Abstract
Optimization of aqueous solutions of the integral membrane protein (IMP) OmpW for NMR structure determination has been monitored with micro-coil NMR, which enables the acquisition of NMR spectra using only micrograms of protein and detergent. The detergent 30-Fos (2-undecylphosphocholine) was found to yield the best 2D [15N,1H]-TROSY correlation NMR spectra of [2H,15N]-labeled OmpW. For the OmpW structure determination we then optimized the 30-Fos concentration, the sample temperature and long-time stability, and the deuteration level of the protein. Some emerging guidelines for reconstitution of β-barrel integral membrane proteins in structural biology are discussed.
Keywords: Structural biology, integral membrane proteins, solubilization in detergent micelles, E. coli outer membrane protein W
The E. coli outer membrane protein W (OmpW) (Jalajakumari and Manning 1990) belongs to a family of small outer membrane proteins responsible for transport of small molecules across the membrane (Bush and Saier 2003). A crystal structure of the 191-amino acid protein OmpW in a mixture of the LDAO (lauryl-dimethylamine-oxide) and OG (n-octyl-β-D-glucoside) detergents at 2.7 Å resolution shows an 8-stranded β-barrel with height ≈ 50 Å and diameter ≈ 17 Å (Hong et al. 2006). OmpW was selected for a structure determination by NMR in solution because significantly longer loops connect the transmembrane β-strands than in the previously investigated, related proteins OmpX (Fernández et al. 2004; Vogt and Schulz 1999) and OmpA (Arora et al. 2001; Fernández et al. 2001, 2003; Pautsch and Schulz 1998, 2000). In addition to the biological interest, studies of OmpW thus promise to extend the present knowledge on the complementarity of X-ray crystal structures and NMR solution structures of this class of integral membrane proteins (IMP) (Fairman et al. 2011; Bill et al. 2011). This communication describes the preparation of a “structure-quality” solution of OmpW, which enabled complete NMR assignments of the polypeptide backbone as a basis for the structure determination (to be published).
The selection of the detergents used to reconstitute OmpW was based on the following previous experiments with β-barrel IMPs. In earlier work we identified DHPC (dihexanoylphosphocholine) as a suitable detergent for reconstitution of the outer membrane protein X (OmpX) for NMR structure determination (Fernandez et al. 2003, 2004). In subsequent screens of a large library of commercially available and newly synthesized detergents for optimal reconstitution of OmpX, DHPC was used as a reference (Zhang et al. 2008). Prior to NMR screening, the detergent library was prescreened using an SDS-PAGE assay, which revealed that phosphocholine derivatives are the most efficient detergents for OmpX refolding. 30-Fos, 34-Fos, 115-Fos, 116-Fos, 138-Fos, 179-Fos, 180-Fos, 182-Fos, 185-Fos, 228-Fos, 229-Fos, Fos-10, Fos-11, Fos-12 (DPC) and Fos-13 (TPC) (chemical structures are shown in Zhang et al. 2008) were then further evaluated using microcoil NMR equipment to record 2D [15N,1H]-TROSY correlation spectra (Pervushin et al. 1997). The eight detergents DHPC, Fos-10, Fos-11, 116-Fos, 115-Fos, 138-Fos, 179-Fos and Fos-30 were thus found to yield high-quality NMR spectra. In a follow-up study with the E.coli outer membrane protein OmpA, the same eight detergents yielded good results. The best two detergents identified in these earlier studies with β-barrel IMPs, i.e., Fos-10 (n-decylphosphocholine) and 30-Fos (2-undecylphosphocholine), were selected for the OmpW project, and DHPC was retained as a reference (Fig. 1, a–c).
Fig. 1.
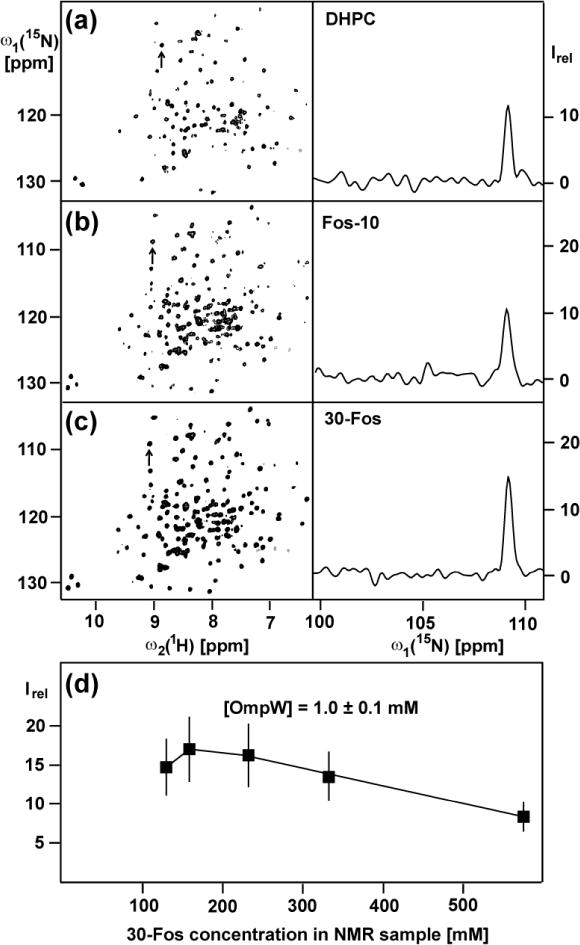
700 MHz 2D [15N,1H]-TROSY correlation NMR spectra of [2H,15N]-labeled OmpW solubilized at 1 mM concentration in 230 mM aqueous solutions of three different detergents at 298 K. (a) DHPC. (b) Fos-10. (c) 30-Fos. For each detergent the left panel shows a contour plot of the 15N–1H correlation peaks, and the right panel displays a cross section along ω1 (15N) of the peak identified with an arrow in the left panel. (d) Plot of the mean intensity of the cross-peaks in the 2D [15N,1H]-TROSY spectra of [2H,15N]-labeled OmpW reconstituted in 30-Fos micelles, Irel, versus the 30-Fos concentration. The error bars represent the standard deviation to the mean taken over all the 15N–1H cross peaks. The following acquisition and processing parameters were used to record the 2D [15N,1H]-TROSY spectra with a 1.7 mm microcoil probehead: data size 100 (t1) × 1024 (t2) complex points; t1max = 35.24 ms; t2max = 86.11 ms; 128 scans per t1 increment; overall measurement time 8 h per experiment. Before Fourier transformation the data matrices were multiplied with an exponential window function in the acquisition dimension, and with a 75º-shifted sine bell window (DeMarco and Wüthrich 1976) in the indirect dimension.
The following criteria were used to evaluate the detergents on the basis of 2D [15N,1H]-TROSY correlation spectra: (i) Completeness of NMR observation of the IMP by comparison of the number of observed backbone 15N-1H correlation peaks with the number expected from the amino acid sequence. (ii) High average peak intensity and uniform distribution of peak intensities. (iii) Analysis of the peak line shapes to support the interpretation of the peak intensity measurements. For OmpW/DHPC mixed micelles, 112 of the expected 185 polypeptide backbone cross-peaks (191 amino acids, 5 Pro) were resolved in the 2D [15N,1H]-TROSY spectra, whereas about 190 peaks were counted for OmpW/Fos-10 and OmpW/30-Fos. The heights of the cross-peaks in the 2D [15N,1H]-TROSY correlation map of OmpW/30-Fos were about 25 % larger than in the spectra of OmpW/Fos-10 mixed micelles. Since the average of the line widths obtained with the two detergents was closely similar, we concluded that the difference in peak heights was due to higher loss of protein during the reconstitution with Fos-10. 30-Fos was therefore selected for further optimization of OmpW solutions for NMR structure determination.
Considering the previous observation that high-quality solution NMR spectra of the related protein OmpX in Fos-10 micelles could be obtained only for a narrow range of detergent concentrations (Stanczak et al. 2009), we investigated the NMR spectra at variable 30-Fos concentrations (Fig. 1d). At 1 mM OmpW concentration the pattern of cross-peaks in 2D [15N,1H]-TROSY spectra was found to be unchanged for 30-Fos concentrations in the range 120 to 600 mM, documenting the structural integrity of the OmpX/30-Fos micelles over this concentration range. However, optimal line widths and peak heights of the 15N–1H signals were observed only for 30-Fos concentrations between about 160 and 230 mM (Fig. 1D). Additional measurements of translational and rotational diffusion rates (Horst et al. 2012; experimental details are given in the Materials and Methods section) showed that the diffusion coefficients Dt and Dr for the OmpW/30-Fos mixed micelles decrease at higher detergent concentrations (Table 1). The monotonous dependence of Dt and Dr on the 30-Fos concentration was used to extrapolate for diffusion coefficients at infinite dilution, Dt,0 and Dr,0, and from these to calculate the hydrodynamic radius, Rh,0 (Stanczak et al. 2009). The resulting Rh,0 values of about 30 Å correspond to an effective rotational correlation time at infinite dilution, τc, of 27 ns. Comparison of the Rh values at different detergent concentrations (Table 1) shows that the hydrodynamic radius of OmpW/30-Fos mixed micelles remains unchanged at 30 ± 1 Å at variable detergent concentrations above about 160 mM. Similar to earlier studies with OmpX reconstituted in Fos-10 micelles (Stanczak et al. 2009), these data document the presence of a homogeneous population of OmpW/30-Fos micelles over a wide range of detergent concentrations. The deterioration of the NMR spectra at 30-Fos concentrations above 230 mM (Fig. 1d) can therefore be traced to increased micro-viscosity due to the higher content of protein-free 30-Fos micelles.
Table 1.
Translational and rotational diffusion coefficients, Dt and Dr, respectively and the hydrodynamic radius, Rh, of 1 mM OmpW at variable 30-Fos concentrations.
| [30-Fos] (mM) | Dt (10−11m2/s)a | Dr (106/s)b | Rh (Å)c |
|---|---|---|---|
| 160 | 4.9 ± 0.1 | 4.5 ± 0.129 ± 1 | |
| 230 | 4.4 ± 0.1 | 3.5 ± 0.130 ± 1 | |
| 330 | 3.2 ± 0.1 | 2.7 ± 0.1 | 30 ± 1 |
Determined by 1H-TRO-STE NMR experiments (Horst et al. 2011)
Determined by TRACT NMR experiment (Lee et al. 2006)
Rh = (3Dt/4Dr)1/2
OmpW solubilized in 230 mM 30-Fos was found to be stable at temperatures up to about 310 K. Increasing the temperature from 298 to 308 K did not significantly influence the cross-peak pattern of the 2D [15N,1H]-TROSY spectra, but there was a two-fold increase of the peak heights (Fig. 2). Exposure of the protein sample to 308 K for 18 days did not change the chemical shifts but there was a loss in the peak intensities of about 40 % (Fig. 2c). This shows that the structure of the OmpW/30-Fos mixed micelles is maintained over this time span, but that part of the protein is lost and probably precipitated out of the solution. A further increase of the temperature to 316 K did not result in further improvement of the NMR spectrum (Fig. 2d), and at this temperature the protein was lost within two days.
Fig. 2.
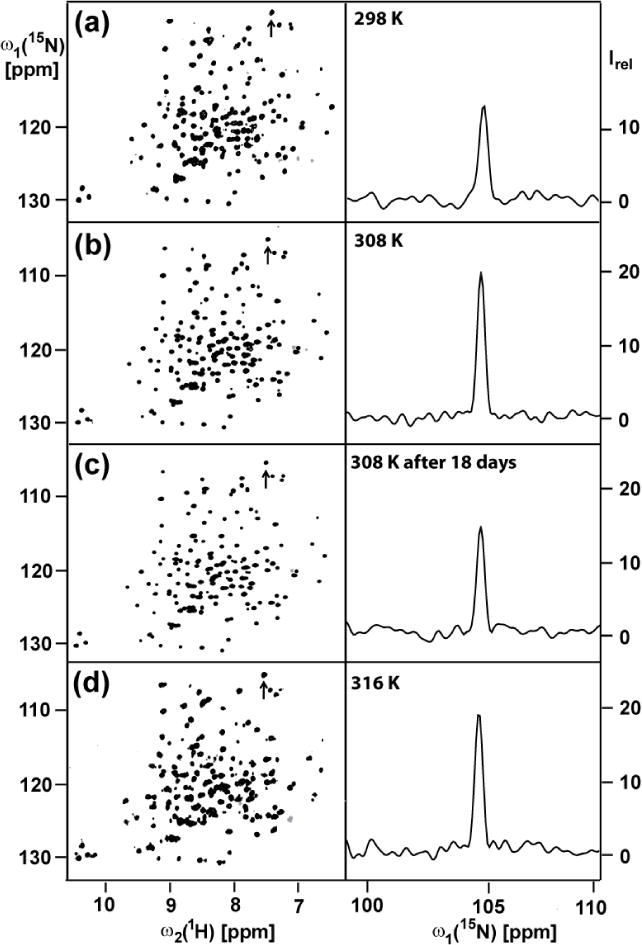
Temperature dependence of the 700 MHz 2D [15N,1H]-TROSY correlation NMR spectrum of a 1 mM aqueous solution of [2H,15N]-labeled OmpW reconstituted in 230 mM 30-Fos detergent. Same presentation as in Fig. 1, a–c. The same acquisition and processing parameters were used as in Fig. 1. The sample temperatures are indicated in the panels on the right. (c) To test for long-time stability, the measurement (b) was repeated after keeping the sample at 308 K for 18 days.
Finally we investigated whether or not it was warranted to go to the extra expense of using the highest possible deuteration level for OmpW, as can be achieved by using deuterated glucose as the sole carbon source in the growth medium. With the increased level of deuteration, line narrowing along ω1 (15N) in the [15N,1H]-TROSY spectra from about 28 ± 6 Hz to 21 ± 5 Hz was achieved, which results in significantly improved spectral resolution and increased peak heights (Fig. 3).
Fig. 3.
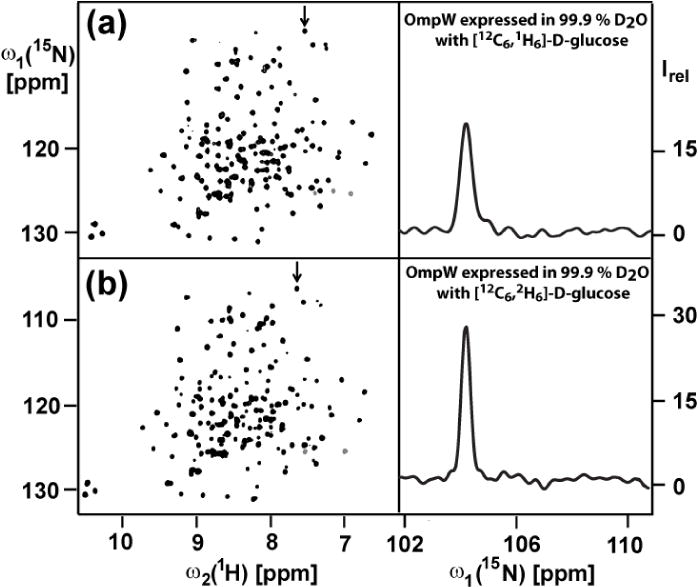
Effect of variable 2H-labeling levels on the 2D [15N,1H]-TROSY spectra at 298 K of 1 mM [2H,15N]-labeled OmpW reconstituted in aqueous solution containing 230 mM 30-Fos. Same presentation as in Figures 1, a–c and 2. (a) Protein expressed with protonated glucose. (b) Protein expressed with deuterated glucose. The same acquisition and processing parameters were used as in Fig. 1, except that 256 scans per t1 increment were accumulated and the overall measurement time per experiment was 16 h.
In conclusion, based on the results from the micro-scale NMR experiments shown in the figures 1–3 and in Table 1, milligram quantities of uniformly 2H,13C,15N-labeled OmpW were prepared for a structure determination at 308 K, using solution conditions of 5 mM phosphate buffer at pH 6.8, 10 mM NaCl, 0.3 % (v/w) NaN3 and 230 mM 30-Fos (to be published). With regard to future work with small β-barrel IMPs, the present work combined with previous experience from studies of OmpX and OmpA (Arora et al. 2001; Fernandez et al. 2001; Horst et al. 2012; Stanczak et al. 2009; Zhang et al. 2008) provides the following guidelines: While short-chain phosphocholine derivatives are clearly indicated as favorable detergents for NMR structure determination, a screen of several from among the best detergents is recommended for each new protein. Here, this is clearly illustrated by the fact that reconstitution with DHPC enabled a complete structure determination of OmpX, although Fos-10 and 30-Fos were subsequently found to be superior detergents for this protein (Zhang et al. 2008), whereas in the more demanding situation of OmpW, DHPC did not yield “structure-quality” reconstitution (Figure 1a–c). Furthermore, once the “best” detergent for a given IMP has been selected, it pays to optimize the solution conditions for the NMR experiments, as shown in the figures 1d, 2 and 3.
Materials and Methods
OmpW precloned in the pET11a plasmid was transformed into E.coli BL-21 (DE3) (Stratagene) competent cells for expression. Transformed cells were first grown at 37 °C in 20 ml of Luria Bertani (LB) medium to an optical density at 600 nm (OD600) above 1.5. 50 ml of unlabeled M9 minimal medium was inoculated with 2 ml of this LB culture and grown at 37 °C to an OD600 ≥ 0.5 The cells were transferred to 200 ml of minimal medium containing 99.9 % D2O (Spectra Gases), 4 g/l [1H,12C]-glucose or [2H,12C]-glucose, and 1 g/l [15N]-ammonium chloride (CIL, > 98 % 15N). At OD600 ≥ 0.5 the cells were spun down and resuspended in 1 l of minimal medium where protein overexpression was induced at OD600 ≥ 0.7 with 1 mM isopropyl-β-D-thiogalactopyranoside at 37 °C. About 5 hrs after induction the cells had reached the stationary growth phase and were harvested by centrifugation at 5000 g and 4 °C for 10 min. The cell pellet was resuspended in TE buffer (20 mM Tris-HCl at pH 7.5, 5 mM EDTA) and sonicated for 25 min., using a Misonix 3000 sonicator. The solution was pelleted for 1 h at 5000 g and 4 °C, and the insoluble pellet was resuspended at room temperature in TE buffer containing 2 % (v/v) Triton C-100, and again pelleted at 5000 g and 4 °C for 30 min. Excess triton was removed by repeating the same procedure with TE buffer. OmpW was recovered from the inclusion bodies by resuspending the pellet in TE buffer containing 6 M urea for 2.5 h at room temperature, and remaining cell debris were removed by centrifugation at 48000 g and 4 °C for 20 min. Purification of OmpW was performed using an ÄKTA purifier (Amersham/GE) with a 5 ml HiTrap Q HP anion exchange column (Amersham/GE). OmpW was repeatedly bound to the column using buffer A (20 mM Tris-HCl at pH 7.5, 6 M urea) and eluted with 6 % (w/v) NaCl. The pooled OmpW fractions were loaded onto a size exclusion column Superdex Hiload 16/60 (Amersham/GE) equilibrated with buffer B (20 mM Tris-HCl at pH 7.5, 6 M urea, 100 mM NaCl) and eluted with the same buffer. The pooled pure OmpW fractions were desalted using a 5 ml HiPrep desalting column (Amersham/GE) and then concentrated to about 12 mg/ml using a Vivaspin-2 concentrator with 10 kDa molecular weight cutoff.
To reconstitute OmpW into detergent micelles, 100 μl of freshly prepared solution containing 12 mg/ml of [2H,15N]-labeled OmpW in 6 M urea was added to 600 μl refolding buffer (20 mM Tris-HCl at pH 7.5, 5 mM EDTA, 600 mM L-Arg, 45 mM detergent) and stirred. The refolding buffer was then exchanged with NMR buffer (5 mM phosphate buffer at pH 6.8, 10 mM NaCl, 0.3 % (v/w) NaN3, 10 mM detergent) by repeated dilution/concentration cycles. “Upconcentration” of the detergent during these dilution/concentration cycles (Stanczak et al. 2009) was taken into account when adjusting the detergent concentration in the NMR sample. For example, 10 mM 30-Fos in the NMR buffer results in approximately 160 mM 30-Fos in the NMR sample (Fig. 4).
Figure 4.
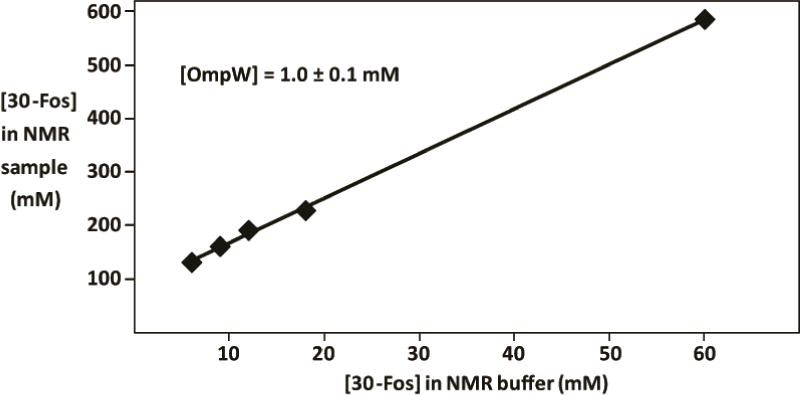
Up-concentration of the detergent 30-Fos during OmpW preparation (see text). The 30-Fos concentration in the NMR sample is plotted along the vertical axis versus the Fos-10 concentration in the NMR buffer along the horizontal axis. The 30-Fos concentrations in the NMR buffer and the NMR sample were measured with microscale 1D 1H NMR experiments. The 1D 1H NMR spectra were collected with the following parameters: complex data size = 16 K, acquisition time = 1.38 s, number of scans = 128, sweep width = 11900 Hz.
NMR experiments were recorded on a Bruker DRX-700 spectrometer equipped with a 1.7 mm TXI microcoil probehead (Bruker, Billerica, MA). 2D [15N,1H]-TROSY correlation experiments (Pervushin et al. 1997) were recorded as described previously (Stanczak et al. 2009). Details of the parameter settings are given in the legends of Figures 1 and 3. The NMR data were processed with the software TOPSPIN 1.3 (Bruker). The analysis of the [15N,1H]-TROSY correlation spectra was performed with XEASY (Bartels et al. 1995).
The rotational diffusion coefficient of the OmpW/30-Fos micelles, Dr, was determined with the TRACT experiment (Lee et al. 2006). 40 relaxation delays ranging from 1 to 32 ms were used, with 256 scans per relaxation delay. The relaxation rates Rα and Rβ used to calculate Dr were obtained by fitting the integrals over the spectral region from 10.0 to 8.7 ppm to a one-parameter exponential decay. The translational diffusion constant, Dt, was measured using transverse relaxation-optimized stimulated echo (1H-TRO-STE) spectroscopy (Horst et al. 2011). 16 diffusion-weighted one-dimensional spectra were recorded in a two-dimensional manner, using a pair of gradient pulses of 4.5 ms duration separated by 50 ms, with gradient strengths ranging from 1 to 50 G cm−1. The TRACT and 1H-TRO-STE data were analyzed with in-house TOPSPIN macros in combination with the program XMGRACE (http://plasma-gate.weizmann.ac.il).
Acknowledgments
This work was supported by the Joint Center for Innovative Membrane Protein Technologies (JCIMPT-complexes, NIH Roadmap Grant P50 GM073197). K.W. is the Cecil H. and Ida M. Green Professor of Structural Biology at The Scripps Research Institute.
References
- Arora A, Abildgaard F, Bushweller JH, Tamm LK. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat Struct Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- Bartels C, Xia T, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- Bill RM, Henderson PJ, Iwata S, Kunji ER, Michael H, Neutze R, Newstead S, Poolman B, Tate CG, Vogel H. Overcoming barriers to membrane protein structure determination. Nat Biotechnol. 2011;29:335–340. doi: 10.1038/nbt.1833. [DOI] [PubMed] [Google Scholar]
- Busch W, Saier MH., Jr The IUBMB-endorsed transporter classification system. Methods Mol Biol. 2003;227:21–36. doi: 10.1385/1-59259-387-9:21. [DOI] [PubMed] [Google Scholar]
- Fairman JW, Noinaj N, Buchanan SK. The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr Opin Struct Biol. 2011;21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco A, Wüthrich K. Digital filtering with a sinusoidal window function: an alternative technique for resolution enhancement in FT NMR. J Magn Reson. 1976;24:201–204. [Google Scholar]
- Fernández C, Hilty C, Bonjour S, Adeishvili K, Pervushin K, Wüthrich K. Solution NMR studies of the integral membrane proteins OmpX and OmpA from Escherichia coli. FEBS Lett. 2001;504:173–178. doi: 10.1016/s0014-5793(01)02742-9. [DOI] [PubMed] [Google Scholar]
- Fernández C, Wüthrich K. NMR solution structure determination of membrane proteins reconstituted in detergent micelles. FEBS Lett. 2003;555:144–150. doi: 10.1016/s0014-5793(03)01155-4. [DOI] [PubMed] [Google Scholar]
- Fernández C, Hilty C, Wider G, Güntert P, Wüthrich K. NMR structure of the integral membrane protein OmpX. J Mol Biol. 2004;336:1211–1221. doi: 10.1016/j.jmb.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Hong H, Patel DR, Tamm LK, van den Berg B. The outer membrane protein OmpW forms an eight-stranded β-barrel with a hydrophobic channel. J Biol Chem. 2006;28:7568–7577. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
- Horst R, Horwich AL, Wüthrich K. Translational diffusion of macromolecular assemblies measured using transverse-relaxation-optimized pulse field gradient NMR. J Am Chem Soc. 2011;133:16354–16357. doi: 10.1021/ja206531c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst R, Stanczak P, Serrano P, Wüthrich K. Translational diffusion measurements by micro-coil NMR in aqueous solutions of the Fos-10 detergent-solubilized membrane protein OmpX. J Phys Chem B. 2012;116:6775–6780. doi: 10.1021/jp212401w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalajakumari MB, Manning PA. Nucleotide sequence of the gene, ompW, encoding a 22 kDa immunogenic outer membrane protein of Vibrio cholera. Nucl Acids Res. 1990;18:2180. doi: 10.1093/nar/18.8.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Hilty C, Wider G, Wüthrich K. Efficient correlation time measurements for macromolecules from NMR relaxation interference. J Magn Reson. 2006;178:72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Pautsch A, Schulz GE. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- Pautsch A, Schulz GE. High-resolution structure of the OmpA membrane domain. J Mol Biol. 2000;298:273–282. doi: 10.1006/jmbi.2000.3671. [DOI] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanczak P, Horst R, Serrano P, Wüthrich K. NMR characterization of membrane protein–detergent micelle solutions by use of microcoil equipment. J Am Chem Soc. 2009;131:18450–18456. doi: 10.1021/ja907842u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt J, Schulz GE. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure. 2010;7:1301–1309. doi: 10.1016/s0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Horst R, Geralt M, Ma X, Hong W, Finn MG, Stevens RC, Wüthrich K. Microscale NMR screening of new detergents for membrane protein structural biology. J Am Chem Soc. 2008;130:7357–7363. doi: 10.1021/ja077863d. [DOI] [PMC free article] [PubMed] [Google Scholar]


