Abstract
Festuca arundinacea Schreb., commonly known as tall fescue, is a major forage crop in temperate regions. Recently, a molecular analysis of different accessions of a world germplasm collection of tall fescue has demonstrated that it contains different species from the genus Festuca and allowed their rapid classification into the three major morphotypes (Continental, Mediterranean and Rhizomatous). In this study, we explored the genetic diversity of 161 accessions of Festuca species from 29 countries, including 28 accessions of INTA (Argentina), by analyzing 15 polymorphic SSR markers by capillary electrophoresis. These molecular markers allowed us to detect a total of 214 alleles. The number of alleles per locus varied between 5 and 24, and the values of polymorphic information content ranged from 0.627 to 0.840. In addition, the accessions analyzed by flow cytometry showed different ploidy levels (diploid, tetraploid, hexaploid and octaploid), placing in evidence that the world germplasm collection consisted of multiple species, as previously suggested. Interestingly, almost all accessions of INTA germplasm collection were true hexaploid tall fescue, belonging to two eco-geographic races (Continental and Mediterranean). Finally, the data presented revealed an ample genetic diversity of tall fescue showing the importance of preserving the INTA collection for future breeding programs.
Keywords: SSR markers, genetic diversity, flow cytometry, tall fescue, genetic resources
The genus Festuca, which belongs to the family Poaceae, contains over 500 species of grasses, varying in ploidy level from diploid (2n = 2x = 14) to dodecaploid (2n = 12x = 84) (Hand et al., 2012a). The most relevant species of Festuca in agriculture is Festuca arundinacea Schreb., commonly known as tall fescue (Hand et al., 2010). Tall fescue is a forage grass widely grown throughout temperate regions. This crop plays a critical role in forage and livestock systems, forming the plant basis for beef and milk production worldwide. This species is also used as turf grass and to preserve the soil (Smarda et al., 2008). Festuca arundinacea Schreb. is an allohexaploid (2n = 6x = 42) species which has a gametophytic self-incompatibility system controlled by two genes designated S and Z (Lundquist, 1962). Tall fescue is evolutionarily close to other Festuca species, including diploid and hexaploid meadow fescue (Festuca pratensis) (Hand et al., 2010). This important crop is also member of a polyploid set of taxa, this including a tetraploid (Festuca arundinacea var. glaucescens Boiss syn. Festuca arundinacea subsp. fenas -Lag.-Arcang.), an octaploid (Festuca arundinacea subsp. atlantigena -St. Yves- Auquier) and a decaploid one (Festuca arundinacea var. letourneuxiana -St. Yves- syn. Festuca arundinacea subsp. cirtensis -St. Yves- J. Gamisans) (Hand et al., 2010). Additionally, there are three major eco-geographic races (morphotypes) commonly described within tall fescue: Continental, Mediterranean and Rhizomatous (Hand et al., 2010).
The phylogenetic relationship among accessions from a world germplasm collection of tall fescue has been recently analyzed using the matK gene and rDNA ITS region (Hand et al., 2012a). These robust analyses allowed classifying the accessions into the three morphotypes and showed the presence of tall fescue sub-species of varying ploidy levels, in addition to other closely related species within this germplasm collection (Hand et al., 2012a). In addition, numerous molecular tools have been developed to explore the genetic diversity of these related plants (Xu et al., 1994; Pasakinskiene et al., 2000; Jones et al., 2002; Mian et al., 2002; Momotaz et al., 2004; Saha et al., 2004; Lauvergeat et al., 2005; Saha et al., 2006; Tehrani et al., 2009; Hand et al., 2012a,b). Particularly, large genetic diversity of tall fescue has been observed using simple sequence repeats (SSR) and single-nucleotide polymorphism (SNP) molecular markers (Hand et al., 2012a,b). In this study, we estimated the genetic diversity and ploidy level of tall fescue from the publicly available National Institute of Agricultural Technology (INTA) germplasm collection (Argentina).
The material used consisted of 133 accessions from the United States Department of Agriculture (USDA) and 28 populations collected in the Pampa Region of Argentina, an area located between 34°10′ S 59°03′ W and 36°11′ S 62°46′ W. The latter are available in the Active Germplasm Bank (AGB) at the National Institute of Agricultural Technology of Argentina. Accessions “718” and “791” were collected in Unquera, Santander, Spain (43°37′ S 5°54′ W) and Ortigueira, La Coruña, Spain (43°65′ S 7°88′ W), respectively.
For the analysis of genetic variability, genomic DNA (75 mg) was extracted from 30 young leaves of the populations (bulk), according to the methodology described by Puecher et al. (2001). We analyzed a set of 10 genomic SSRs from Lolium: LPSSRK10F08, LPSSRK10H05, LPSSRH01A07, LPSSRH07G05, LPSSRH03F03, LPSSRK02E02, LPSSRK03A02, LPSSRK03B03, LPSSRH01H06, and LPSSRK02E08 (Jones et al., 2001; Forster et al., 2004), and a set of 30 EST-SSRs from tall fescue: NFFa002, NFFa004, NFFa009, NFFa015, NFFa019, NFFa021, NFFa023, NFFa024, NFFa027, NFFa030, NFFa031, NFFa034, NFFa036, NFFa039, NFFa041, NFFa045, NFFa047, NFFa048, NFFa049, NFFa052, NFFa058, NFFa059, NFFa061, NFFa064, NFFa066, NFFa068, NFFa069, NFFa073, NFFa074, and NFFa075 (Saha et al., 2004). PCR amplifications were performed in a final volume of 20 μL in the presence of 75 ng DNA, 1 U of Taq polymerase (Platinum Taq DNA Polymerase, Invitrogen), 2.5 mM MgCl2, 0.2 mM of each dNTP, 2 μL of 10 X PCR Buffer (Invitrogen) and 0.5 mM of each primer. The PCR protocol was: 1 cycle at 94 °C for 3 min, 40 cycles at 94 °C for 30 s, 55–65 °C (see Table 1) for 1 min, and 72 °C for 2 min. SSR fragments were detected by a Genetic Analyzer ABI 3130 (Applied Biosystems). Each PCR was performed six times.
Table 1.
SSR marker properties following screening of 161 tall fescue accessions.
| Locus | Alleles per locus | Allele size (bp) | PIC | T (°C) |
|---|---|---|---|---|
| NNFa002 | 13 | 280–316 | 0.627 | 62 |
| NNFa015 | 17 | 143–215 | 0.729 | 60 |
| NNFa019 | 13 | 122–182 | 0.671 | 60 |
| NNFa023 | 13 | 156–207 | 0.81 | 62 |
| NNFa024 | 5 | 190–208 | 0.743 | 62 |
| NNFa031 | 16 | 220–380 | 0.709 | 65 |
| NNFa034 | 15 | 161–236 | 0.744 | 62 |
| NNFa041 | 11 | 175–210 | 0.751 | 60 |
| NNFa048 | 16 | 244–310 | 0.761 | 60 |
| NNFa058 | 18 | 85–246 | 0.764 | 60 |
| NNFa064 | 11 | 145–204 | 0.794 | 60 |
| NNFa066 | 12 | 274–329 | 0.721 | 62 |
| LPSSRK03B03 | 24 | 241–320 | 0.791 | 55 |
| LPSSRK10H05 | 16 | 200–310 | 0.7 | 55 |
| LPSSRK02E08 | 14 | 153–227 | 0.84 | 55 |
Phylogenetic and genetic diversity analyses were done using Genemapper 3.4 (Applied Biosystems). A phylogenetic tree was constructed as previously described (Ayub et al., 2007; Soto et al., 2010, 2011, 2012a,b). The root tip squash method described by Ahloowalia (1965) was used to determine chromosome number. Three plants were studied with this protocol: Ryegrass (Lolium perenne) CV. Florida (Gentos) = 2x, Festuca arundinacea subsp. Fenas (Lagsca) Arcangeli Segovia- Spain (GenBank accession number: AF532951) = 4x, and Festuca arundinacea Encore variety (Marathon II®) = 6x. These plants were used as controls for the flow cytometry assays. The ploidy level of all accessions was determined using a flow cytometer (Partec, CA) according to Hand et al. (2010).
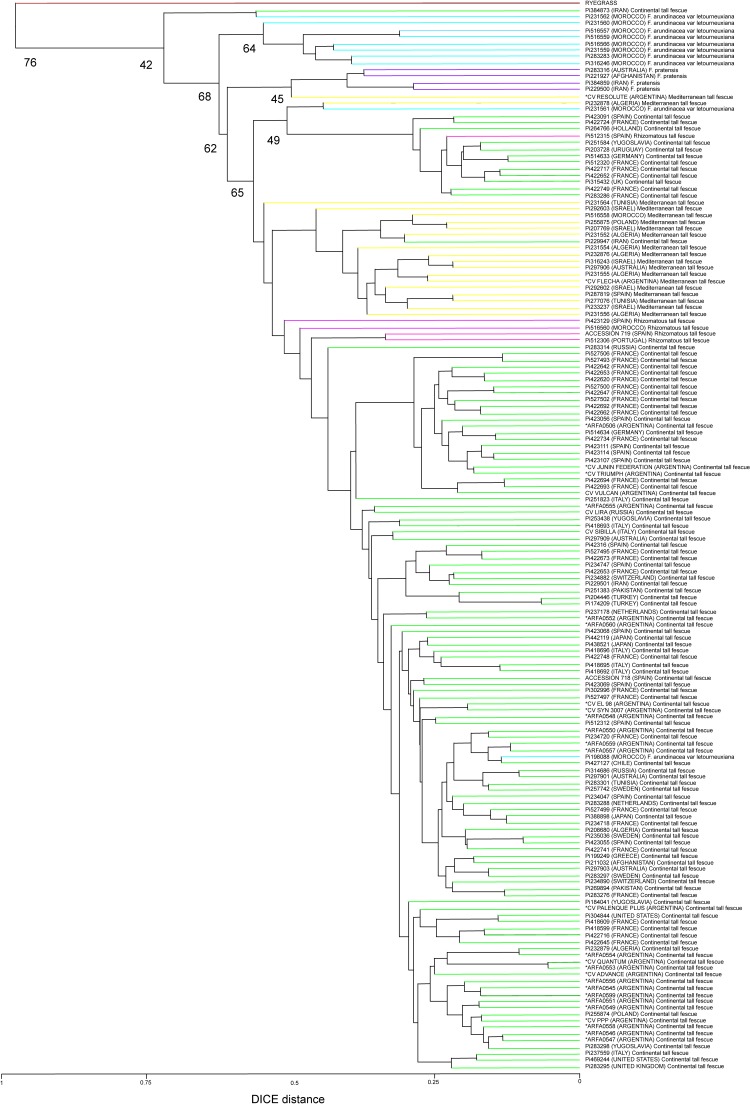
We selected 12 EST-SSRs derived from F. arundinacea and 3 SSRs derived from Lolium due to their high levels of polymorphism. The 15 SSRs selected showed 214 alleles, with band sizes of 122 bp-380 bp and multiple products per SSR (ranging from 5 to 24), at an average of 14.26 alleles per locus (Table 1). In addition, we observed high Polymorphic Information Content (PIC) values: 0.627–0.84 (Table 1). In the dendrogram, Ryegrass was used as an external control (outgroup) because this species is related to the genus Festuca. Ryegrass was the most divergent cluster, with a genetic distance of 1 (+ 1). As described by Hand et al. (2012a), the dendrogram showed an association between the accessions previously assigned to Festuca arundinacea var. letourneuxiana, Festuca pratensis and tall fescue (Continental, Mediterranean and Rhizomatous) (Figure 1).
Figure 1.
UPGMA dendrogram showing the relationships among 161 accessions of Festuca from different countries. Bootstrap percentages are indicated at the branch points. Tree topologies obtained using UPGMA, Neighbor joining, Minimum evolution and Maximum parsimony methods were identical. Ryegrass (red), Continental tall fescue (green), Mediterranean tall fescue (yellow), Rhizomatous tall fescue (pink), Festuca arundinacea var letourneuxiana (blue) and Festuca pratensis (violet). Accessions from INTA germplasm collection (*).
Although using a set of SRR molecular markers different from that used by Hand et al. (2012a) our evolutionary analysis supports the current classification. Only few accessions were in incongruent position: Pi384873 (Iran) Continental tall fescue, Pi198088 (Morocco) Festuca arundinacea var letourneuxiana, Pi229947 (Iran) Continental tall fescue, Pi512315 (Spain) Rhizomatous accessions within Festuca arundinacea var. letourneuxiana, Continental tall fescue, Mediterranean tall fescue and Continental tall fescue groups, respectively (Figure 1). In addition, CV. RESOLUTE (Argentina) Mediterranean tall fescue, Pi232878 (Algeria) Mediterranean tall fescue and Pi231561 (Morocco) Festuca arundinacea var. letourneuxiana, previously described by Hand et al. (2012a), were not found within a defined cluster (Figure 1). With the exception of CV. RESOLUTE, the accessions of tall fescue belonging to the INTA germplasm collection formed a cluster with Continental or Mediterranean tall fescue (Figure 1), suggesting that these populations can be used in breeding programs.
The genetic variation within and between populations was analyzed using 22 populations representing different clusters, and employing the 15 polymorphic SSRs listed in Table 1. We found 153 alleles (85–380 bp) with 14 alleles per locus. The DICE similarity coefficient computed for the 660 individuals (30 individuals per population) was 0.12 to 0.8. The cophenetic correlation was r = 0.75. AMOVA (Excoffier et al., 1992) analysis revealed a 71% variance within populations and a 29% variance among populations, with a statistically significant variance in heterogeneity among populations (p < 0.0001). Our data place in evidence a large genetic diversity among and within the populations tested.
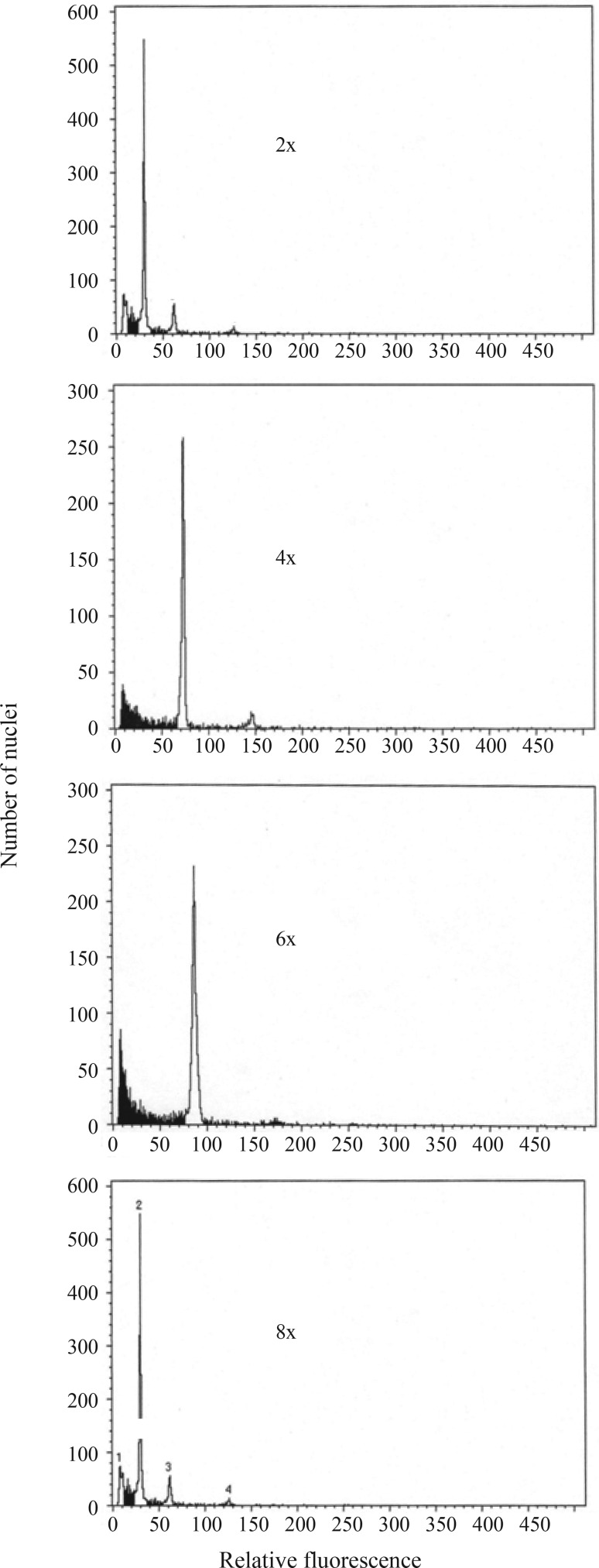
Ploidy level information is also important for bio-diversity and evolutionary studies, and flow cytometry is an efficient, rapid and convenient method to estimate DNA content in plants. The flow cytometry data obtained herein were validated by chromosome counts from root tip cells. Our results suggest that the tall fescue accessions analyzed have four ploidy levels: Pi384873 (Iran) Continental tall fescue (diploid), Pi423129 (Spain) Rhizomatous tall fescue (tetraploid), Festuca pratensis and the three major morphotypes of tall fescue (hexaploid), and Festuca arundinacea var. letourneuxiana (octaploid) (Figure 2). Festuca pratensis, the three major morphotypes of tall fescue, and Festuca arundinacea var. letourneuxiana were classified as diploid, hexaploid and decaploid, respectively (Hand et al., 2012a). Our results, thus, indicate that the ploidy pattern within Festuca accessions is more complex than expected.
Figure 2.
Flow cytometry fluorescence intensities of Pi384873 from Iran (2x), Pi423129 from Spain (4x), Pi221927 from Afghanistan (6x) and Pi231562 from Morocco (8x). Ploidy levels were analyzed for all accessions. The figure shows only one representative example of each ploidy level.
The selection of Festuca arundinacea Schreb. varieties was intensely based on phenotypic appearance, and this breeding method is accompanied with problems, such as decreased forage and seed yield. To overcome this, a supplementary study based on molecular markers for a more precise evaluation of genetic variability at the genotype level is desireable. The estimation of genetic variation among and within populations is, therefore, a useful tool to predict potential genetic gain in breeding programs which make use of a genetic structure analysis of germplasm collections. Among DNA markers, SSRs or microsatellites, which are abundant, codominant and hypervariable, are being extensively used in genetic mapping, phylogenetic studies, and marker-assisted selection (Hand et al., 2012a). In this study, we used SSR markers and capillary electrophoresis as tools to assess the genetic variation and determine the relationships among different Festuca accessions from a wide range of geographical origins. These accessions are available in international collections (USDA and AGB), and the high quality and easily reproducible data presented in this work can be used to select diverse parents in breeding programs. They are also useful for maintaining the genetic variation in germplasm, which is crucial in utilizing the genetic potential of these genotypes for improvement of traits needed for adaptation to different conditions. In addition, the results from the flow cytometric analysis of DNA content in the Festuca accessions suggest that the genetic structure of fescue is more complex than previously thought. Thus, analyzing the DNA content of plants should be an easy way for creating new cultivars in this economically important forage grass. Furthermore, the recent identification and characterization of SNP markers in tall fescue morphotypes suggests that the SNP collection could be used for cultivar identification, genetic linkage map construction, genome-wide association studies and genomic selection in this important crop (Hand et al., 2012b). Finally, the results shown in this study indicate that Festuca arundinacea Schreb. has a high level of genetic diversity within the INTA germplasm collection and, as a consequence, represents valuable material for future breeding programs.
Acknowledgments
We thank Dr. John Foster for the information about the efficiency of cross-amplification and ortholocus detection by perennial ryegrass genomic DNA-derived SSR markers in tall fescue. In memory of Ing. Agr. Raul Rios (Buenos Aires, Argentina, 1952–2010).
Footnotes
Associate Editor: Dario Grattapaglia
References
- Ahloowalia BS. A root tip squash technique for screening chromosome number in Lolium. Euphytica. 1965;14:170–172. [Google Scholar]
- Ayub ND, Pettinari MJ, Méndez BS, López NI. The polyhydroxyalkanoate genes of a stress resistant Antarctic Pseudomonas are situated within a genomic island. Plasmid. 2007;58:240–248. doi: 10.1016/j.plasmid.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse P, Quattro J. Analysis of molecular variance inferred from genetic distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster JW, Jones ES, Batley J, Smith KF. Molecular marker based genetic analysis of pasture and turf grasses. Dev Plant Breed. 2004;11:197–238. [Google Scholar]
- Hand ML, Cogan NO, Stewart AV, Forster JW. Evolutionary history of tall fescue morphotypes inferred from molecular phylogenetics of the Lolium-Festuca species complex. BMC Evol Biol. 2010;10:e303. doi: 10.1186/1471-2148-10-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand ML, Cogan NO, Forster JW. Molecular characterisation and interpretation of genetic diversity within globally distributed germplasm collections of tall fescue (Festuca arundinacea Schreb.) and meadow fescue (F. pratensis Huds.) Theor Appl Genet. 2012a;124:1127–1137. doi: 10.1007/s00122-011-1774-6. [DOI] [PubMed] [Google Scholar]
- Hand ML, Cogan NO, Forster JW. Genome-wide SNP identification in multiple morphotypes of allohexaploid tall fescue (Festuca arundinacea Schreb) BMC Genomics. 2012b;13:e219. doi: 10.1186/1471-2164-13-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ES, Dupal MP, Kölliker R, Drayton MC, Forster JW. Development and characterisation of simple sequence repeat (SSR) markers for perennial ryegrass (Lolium perenne L.) Theor Appl Genet. 2001;102:405–415. [Google Scholar]
- Jones ES, Mahoney NL, Hayward MD, Armstead IP, Jones JG, Humphreys MO, King IP, Kishida T, Yamada T, Balfourier F, et al. An enhanced molecular marker based genetic map of perennial ryegrass (Lolium perenne) reveals comparative relationships with other Poaceae genomes. Genome. 2002;45:282–295. doi: 10.1139/g01-144. [DOI] [PubMed] [Google Scholar]
- Lauvergeat V, Barre P, Bonnet M, Ghesquire M. Sixty simple sequence repeat markers for use in the Festuca-Lolium complex of grasses. Mol Ecol Notes. 2005;5:401–405. [Google Scholar]
- Lundquist A. The nature of the two-loci incompatibility system in grasses II. Number of alleles at the incompatibility loci in Festuca pratensis Huds. Hereditas. 1962;48:169–181. [Google Scholar]
- Mian M, Hopkins A, Zwonitzer J. Determination of genetic diversity in tall fescue with AFLP markers. Crop Sci. 2002;42:944–950. [Google Scholar]
- Momotaz A, Forster W, Yamada T. Identification of cultivars and populations of Lolium, Festuca and Festulolium hybrids through the detection of simple sequence repeat polymorphism. Plant Breeding. 2004;123:1–7. [Google Scholar]
- Pasakinskienè I, Griffiths C, Bettany A, Pasakinskienè V, Humphreys M. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica. 2000;28:9–17. [Google Scholar]
- Puecher DI, Katsiotis A, Leggett JM, Loukas M, Tsakas S. Genetic variability measures among Bromus catharticus Vahl. Populations and cultivars with RAPD and AFLP markers. Euphytica. 2001;121:229–236. [Google Scholar]
- Saha MC, Mian RMA, Eujay I, Zwonitzer JC, Wang L, May GD. Tall fescue EST-SSR markers with transferability across several grass species. Theor Appl Genet. 2004;101:783–791. doi: 10.1007/s00122-004-1681-1. [DOI] [PubMed] [Google Scholar]
- Saha MC, Cooper JD, Mian RMA, Zwonitzer JC, Chekhovskiy K, Hopkins AA. An SSR- and AFLP-based genetic linkage map of tall fescue (Festuca arundinacea Schreb.) Theor Appl Genet. 2005;110:323–336. doi: 10.1007/s00122-004-1843-1. [DOI] [PubMed] [Google Scholar]
- Smarda P, Bures P, Horová L, Foggi B, Rossi G. Genome size and GC content evolution of Festuca: Ancestral expansion and subsequent reduction. Ann Bot. 2008;101:421–433. doi: 10.1093/aob/mcm307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto G, Fox R, Ayub N, Alleva K, Guaimas F, Erijman EJ, Mazzella A, Amodeo G, Muschietti J. TIP5;1 is an aquaporin specifically targeted to pollen mitochondria and is probably involved in nitrogen remobilization in Arabidopsis thaliana. Plant J. 2010;64:1038–1047. doi: 10.1111/j.1365-313X.2010.04395.x. [DOI] [PubMed] [Google Scholar]
- Soto G, Stritzler M, Lisi C, Alleva K, Pagano ME, Ardila F, Mozzicafreddo M, Cuccioloni M, Angeletti M, Ayub ND. Acetoacetyl-CoA thiolase regulates the mevalonate pathway during abiotic stress adaptation. J Exp Bot. 2011;62:5699–5711. doi: 10.1093/jxb/err287. [DOI] [PubMed] [Google Scholar]
- Soto G, Alleva K, Amodeo G, Muschietti J, Ayub ND. New insight into the evolution of aquaporins from flowering plants and vertebrates: Orthologous identification and functional transfer is possible. Gene. 2012;503:165–176. doi: 10.1016/j.gene.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Soto G, Setten L, Lisi C, Maurelis C, Mozzicafreddo M, Cuccioloni M, Angeletti M, Ayub ND. Hydroxybutyrate prevents protein aggregation in the halotolerant bacterium Pseudomonas sp. CT13 under abiotic stress. Extremophiles. 2012;16:455–462. doi: 10.1007/s00792-012-0445-0. [DOI] [PubMed] [Google Scholar]
- Tehrani MS, Mardi M, Sahebi J, Catalán P, Díaz-Pérez A. Genetic diversity and structure among Iranian tall fescue populations based on genomic-SSR and EST-SSR marker analysis. Plant Syst Evol. 2009;282:57–70. [Google Scholar]
- Xu W, Sleper D, Krause G. Genetic diversity of tall fescue germplasm based on RFLPs. Crop Sci. 1994;34:246–252. [Google Scholar]