Abstract
The insect fat body is a multifunctional organ analogous to the vertebrate liver. The fat body is involved in the metabolism of juvenile hormone, regulation of environmental stress, production of immunity regulator-like proteins in cells and protein storage. However, very little is known about the molecular mechanisms involved in fat body physiology in stingless bees. In this study, we analyzed the transcriptome of the fat body from the stingless bee Melipona scutellaris. In silico analysis of a set of cDNA library sequences yielded 1728 expressed sequence tags (ESTs) and 997 high-quality sequences that were assembled into 29 contigs and 117 singlets. The BLAST X tool showed that 86% of the ESTs shared similarity with Apis mellifera (honeybee) genes. The M. scutellaris fat body ESTs encoded proteins with roles in numerous physiological processes, including anti-oxidation, phosphorylation, metabolism, detoxification, transmembrane transport, intracellular transport, cell proliferation, protein hydrolysis and protein synthesis. This is the first report to describe a transcriptomic analysis of specific organs of M. scutellaris. Our findings provide new insights into the physiological role of the fat body in stingless bees.
Keywords: fat body, gene expression, Melipona, stingless bees, transcriptome
The insect fat body is a diffuse organ that fills the body cavity and consists of mesodermal cells known as trophocytes or fat body cells (Arrese and Soulages, 2010; Roma et al., 2010). In addition to trophocytes, bees contain oenocytes, another cell type of ectodermal origin found scattered throughout the fat body (Paes de Oliveira and Cruz-Landim, 2003; Martins et al., 2011a,b; Price et al., 2011).
The insect fat body is a multifunctional organ with a role in a variety of metabolic processes, including: a) storage of proteins, lipids and carbohydrates, which are precursors for metabolism in other organs, b) regulation of chemical compounds released in the haemolymph, c) synthesis of vitellogenin and d) synthesis of antimicrobial peptides that act in the innate immune response (Cruz-Landim, 1985; Tzou et al., 2002; Andrade et al., 2010, 2011; Ottaviani et al., 2011).
Gene expression, assessed via transcriptomic analysis, can provide insights into animal biology and physiology since changes in gene expression may reflect genetic responses to environmental stimuli, as well as immune responses. Expressed sequence tags (ESTs) provide a fast, accurate tool for gene identification and are widely used in functional genomic studies (Adams et al., 1991; Zweiger and Scott, 1997; Martins et al., 2011b).
Melipona scutellaris is a stingless bee found in northeastern Brazil. Populations of this bee have decreased substantially as a consequence of human activity (Kerr et al., 1996) and studies of this species are important for its maintenance.
Workers of M. scutellaris were obtained from the stingless bee colony at the Institute of Genetics and Biochemistry, Federal University of Uberlandia, State of Minas Gerais, Brazil. Foraging workers were identified by the presence of pollen in the pollen sacs and were collected when returning to the hive. The abdomens of 15 foragers were opened with forceps while submerged in insect saline solution (0.1 M NaCl, 0.1 M KCl, 0.1 M CaCl2) and the fat body layers under the tergites and sternites were separated, transferred to liquid nitrogen and stored at −80 °C.
mRNA was purified using a Micro-FastTrack 2.0 mRNA isolation kit (Invitrogen) and used to synthesize the first strand of cDNA. A cDNA library of the M. scutellaris fat body was constructed using a SuperScript Plasmid System kit with Gateway Technology for cDNA Synthesis and Cloning (Invitrogen). The clones were sequenced using DYEnamic ET Dye Terminator Cycle Sequencing for MegaBace DNA Analysis Systems kits (GE Healthcare) according to the manufacturers recommendations and analyzed with a MegaBace 1000 sequencer (Amersham Biosciences).
The computer program EGassembler from the Human Genome Center was used to trim the vector and mitochondrial sequences. Phred (Ewing and Green, 1998; Ewing et al., 1998) and Base Caller Cimarron v.3.12 softwares were used to identify high-quality sequences (Phred > 20 and ≥150 base pairs) that were assembled using CAP3. The high-quality sequences were compared using the algorithms BLAST X and BLAST N v.2.2.25 with an initial E-value threshold of 10−6 (Altschul et al., 1990, 1997). Gene Ontology and FatiGO tools were used to assign possible biological functions to the fat body transcripts based on comparison with Drosophila proteins, as described by Al-Shahrour et al. (2004).
A total of 2.26 × 105 clones were obtained from the fat body cDNA library. The length of randomly cloned cDNAs was determined by agarose gel electrophoresis; the fragment size varied from 150 to 2000 base pairs (data not shown).
The fat body cDNA sequences yielded 1728 ESTs and 997 high quality reads longer than 150 bp, totaling 197,904 nucleotides. Of the 997 high quality reads, 392 (39.3%) ESTs were from mitochondrial genes and the remaining 605 (60.7%) were edited and assembled into 29 contigs and 117 singlets. Seventy-one percent of the ESTs showed matches with the transcriptome shotgun assembly from Melipona quadrifasciata obtained by Woodard et al. (2011). The ESTs identified here were deposited in dbEST under accession numbers HO000185-HO000320 and HO000363-HO000419.
Nearly 40% of the M. scutellaris fat body ESTs were of mitochondrial origin and are likely also found in the fat body of Aedes aegypti (Feitosa et al., 2006). The number of mitochondria per cell increases 700 fold in newly emerged adult insects (Kurella et al., 2001), suggesting that the mitochondria of fat body cells are an important source of energy for adult bees. In bees, the fat body plays a role in protein synthesis and storage (Ivanova and Staikova, 2007) and, in agreement with this, we found transcripts involved in protein synthesis.
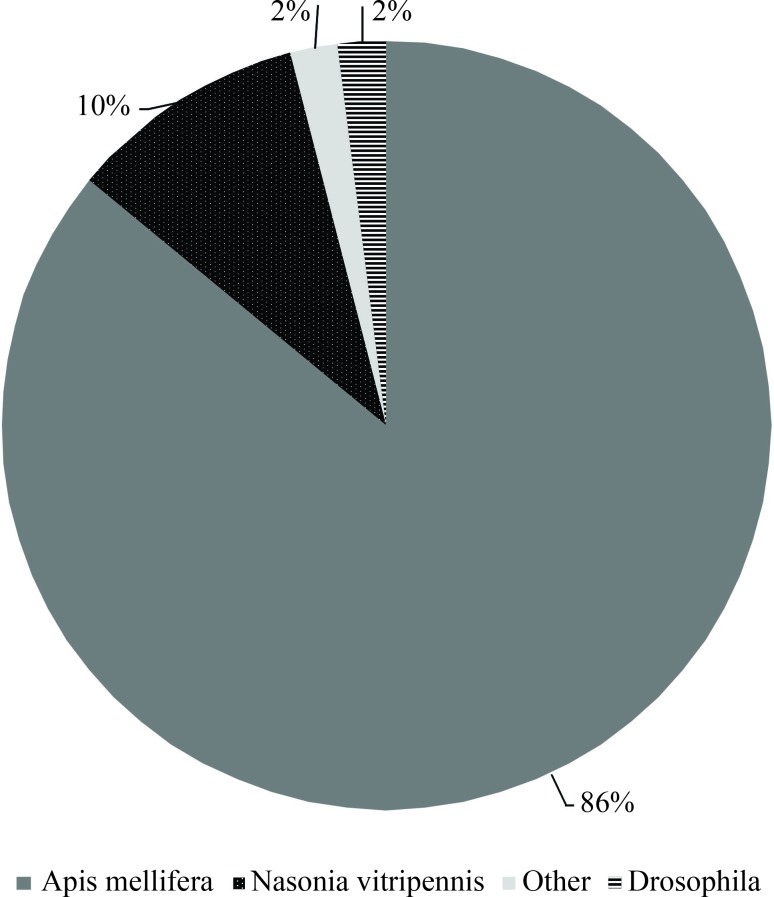
The M. scutellaris fat body ESTs showed 86% similarity with Apis mellifera genes and the remaining 14% with Nasonia vitripennis, Drosophila melanogaster and others (Figure 1). Of the two hymenopteran species, A. mellifera is phylogenetically closer to M. scutellaris than N. virtripennis (The Honeybee Genome Sequencing Consortium, 2006; Nasonia Genome Working Group, 2010).
Figure 1.
Transcript distribution of the best matches of the Melipona scutellaris fat body cDNA library. The best matches were listed based on the insect species without considering E-values.
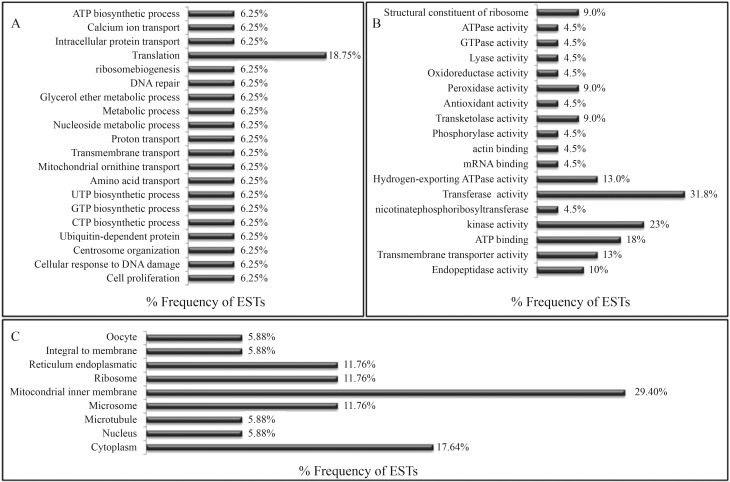
The comparison of contigs and singlets with their respective orthologs in D. melanogaster showed that the transcripts identified in M. scutellaris fat bodies were associated with known biological processes, cellular components and molecular functions (Figure 2). The main functions of these proteins were anti-oxidation, phosphorylation, metabolism, detoxification, transmembrane transport, intracellular transport, cell proliferation, protein hydrolysis and protein synthesis. The contigs were assembled from overexpressed genes and most were found to encode proteins with roles in cell metabolism (Table 1). These data agree with the physiological role of the insect fat body (Price et al., 2011).
Figure 2.
Transcript distribution of Melipona scutellaris fat body cDNA library. Protein functions were assigned based on homology with Drosophila genes. (A) Biological process. (B) Molecular function. (C) Cell component.
Table 1.
Best matched Melipona scutellaris contigs from BlastN analysis. The Melipona contig ID is the access number in GenBank. Only sequences that matched the insect sequence are shown.
| Melipona contig ID | Best match | E-value | Identity (%) |
|---|---|---|---|
| HO000238.1 | PREDICTED: similar to CG7530-PA, isoform A [Apis mellifera] | 4.00E-18 | 90 |
| HO000202.1 | PREDICTED: similar to CG4692-PB, isoform B [Apis mellifera] | 1.00E-46 | 68 |
| HO000240.1 | PREDICTED: similar to translationally controlled tumor protein [Nasonia vitripennis] | 9.00E-15 | 76 |
| HO000411.1 | PREDICTED: similar to F27C1.2a [Apis mellifera] | 1.00E-10 | 54 |
| HO000230.1 | PREDICTED: similar to CG10672-PA [Apis mellifera] | 2.00E-19 | 63 |
| HO000262.1 | PREDICTED: similar to CG3271-PB, isoform B [Apis mellifera] | 3.00E-11 | 79 |
| HO000263.1 | PREDICTED: Apis mellifera putative fatty acyl-CoA reductase CG5065-like | 3.00E-33 | 72 |
| HO000317.1 | Heat shock protein 90 [Apis mellifera] | 3.00E-16 | 71 |
| HO000312.1 | PREDICTED: similar to R04B5.5 [Apis mellifera] | 2.00E-71 | 100 |
| HO000199.1 | PREDICTED: similar to sentrin/sumo-specific protease senp7 [Nasonia vitripennis] | 5.00E-10 | 58 |
| HO000273.1 | PREDICTED: similar to serine/threonine-protein kinase rio1 [Nasonia vitripennis] | 4.00E-18 | 64 |
| HO000373.1 | Farnesoic acid o-methyltransferase-like isoform 1 protein [Melipona scutellaris] | 4.00E-34 | 100 |
| HO000393.1 | PREDICTED: similar to microsomal glutathione S-transferase-like CG1742-PA, isoform A isoform 1 [Apis mellifera] |
4.00E-54 | 70 |
| HO000389.1 | GJ13450 [Drosophila virilis] | 6.00E-06 | 53 |
The BLASTX and BLASTN analyses identified sequences homologous to proteins associated with the immune response (Toll proteins, kinases and cytochrome P450), resistance to insecticides, cell cycle control, juvenile hormone metabolism and resistance to environmental stress.
The insect fat body is an organ of the immune system and we identified ESTs related to the immune response in the fat bodies of M. scutellaris, e.g., Toll and other genes, as well as genes for cytochrome P450 and kinases. Martins et al. (2011a) found P450 in A. aegypti oenocytes, a cell type scattered amongst trophocytes in bees. Insects are resistant to infection by microorganisms, although an acquired immune system is lacking. The immune response of insects consists of an innate immune system in which microorganisms and molecules are recognized by specific receptors, thereby activating the cellular immune response associated with phagocytosis, encapsulation and humoral responses (Ribeiro and Brehélin, 2006; Andrade et al., 2010). Hemocytes are cells that play a role in the cellular immune response of insects and have been described in M. scutellaris larvae (Amaral et al., 2010).
Insects can recognize specific microbial markers known as pathogen-associated molecular patterns (PAMPs). PAMPs are recognized by pattern recognition receptors (PRR) that mediate cellular immune responses such as phagocytosis and the serine-proteinase cascade that activates melanization and/or the release of antimicrobial substances (Kavanagh and Reeves, 2004). A PRR found in insects is Toll, a transmembrane receptor with a leucine-rich extracellular domain and an intracellular region similar to the interleukin-1 receptor (Leclerc and Reichhart, 2004). Toll receptors recognize lipids, carbohydrates, peptides and nucleic acids from different invaders (Akira et al., 2006).
Amongst the M. scutellaris fat body ESTs, 23% were for kinases, mainly mitogen-activated protein kinases (MAP-kinases). Protein kinases (PKs) play a central role in signal transduction, including the transmission of environmental stimuli, the coordination of intracellular processes and in invertebrate defense against pathogens (Kim et al., 2002; Wojda et al., 2004; Chen Chih et al., 2007). In addition, MAP-kinases, such as p42/44 ERK, p38 MAPK, JNK, PKA, PKB, PKC and Akt are involved in the control of cell apoptosis (Cross et al., 2000).
Among the transcripts identified in this study we found an EST that encoded cytochrome P450 (CYP6g2), a family of enzymes with multiple functions (Mansuy, 1998). Although numerous P450 isoforms have been identified in insects, with an average of 80–120 isoforms per individual (Tijet et al., 2001), A. mellifera has only 46 genes that encode P450 proteins (Claudianos et al., 2006). In Drosophila, the genes Cyp12d1, Cyp6g1 and Cyp6g2 are associated with resistance to DDT, neonicotinoids and growth regulators (Daborn et al., 2007). In the M. scutellaris fat body, the EST that encoded for P450 was an ortholog of D. melanogaster CYP6g2, which suggested a function in resistance to insecticides in this stingless bee.
Heat shock proteins (HSPs) have various functions in combating environmental stress and a variety of agents, such as temperature, dehydration, chemicals, heavy metals and other xenobiotics can regulate the expression of HSP (Sun and MacRae, 2005; Rinehart et al., 2006; Benoit et al., 2010). Several ESTs for HSPs were detected in the M. scutellaris fat body transcriptome. Two of these ESTs showed high similarity (92%; E-value = 7 × 10−77) with cytosolic HSP90 isoforms that have been associated with caste differentiation in A. mellifera (Xu et al., 2010). Some genes associated with caste differentiation in honeybees have been identified (Pinto et al., 2002; Cristino et al., 2006; Barchuk et al., 2007; Mackert et al., 2010), and caste differentiation in stingless bees has also been widely studied (Kerr, 1947; Kerr and Nielsen, 1966; Bonetti, 1984, 1995; Santana et al., 2006; Vieira et al., 2008).
An EST found in the fat body of M. scutellaris was similar to a putative farnesoic acid O-methyl-transferase (FAMeT). This putative enzyme catalyzes the synthesis of methylfarnesoate from farnesoic acid in the biosynthetic pathway of juvenile hormone in the corpora allata. Vieira et al. (2008) found that FAMeT of M. scutellaris (MsFAMeT) has different expression levels in castes, suggesting that this enzyme may be associated with the metabolism of juvenile hormone in this stingless bee. These findings suggest either that this enzyme is ambiguous in Melipona and can participate in other biochemical pathways or that it is not specific for juvenile hormone synthesis. Although no ESTs encoding for enzymes that degrade juvenile hormone were detected in the M. scutellaris fat body, RT-PCR and qPCR detected mRNAs for juvenile hormone esterase and juvenile hormone epoxide hydrolase (data not shown), which confirms the expression of these genes in this fat body.
In conclusion, we have identified 1728 ESTs from the fat body of M. scutellaris. In silico analysis of these ESTs has provided new insights into the physiological roles of this tissue in phenomena such as innate immunity, cellular proliferation, resistance to insecticides and environmental stress, and caste differentiation in stingless bees.
Acknowledgments
This work was supported by grants from the Brazilian research agencies Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG - EDT 522/07 to JES and CBB-APQ-02128-10 to AMB), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coorde-nação de Aperfeiçoamento de Pessoal de Ensino Superior (CAPES). We thank Kevin Cloonan (University of California, Davis) for critically reviewing the English version of this manuscript.
Footnotes
Associate Editor: Klaus Hartfelder
References
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: A web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral IMR, Neto JFM, Pereira GB, Franco MB, Beletti ME, Kerr WE, Bonetti AM, Ueira-Vieira C. Circulating hemocytes from larvae of Melipona scutellaris (Hymenoptera, Apidae, Meliponini): Cell types and their role in phagocytosis. Micron. 2010;41:123–129. doi: 10.1016/j.micron.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Andrade GS, Serrão JE, Zanuncio JC, Zanuncio TV, Leite GLD, Polanczyk RA. Immunity of an alternative host can be overcome by higher densities of its parasitoids Palmistichus elaeisis and Trichospilus diatraeae. PloS One. 2010;5:e13231. doi: 10.1371/journal.pone.0013231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Soulages JL. Insect fat body: Energy, metabolism and regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo DO, Zanuncio JC, Delabie JHC, Serrão JE. Temporal variation of vitellogenin synthesis in Ectatomma tuberculatum (Formicidae, Ectatomminae) workers. J Insect Physiol. 2011;57:972–977. doi: 10.1016/j.jinsphys.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simões ZLP, Maleszka R. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev Biol. 2007;7:e70. doi: 10.1186/1471-213X-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit JB, Lopez-Martinez G, Phillips ZP, Patrick KR, Denlinger DL. Heat shock proteins contribute to mosquito dehydration tolerance. J Insect Physiol. 2010;56:151–156. doi: 10.1016/j.jinsphys.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti AM. Efeitos do hormônio juvenil no desenvolvimento ovariano de Melipona quadrifasciata. Rev Bras Biol. 1984;44:509–516. [Google Scholar]
- Bonetti AM, Kerr WE, Matusita SH. Effects of juvenile hormones I, II and III, in single and fractionated dosage in Melipona bees. Rev Bras Biol. 1995;55:113–120. [PubMed] [Google Scholar]
- Chen Chih WR, Shaio MF, Cho WL. A p38 MAP kinase regulates the expression of the Aedes aegypti defensin gene in mosquito cells. Insect Mol Biol. 2007;16:389–399. doi: 10.1111/j.1365-2583.2007.00734.x. [DOI] [PubMed] [Google Scholar]
- Claudianos C, Ranson H, Johnson R, Biswas S, Schuler M, Berenbaum M, Feyereisen R, Oakeshott J. A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol Biol. 2006;15:615–636. doi: 10.1111/j.1365-2583.2006.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristino A, Nunes F, Lobo C, Bitondi MMG, Simões ZLP, Da Fontoura Costa L, Lattorff H, Moritz R, Evans J, Hartfelder K. Caste development and reproduction: A genome wide analysis of hallmarks of insect eusociality. Insect Mol Biol. 2006;15:703–714. doi: 10.1111/j.1365-2583.2006.00696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross TG, Scheel-Toellner D, Henriquez NV, Deacon E, Salmon M, Lord JM. Serine/threonine protein kinases and apoptosis. Exp Cell Res. 2000;256:34–41. doi: 10.1006/excr.2000.4836. [DOI] [PubMed] [Google Scholar]
- Cruz-Landim C. Histological and cytological studies on the fat body of the queen honeybee abdomen during the active oviposition phase. Rev Bras Biol. 1985;45:221–232. [Google Scholar]
- Daborn PJ, Lumb C, Boey A, Wong W, Batterham P. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem Mol Biol. 2007;37:512–519. doi: 10.1016/j.ibmb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier LD, Wendl MC, Green P. Base-calling of automated sequencer traces usingPhred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Feitosa FM, Calvo E, Merino EF, Durham AM, James AA, Bianchi AG, Marinotti O, Capurro ML. A transcriptome analysis of the Aedes aegypti vitellogenic fat body. J Insect Sci. 2006;6:1–26. doi: 10.1673/1536-2442(2006)6[1:ATAOTA]2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova E, Staikova T. Stage specificity in the expression of proteins of honey bee fat body (Apis mellifera L.) in the course of ontogenesis. J Cell Mol Biol. 2007;6:129–135. [Google Scholar]
- Kavanagh K, Reeves EP. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol Rev. 2004;28:101–112. doi: 10.1016/j.femsre.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Kerr WE. Estudo Sobre o Genero Melipona. USP Editora; São Paulo: 1947. p. 67. [Google Scholar]
- Kerr WE, Nielsen RA. Evidences that genetically determined Melipona queens can become workers. Genetics. 1966;54:859–866. doi: 10.1093/genetics/54.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr WE, Carvalho GA, Nascimento VA. Abelha Uruçu: Biologia, Manejo e Conservação. Fundação Acangaú; Belo Horizonte: 1996. p. 144. [Google Scholar]
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan M-W, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- Kurella M, Hsiao L-L, Yoshida T, Randall JD, Chow G, Sarang SS, Jensen RV, Gullans SR. DNA microarray analysis of complex biologic processes. J Am Soc Nephrol. 2001;12:1072–1078. doi: 10.1681/ASN.V1251072. [DOI] [PubMed] [Google Scholar]
- Leclerc V, Reichhart JM. The immune response of Drosophila melanogaster. Immunol Rev. 2004;198:59–71. doi: 10.1111/j.0105-2896.2004.0130.x. [DOI] [PubMed] [Google Scholar]
- Mackert A, Hartfelder K, Bitondi MMG, Simões ZLP. The juvenile hormone (JH) epoxide hydrolase gene in the honey bee Apis mellifera genome encodes a protein which has negligible participation in JH degradation. J Insect Physiol. 2010;56:1139–1146. doi: 10.1016/j.jinsphys.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Mansuy D. The great diversity of reactions catalyzed by cytochromes P450. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;121:5–14. doi: 10.1016/s0742-8413(98)10026-9. [DOI] [PubMed] [Google Scholar]
- Martins GF, Guedes BAM, Silva LM, Serrão JE, Fortes-Dias CL, Ramalho-Ortigão JM, Pimenta PFP. Isolation, primary culture and morphological characterization of oenocytes from Aedes aegypti pupae. Tissue Cell. 2011a;43:83–90. doi: 10.1016/j.tice.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Martins GF, Ramalho-Ortigão JM, Lobo NF, Severson DW, McDowell MA, Pimenta PFP. Insights into the transcriptome of oenocytes from Aedes aegypti pupae. Mem Inst Oswaldo Cruz. 2011b;106:308–315. doi: 10.1590/s0074-02762011000300009. [DOI] [PubMed] [Google Scholar]
- Nasonia Genome Working Group Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani E, Malagoli D, Franceschi C. The evolution of the adipose tissue: A neglected enigma. Gen Comp Endocrinol. 2011;174:1–4. doi: 10.1016/j.ygcen.2011.06.018. [DOI] [PubMed] [Google Scholar]
- Paes de Oliveira VT, Cruz-Landim C. Size of fat body trophocytes and the ovarian development in workers and queens of Melipona quadrifasciata anthidioides. Sociobiology. 2003;41:701–709. [Google Scholar]
- Pinto LZ, Hartfelder K, Bitondi MMG, Simões ZLP. Ecdysteroid titers in pupae of highly social bees relate to distinct modes of caste development. J Insect Physiol. 2002;48:783–790. doi: 10.1016/s0022-1910(02)00103-8. [DOI] [PubMed] [Google Scholar]
- Price DP, Seo SS, Kim JY, Park SY. Anti-apoptotic protein TCTP controls the stability of the tumor suppressor p53. FEBS Lett. 2011;585:29–35. doi: 10.1016/j.febslet.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Ribeiro C, Brehélin M. Insect haemocytes: What type of cell is that? J Insect Physiol. 2006;52:417–429. doi: 10.1016/j.jinsphys.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Rinehart JP, Hayward SAL, Elnitsky MA, Sandro LH, Lee RE, Jr, Denlinger DL. Continuous up-regulation of heat shock proteins in larvae, but not adults, of a polar insect. Proc Natl Acad Sci USA. 2006;103:14223–14227. doi: 10.1073/pnas.0606840103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roma GC, Bueno OC, Camargo-Mathias MI. Morphophysiological analysis of the insect fat body: A review. Micron. 2010;41:395–401. doi: 10.1016/j.micron.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Santana FA, Nunes FMF, Vieira CU, Machado MAMS, Kerr WE, Silva WA, Jr, Bonetti AM. Differentially displayed expressed sequence tags in Melipona scutellaris (Hymenoptera, Apidae, Meliponini) development. An Acad Bras Ciênc. 2006;78:69–75. doi: 10.1590/s0001-37652006000100008. [DOI] [PubMed] [Google Scholar]
- Sun Y, MacRae TH. The small heat shock proteins and their role in human disease. FEBS J. 2005;272:2613–2627. doi: 10.1111/j.1742-4658.2005.04708.x. [DOI] [PubMed] [Google Scholar]
- The Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijet N, Helvig C, Feyereisen R. The cytochrome P450 gene superfamily in Drosophila melanogaster: Annotation, intron-exon organization and phylogeny. Gene. 2001;262:189–198. doi: 10.1016/s0378-1119(00)00533-3. [DOI] [PubMed] [Google Scholar]
- Tzou P, De Gregorio E, Lemaitre B. How Drosophila combats microbial infection: A model to study innate immunity and host-pathogen interactions. Curr Opin Microbiol. 2002;5:102–110. doi: 10.1016/s1369-5274(02)00294-1. [DOI] [PubMed] [Google Scholar]
- Vieira CU, Bonetti AM, Simões ZLP, Maranhão AQ, Costa CS, Costa MCR, Siquieroli ACS, Nunes FMF. Farnesoic acid O-methyl transferase (FAMeT) isoforms: Conserved traits and gene expression patterns related to caste differentiation in the stingless bee, Melipona scutellaris. Arch Insect Biochem Physiol. 2008;67:97–106. doi: 10.1002/arch.20224. [DOI] [PubMed] [Google Scholar]
- Wojda I, Kowalski P, Jakubowicz T. JNK MAP kinase is involved in the humoral immune response of the greater wax moth larvae Galleria mellonella. Arch Insect Biochem Physiol. 2004;56:143–154. doi: 10.1002/arch.20001. [DOI] [PubMed] [Google Scholar]
- Woodard SH, Fischman BJ, Venkat A, Hudson ME, Varala K, Cameron SA, Clark AG, Robinson GE. Genes involved in convergent evolution of eusociality in bees. Proc Natl Acad Sci USA. 2011;108:7472–7477. doi: 10.1073/pnas.1103457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PJ, Xiao JH, Xia QY, Murphy B, Huang DW. Apis mellifera has two isoforms of cytoplasmic HSP90. Insect Mol Biol. 2010;19:593–597. doi: 10.1111/j.1365-2583.2010.01015.x. [DOI] [PubMed] [Google Scholar]
- Zweiger G, Scott RW. From expressed sequence tags to epigenomics: An understanding of disease processes. Curr Opin Biotechnol. 1997;8:684–687. doi: 10.1016/s0958-1669(97)80119-x. [DOI] [PubMed] [Google Scholar]
Internet Resources
- EGassembler from Human Genome Center, http://egassembler.hgc.jp (accessed June 4, 2011).
- Gene Ontology, http://www.geneontology.org (accessed June 11, 2011).