded localities for C. spectabilis in the Brazilian state of Acre in the southwestern Amazon basin (Guilherme and Santos, 2009). The available data on the distribution of C. obrieni and C. spectabilis indicate a pattern different from that of other birds dependent on bamboo habitats, such as Syndactyla ucayalae, Anabazenops dorsalis, Cercomacra manu, Drymophila devillei, and Ramphotrigon megacephalum, whose stronghold lies in southwestern Amazonia (Kratter, 1997). When they occur outside this area, these species tend to be associated with minor tracts of bamboo habitats, which form a diagonal from southwestern Amazonia, passing through the states of Rondônia and Mato Grosso to southeastern Pará, in the region of Carajás, reaching even the Belém area of endemism in easternmost Amazonia (Parker et al., 1997; Lees et al., in press). Unlike those aforementioned species, which are locally but continuously distributed in this area, the distributions of C. obrieni and C. spectabilis appear to be truly disjunct, with the latter restricted to western Amazonia and the former to its eastern portion (Kratter, 1997). If this patterns holds, the allopatry of C. obrieni and C. spectabilis may help explain the comparative divergent phenotypes and genotypes when compared to another phylogenetically close sister species pair of Celeus, such as C. undatus and C. grammicus.
The evidence suggests that in the past, these open bamboo forests were more widely distributed than in the present day, extending eastwards towards the Cerrado, with offshoots that reached as far south as the Atlantic Forest of southeastern Brazil. This major corridor of open forest habitats would provide a historic connection between the geographic ranges of the two species. According to the molecular clock analysis, the cladogenetic event that led to the separation of C. obrieni and C. spectabilis took place during the climatic fluctuations and associated habitat modifications of the mid Pleistocene. It is worthy of note that the 2.1% nucleotide substitution rate used in the present study is based on average across many unrelated taxa (Weir and Schluter, 2008), therefore, such conclusions must be interpreted with caution.
The climate cycles and related shifts in the distribution of habitats that occurred during the Pleistocene have been identified by several biogeographers (Prance, 1987; Haffer, 2001; Silva and Bates, 2002) as the primary factor driving recent speciation processes in both forested and savanna environments in South America. In particular, the climatic fluctuations of the mid-Pleistocene would have provoked a reduction and fragmentation of the forest cover, leading to the isolation of the ancestral populations of C. obrieni and C. spectabilis. The geomorphological evidence indicates that a single major refuge would have formed in the center of the present-day distribution of the Cerrado biome during this period, when even gallery forests may have shrunk to the point of interrupting the connection with adjacent areas of forest (Ab’Saber, 1983). The subsequent warming of the climate would have led to an expansion of the forest cover, expanding from the refugia to more peripheral areas, and re-establishing the connectivity of the gallery forests.
Because the latest Brazilian checklist of endangered species was completed in 2003, before Kaempfer’s Woodpecker was rediscovered, it is currently not listed as a threatened species in Brazil, but the IUCN lists it globally as critically endangered (BirdLife International, 2011). However, the recent extensions reported in the known geographic range of the species, which now includes Mato Grosso (Dornas et al., 2011), may contribute to a re-evaluation of the species’ status in the near future (IUCN, 2010). On the other hand, Pinheiro et al. (2012) have concluded that the specialization of C. obrieni for foraging in G. paniculata bamboo patches within the Cerrado may determine its low population densities and its high degree of intolerance of anthropogenic impacts. This would make the species vulnerable to extinction, especially in the current context of agricultural expansion in the Brazilian Cerrado, where large tracts of natural habitat are being converted into soybean and rice plantations (Silva and Bates, 2002). The destruction of local ecosystems has accelerated considerably in recent years (Klink and Machado, 2005). Over the short term, the development of effective conservation strategies for C. obrieni will depend on the collection of more reliable data on population size and viability, for which molecular markers will provide a fundamentally important analytical tool.
Acknowledgments
We thank anonymous reviewers for contributions to earlier versions of the manuscript. L.S.A. received a graduate fellowship from CNPq during the study. A.A., M.V., I.S., and H.S. are supported by productivity fellowships from CNPq.
Footnotes
Associate Editor: Fabrício Rodrigues dos Santos
References
- Ab’Saber AN. O domínio dos cerrados: Introdução ao conhecimento. Rev Serv. 1983;111:41–55. [Google Scholar]
- Assis CP, Raposo MA, Parrini R. Validação de Poospiza cabanisi Bonaparte, 1850 (Passeriformes, Emberizidae) Rev Bras Ornitol. 2006;15:103–112. [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Benz BW, Robbins MB. Molecular phylogenetics, vocalizations, and species limits in Celeus woodpeckers (Aves, Picidae) Mol Phylogenet Evol. 2011;61:29–44. doi: 10.1016/j.ympev.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Dornas T, Valle NC, Hidasi J. Celeus obrieni: Dois novos registros históricos para o estado de Goiás. Atual Ornitol. 2009;147:18–19. [Google Scholar]
- Dornas T, Leite GA, Pinheiro RT, Crozariol MA. Primeiro registro do criticamente ameaçado pica-pau-do-parnaíba Celeus obrieni no Estado do Mato Grosso (Brasil) e comentários sobre distribuição geográfica e conservação. Cotinga. 2011;33:91–93. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:e214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evol. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Guilherme E, Santos MP. Birds associated with bamboo forests in eastern Acre, Brazil. Bull Br Ornithol Club. 2009;129:229–240. [Google Scholar]
- Hackett SJ. Molecular phylogenetics and biogeography of Tanagers in the genus Ramphocelus (Aves) Mol Phylogenet Evol. 1996;5:368–382. doi: 10.1006/mpev.1996.0032. [DOI] [PubMed] [Google Scholar]
- Haffer J. Hypotheses to explain the origin of species in Amazonia. In: Vieira ICG, Silva JMC, Oren DC, D’Incao MA, editors. Biological and Cultural Diversity in Amazonia. Museu Paraense Emílio Goeldi; Belém: 2001. pp. 45–118. [Google Scholar]
- Hall TA. BIOEDIT: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidasi JL, Mendonça GA, Blamires D. Primeiro registro documentado de Celeus obrieni (Picidae) para o estado de Goiás, Brasil. Revista Brasileira de Ornitologia. 2008;16:373–375. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Klink CA, Machado RB. A conservação do Cerrado Brasileiro. Megadiversidade. 2005;1:147–155. [Google Scholar]
- Kratter AW. Bamboo specialization by Amazonian birds. Biotropica. 1997;29:100–110. [Google Scholar]
- Kratter AW. The nests of two bamboo specialists: Celeus spectabilis and Cercomacra manu. Biotropica. 1998;69:37–44. [Google Scholar]
- Lacerda DR, Marini MÂ, Santos FR. Mitochondrial DNA corroborates the species distinctiveness of the Planalto (Thamnophilus pelzelni Hellmayr, 1924) and the Sooretama (T. ambiguous Swainson, 1825) Slaty-antshrikes (Passeriformes, Thamnophilidae) Braz J Biol. 2007;67:873–882. doi: 10.1590/s1519-69842007000500010. [DOI] [PubMed] [Google Scholar]
- Lees AC, Moura NG, Santana A, Aleixo A, Barlow J, Berenguer E, Ferreira J, Gardner TA. Paragominas: A quantitative baseline in ventory of an eastern amazonian avifauna. Rev Brasil Ornitol. 2012;20:93–118. [Google Scholar]
- Librado P, Rozas J. DnaSP v. 5.10: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Palumbi S, Martin A, Romano S, McMillian WO, Stice L, Grabowski G. The Simple Fool’s Guide to PCR. University of Hawaii Press; Honolulu: 1991. p. 45. [Google Scholar]
- Parker TA, Stotz DF, Fitzpatrick JW. Notes on avian bamboo specialists in southwestern Amazonian Brazil. Ornithol Monogr. 1997;48:543–547. [Google Scholar]
- Pinheiro RT, Dornas T. New records and distribution of Kaempfer’s Woodpecker Celeus obrieni. Rev Bras Ornitol. 2008;16:167–169. [Google Scholar]
- Pinheiro RT, Dornas T, Leite GA, Crozariol MA, Marcelino DG, Corrêa AG. Novos registros do pica-pau-do-parnaíba Celeus obrieni e status conservação no estado de Goiás, Brasil. Rev Bras Ornitol. 2012;20:59–64. [Google Scholar]
- Posada D. jModelTest: Phylogenetic Model Averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Prado AD. Celeus obrieni: 80 anos depois. Atual Ornitol. 2006;134:4–5. [Google Scholar]
- Prance GT. Biogeography of neotropical plants. In: Whitmore TC, Prance GT, editors. Biogeography and Quaternary History in Tropical America. Clarendon Press; Oxford: 1987. pp. 46–65. [Google Scholar]
- Prychitko TM, Moore WS. The utility of DNA sequences of an intron from the b-fibrinogen gene in phylogenetic analysis of Woodpeckers (Aves, Picidae) Mol Phylogenet Evol. 1997;8:193–204. doi: 10.1006/mpev.1997.0420. [DOI] [PubMed] [Google Scholar]
- Rêgo PS, Araripe J, Silva WAG, Albano C, Pinto T, Campos A, Vallinoto M, Sampaio I, Schneider H. Population genetic studies of mitochondrial pseudo-control region in the endangered Araripe Manakin (Antilophia bokermanni) Auk. 2010;127:335–342. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. A Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 1989. Molecular Cloning. [Google Scholar]
- Sanger F, Nichlen S, Coulson AR. DNA Sequencing with chain-termination inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5468. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MPD, Vasconcelos MF. Range extension for Kaempfer’s Woodpecker Celeus obrieni in Brazil, with the first male specimen. Bull Br Ornithol Club. 2007;127:249–252. [Google Scholar]
- Santos MPD, Cerqueira PV, Soares LMS. Avifauna em seis localidades no Centro-Sul do Estado do Maranhão, Brasil. Ornithologia. 2010;4:49–65. [Google Scholar]
- Schneider H. Método de Análise Filogenética: Um Guia Prático. Editora Holos and Sociedade Brasileira de Genética Press; Ribeirão Preto: 2007. p. 200. [Google Scholar]
- Short LL. Woodpeckers of the World. Delaware Museum of Natural History; Greenville: 1982. p. 676. [Google Scholar]
- Silva JMC, Bates JM. Biogeographic patterns and conservation in the South American Cerrado: A tropical savanna hotspot. BioScience. 2002;52:225–233. [Google Scholar]
- Silveira LF, Olmos F. Quantas espécies de aves existem no Brasil? Conceitos de espécie, conservação e o que falta descobrir. Rev Bras Ornitol. 2007;15:289–296. [Google Scholar]
- Sorenson MD, Ast JC, Dimcheff DE, Yuri T, Mindell DP. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol Phylogenet Evol. 1999;12:105–114. doi: 10.1006/mpev.1998.0602. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Beta Version v.4.10b. Sinauer Associates; Sundeland, Massachusetts: 2002. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods) [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software ver. 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias JA, Butchart SHM, Collar NJ. Lost and found: A gap analysis for the Neotropical avifauna. Neotrop Birding. 2006;1:4–22. [Google Scholar]
- Webb DM, Moore WS. A phylogenetic analysis of woodpeckers and their allies using 12S, Cyt b, and COI nucleotide sequences (class Aves; order Piciformes) Mol Phylogenet Evol. 2005;36:233–248. doi: 10.1016/j.ympev.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Weir JT, Schluter D. Calibrating the avian molecular clock. Mol Ecol. 2008;17:2321–2328. doi: 10.1111/j.1365-294X.2008.03742.x. [DOI] [PubMed] [Google Scholar]
- Winkler H, Christie DA. Family Picidae (Woodpeckers) In: del Hoyo J, Elliott A, Sargatal J, editors. Handbook of the Birds of the World. Vol. 7. Lynx Edicions; Barcelona: 2002. pp. 296–555. Jacamars to Woodpeckers. [Google Scholar]
- Whittaker A, Oren DC. Important ornithological records from the rio Juruá, western Amazonia, including twelve additions to the Brazilian avifauna. Bull Br Ornithol Club. 1999;119:235–260. [Google Scholar]
Internet Resources
- BirdLife International Celeus obrieni. 2011 IUCN 2011. IUCN Red List of Threatened Species Version 2011.2, http://www.iucnredlist.org (June 29, 2012).
- CBRO Comitê Brasileiro de Registros Ornitológicos. (10a Edição) 2011 http://www.cbro.org.br (June 29, 2012).
- IUCN Celeus obrieni. 2010 IUCN 2010. IUCN Red List of Threatened Species Version 2010.4., http://www.iucnredlist.org (June 29, 2012).
- Remsen JV, Cadena CD, Jaramillo A, Nores M, Pacheco JF, Robbins MB, Schulenberg TS, Stiles FG, Stotz DF, Zimmer KJ. A classification of the bird species of South America. 2012 AOU, http://www.museum.lsu.edu/~Remsen/SACCBaseline.html (June 29, 2012).