Abstract
Background and objective
In 2008 we developed a shared health research information network (SHRINE), which for the first time enabled research queries across the full patient populations of four Boston hospitals. It uses a federated architecture, where each hospital returns only the aggregate count of the number of patients who match a query. This allows hospitals to retain control over their local databases and comply with federal and state privacy laws. However, because patients may receive care from multiple hospitals, the result of a federated query might differ from what the result would be if the query were run against a single central repository. This paper describes the situations when this happens and presents a technique for correcting these errors.
Methods
We use a one-time process of identifying which patients have data in multiple repositories by comparing one-way hash values of patient demographics. This enables us to partition the local databases such that all patients within a given partition have data at the same subset of hospitals. Federated queries are then run separately on each partition independently, and the combined results are presented to the user.
Results
Using theoretical bounds and simulated hospital networks, we demonstrate that once the partitions are made, SHRINE can produce more precise estimates of the number of patients matching a query.
Conclusions
Uncertainty in the overlap of patient populations across hospitals limits the effectiveness of SHRINE and other federated query tools. Our technique reduces this uncertainty while retaining an aggregate federated architecture.
Keywords: Algorithms; Medical Record Linkage; Search Engine; Medical Records Systems, Computerized; Hospital Shared Services
Background and significance
Clinical data repositories are becoming increasingly important tools for a variety of types of clinical research, including clinical trial recruitment, epidemiology studies, pharmacovigilance monitoring, and comparative effectiveness research. However, scientists have traditionally only been able to access clinical data collected by their own institutions. Therefore, in 2008 we developed a Shared Health Research Information Network (SHRINE), which for the first time enabled research queries across the full patient populations of four Boston hospitals.1 SHRINE builds on other systems that use a federated architecture instead of a large central database.2–7 By keeping patient data within local databases at each hospital, SHRINE can broadcast a query across the network and reveal the number of matching patients at each hospital without requiring any single patient's details to leave an institution. This minimizes hospitals' concern about sharing patient data and allows them to comply with federal and state privacy laws, while at the same time giving researchers access to the largest possible patient population. SHRINE has been cited in high-profile clinical studies and has tremendous potential to transform clinical research.8–11
A significant limitation of SHRINE, as well as any federated network returning aggregate counts, is that the sum of the counts in the federated system is not necessarily the same as what the result would be if the query were run against a combined, central database. This is because the patient populations of each hospital are not mutually exclusive. Patients often receive care from multiple hospitals, especially when those hospitals are geographically close, such as the original four Boston hospitals in SHRINE. As a result, some of the patients being counted by one hospital can be the same patients being counted by another. If those counts are simply added, SHRINE will overestimate the total number of patients in the network. In addition, no single hospital might have the complete medical record of a patient if that patient is also being treated by another hospital. Therefore, local hospital databases might be missing information about their patients, which could result in SHRINE underestimating the number of patients who match a query compared to what would be found in a combined, central database.
In this paper, we show how these problems can be addressed by knowing which patients receive care from more than one hospital. Several methods have been developed to link patients records in different hospital databases that require sharing patient data among the hospitals.12–15 However, in 2002 Grannis demonstrated a way of creating these linkages using data that has been de-identified with a one-way hash function.16 These data are combinations of demographic variables, such as social security number, name, year of birth, and gender. With multiple variables included in the hash, the specificity can approach 100%. The sensitivity can then be increased by generating multiple hash values for each patient using different combinations of variables. If any single hash value matches between patients at two hospitals, a linkage can be created between those patient records. Each hospital could send a one-time list of the hash values for all of its patients to a trusted third party, which will compare the values and generate a random code for each distinct patient. We will demonstrate that once this initial linkage is complete, SHRINE can return more precise estimates of the number of patients matching a query, while still sharing only aggregate counts and not the individual patient codes.
Materials and methods
Partitioning patient populations according to their overlap with other hospitals
We build on Grannis’ idea by proposing that the trusted third party, in addition to returning to the hospitals the random codes that have been assigned to their patients, also return the list of all hospitals where those patients receive care. For example, if patient P1 is in the databases of hospitals H1 and H3; patient P2 is in the databases of hospitals H1, H2, and H3; and patient P3 is only in the database of H3; then H1 will be returned the set {(P1,{H1,H3}), (P2,{H1,H2,H3})}, H2 will be returned {(P2,{H1,H2,H3})}, and H3 will be returned {(P1,{H1,H3}), (P2,{H1,H2,H3}), (P3,{H3})}.
This can be done more succinctly by also generating a random code for each possible list of hospitals. For h hospitals, there are 2h−1 potential combinations. The empty set is not counted because a patient has to be in at least one hospital's database. Theoretically, the number of combinations grows exponentially as the number of hospitals in the network increases; however, most of these combinations will be associated with no patients. There are two reasons for this. First, although there are many patients who receive care from two or three hospitals, there will be essentially none who receive care from, say, 100 different hospitals. Second, the number of combinations of hospitals associated with at least one patient cannot be any larger than the total number of patients. Since there are a finite number of patients in the world (7 billion), and a much smaller number in any one hospital network, this sets an upper bound.
In the case where there is no overlap in patient populations among the hospitals in the network, there will be h combinations, with each containing a single hospital. In networks where the hospitals are geographically distant, located in different countries, or see different populations (eg, pediatrics vs adults), the overlap may be very small, with the number of combinations close to h. The lower bound is one combination, which occurs either when there is a single hospital in the network or when every patient exists in every hospital's database.
Let c be the actual number of hospital combinations with at least one patient. In the above example, there are c=3 combinations represented by the set {(C1,{H1,H2}), (C2,{H1,H2,H3}), (C3,{H3})}. If the trusted third party first returns this set to the hospitals, then it can subsequently return hospital H1 the patient set {(P1,C1),(P2,C2)}, return H2 the set {(P2,C2)}, and return H3 the set {(P1,C1), (P2,C2),(P3,C3)}. This enables each hospital to group its patient population into c partitions, with some partitions within a given hospital having no patients.
Determining bounds on aggregate federated queries without partitioning
Currently in SHRINE, a query returns a single aggregate count from each hospital of the number of matching patients. The process for determining the lower and upper bounds on the actual number of patients in the network depends on the type of query. In this section, we first describe how to do this without any information about which patients exist in multiple hospitals’ databases.
In a Type 1 query, hospitals do not have to exchange data about a patient in order for that patient to be identified as a match in a federated query. For example, in a search for patients with diabetes OR hypertension, as long as one hospital has a record of one of those diagnoses, then the patient will be counted in the aggregate total from all hospitals. Compared to a single combined repository, the federated network will find the same set of patients. The aggregate total will never underestimate the actual number of patients, but it will overestimate it if two hospitals include the same patient in their individual aggregate counts.
For h hospitals returning counts n1 through nh, the upper bound on the actual number of patients is sum(ni), where 1≤i≤h. This case occurs when there is no overlap in the patient populations of the hospitals—the count from each hospital represents a distinct population, and therefore the sum of those counts equals the total number of patients. The lower bound is max(ni) because the count from each individual hospital guarantees that there are at least that number of matching patients in the network, though it could be the case that the matching patients at a single hospital represent the superset of the matching patients at all other hospitals.
In a Type 2 query, individual hospitals can, with certainty, identify some matching patients, but for other patients the required data facts might be spread across multiple hospitals. For example, in a search for patients with diabetes AND hypertension, hospital H1 can confirm that patient P1 matches the query if P1 has both diagnoses in H1's database. However, if patient P2's diabetes diagnosis only exists in H1's database and P2's hypertension diagnosis only exists in H2's database, then neither hospital will identify P2 as a match. Type 2 queries can therefore underestimate the actual number of matching patients. They can also overestimate the number of matching patients in the same way as Type 1 queries. The lower bound is still max(ni), but the upper bound is more difficult to calculate. Without additional information beyond the aggregate count from each hospital, the upper bound is infinite because it is impossible to know how many patients were missed by the query. However, an upper bound can be determined by splitting the Type 2 query into separate Type 1 queries. For example, one Type 1 query can be used to calculate an upper bound on the number of patients in the network with diabetes, and another Type 1 query can be used to calculate an upper bound on the number of patients in the network with hypertension. The actual number of patients who have both diabetes and hypertension cannot be greater than the smaller of the upper bounds of the two Type 1 queries.
In a Type 3 query, individual hospitals cannot be certain that any patient matches a query without additional information from the other hospitals. For example, in a search for patients with diabetes AND NOT hypertension, hospital H1's database contains P2's diabetes diagnosis but has no record that P2 also has hypertension—a fact that only H2 knows. As a result, the patients that H1 identifies in its local search might be false matches, and the actual count might be zero. Therefore, the lower bound on the overall federated Type 3 query is zero. Splitting the Type 3 query into separate Type 1 queries might help. For example, if the lower bound on the number of patients with diabetes in a network is 1000 and the upper bound on the number of patients with hypertension is 800, then there are at least 200 patients with diabetes and not hypertension. The upper bound is calculated in the same way as a Type 2 query—in this example, there can be no more than 1000 patients with diabetes and not hypertension, which would happen if all the patients with hypertension were patients without diabetes.
Determining bounds on aggregate federated queries with partitioning
Partitioning removes the uncertainty around which hospitals contain data about a patient. SHRINE can take advantage of this by splitting a query into c separate queries that each run against a different partition, and then adding the results. In other words, each hospital is asked for not one overall aggregate count, but rather one aggregate count for each of the c partitions. The SHRINE ‘aggregator’, which combines results from each hospital, calculates the bounds for each partition. It then returns the lower bound for the overall query as the sum of the lower bounds for the partitions, and it returns the upper bound for the overall query as the sum of the upper bounds for the partitions. This works because the partitions represent mutually exclusive patient populations. There is no risk of double counting patients by adding the partition counts.
As an example, consider two hospitals, H1 and H2 with three partitions: {(C1,{H1}), (C2,{H2}), (C3,{H1,H2})}. Suppose H1 has 1000 patients with diabetes and H2 has 800 patients with diabetes. Without partitioning, the upper bound on the total number of patients with diabetes is 1000+800=1800 patients. However, because all 800 patients from H2 can also be at H1, the lower bound is 1000. Suppose we know from partitioning that 900 of H1's patients are in C1 and the other 100 are in C3, and 750 of H2's patients are in C2 and the other 50 are in C3. That tells us that most of the patients with diabetes in the network have data at only one hospital. Specifically, the bounds for C1, C2, and C3 are (900, 900), (750, 750), and (100, 150), respectively, giving an overall bounds of (1750, 1800). There is no change to the overall upper bounds. However, by knowing how many patients exist at only one hospital, the overall lower bounds can be increased. With partitioning, in this example the range between the lower and upper bounds fell from 800 to 50, and the number of patients guaranteed to be in the network increased by 75%.
Note that with this method, it is possible to greatly improve the bounds of a query without requiring hospitals to share any information about which specific patients match the query. They are still only returning aggregate counts. The initial process of generating hash values from patient demographics is only used to enable hospitals to partition their local databases. No central master patient index is stored, and no data about individual patients are returned to the user.
Estimating the exact number of patients who match a query
With partitioning, the exact number of matching patients can be determined for the partitions that correspond to a single hospital. However, only a range can still be determined for the other partitions when only aggregate counts are available. A more precise estimate is possible if we allow hospitals associated with a given partition to share some information about individual patients in that same partition. Note that within a given partition, every patient receives care from every hospital associated with that partition. Therefore, this data sharing will not provide any hospital with information about patients the hospital does not already treat.
We will start with Type 1 queries. Suppose there are three hospitals, and one of the partitions is (C1,{H1,H2}). Now consider an investigator from H3 who broadcasts a federated search for patients with diabetes. In addition to their individual counts for C1, hospitals H1 and H2 can work together to estimate the actual number of matching patients in C1, and then return that count to H3. Note that just because H1 and H2 have the same patients in C1, that does not mean they have the same data about those patients. Patient P1 could be treated at both H1 and H2, but only H1 might have the diagnosis of diabetes in its database. If H1 and H2 share with each other their lists of matching patients, they can determine the exact number of patients in C1 with a diagnosis of diabetes, without the risk of double-counting anyone.
This can be done in different ways depending on the policies the hospitals want to adopt. For example, one hospital, H1, can be designated as the aggregator. In this scenario, H2 sends its list of patient codes to H1, and then H1 returns the number of distinct matching patients to H3. Another method would be that H2 sends odd patient codes to H1, and H1 sends even patient codes to H2. Each hospital calculates a partial count, which they independently send to H3.
A problem with this technique is that some queries might require hospitals to exchange very large numbers of patient codes. There could be a practical limitation in terms of performance and bandwidth. An alternative is to use sampling. Suppose H1 has 1000 matching patients in C1, and H2 has 600. H1 sends 10 randomly selected patient codes from its list of matches to H2, and H2 sends 10 random matches to H1. Both hospitals then tell the other which patients are also local matches. For example, H2 returns the three codes it finds to H1, and H1 returns four to H2. Next, H1 uses that information to estimate that 30% of its 1000 matches (300) are also matches at H2, while 70% (700) are unique to H1. Similarly, H2 estimates that 40% of its 600 matches (240) are also matches with H1, while 60% (360) are unique to H2. To resolve discrepancies, they average their estimates, which in this case would be that (300+240)/2=270 matching patients are at both H1 and H2. Finally, the hospitals return the total estimate of 700+360+270=1330 matching patients to H3. For this sampling technique to work, each hospital with matching patients in a partition will have to share data with all the other hospitals in the partition. Thus, with h hospitals and a sample size of s patients, up to h2s patient code exchanges will be required per partition. Even with a small sample size, a large number of patient code exchanges might be necessary to run an entire query.
In Type 1 queries, hospitals only have to share data about patients who matched the query. In Type 2 or Type 3 queries, in order to estimate the number of matching patients in a partition, hospitals will need to share data about patients who potentially do not match the query. For example, in order for H1 and H2 to determine how many patients in C1 have diabetes AND hypertension, while running the query, they will each need to share information with each other about all patients in C1 who have either diabetes OR hypertension. That way they can identify patients with different diagnoses at different hospitals.
Special cases
Since small aggregate counts are potentially identifiable, SHRINE includes an obfuscation algorithm that adds a small random number to the actual count and returns any count less than 10 as ‘<10’.17 With partitioning, it is possible that individual partitions either have fewer than 10 total patients, or they are so small that obfuscation significantly affects the results. Even if the overall total count (the sum of the counts from each partition) presented to a user in SHRINE is large, hospitals might not want to share small un-obfuscated counts ‘behind-the-scenes’ with the query aggregators. One approach is to merge all small partitions into a single partition. This affects how bounds are calculated for that partition because it is no longer known exactly which hospitals have data about those patients. However, it would prevent the patients from being ‘lost’ from the system.
A similar approach of merging partitions can be used when the number of partitions is so large that it is affecting performance. In the extreme case, all partitions associated with more than one hospital can be merged into a single partition. This leaves each hospital with exactly two partitions—the patients who are treated only at that hospital, and the patients who are also treated by at least one other hospital. For networks where there is little overlap in patients between hospitals, this might be the preferred partitioning method.
Unless hospitals repeat the initial step of using hash functions to identify which patients exist at multiple hospitals, they will soon face the problem of new patients entering the network. A special partition will need to be created for these patients. As with merged partitions, the lack of information about whether these patients are also at other hospitals affects the bounds. The more frequently hospitals update their partitions, the smaller this special partition will be.
Simulations
Several parameters of a network determine how much benefit partitioning provides: the number of hospitals; the probability that a patient at one hospital is also in the database of another (patient overlap); the prevalence of the medical concepts being queried; and the probability that a data fact in one hospital (eg, a patient's diagnosis of diabetes) is also recorded in the database of another hospital where that patient receives care (data fact overlap). We demonstrate the effect of these parameters by generating a simulated hospital network consisting of one million patients randomly distributed across five hospitals, with a 20% patient overlap between any two hospitals, a concept with 1% prevalence, and a 10% data fact overlap. Lower and upper bounds of the number of patients returned by a Type 1 federated query with and without partitioning were calculated. An estimate of the actual number of patients that match the query was determined using the sampling technique described above with a sample size of 10. Starting with this initial configuration, each parameter was then modified, one at a time, generating a new simulated network for each combination of parameters. This process was repeated 10 times, and the mean values from the different simulations were calculated. Note that in all simulations, the total number of patients in the network was fixed. Therefore, when the number of hospitals was increased, the number of patients in any one hospital decreased; and when the patient overlap increased, the number of patients in each hospital also increased. The code used for the simulations is available on the i2b2 Community Wiki as a Related Project called Federated Query Simulations (https://community.i2b2.org/wiki/display/Federated).
Results
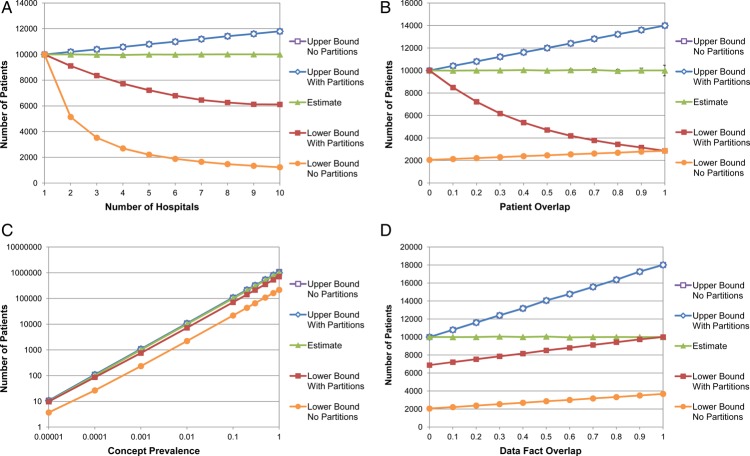
Table 1 and figure 1 illustrate the upper and lower bounds, with and without partitioning, of a federated aggregate query against simulated hospital networks. Also shown is the estimate of the exact number of patients using sampling. The actual number of matching patients in each simulation is 10 000, except in figure 1C, where the concept prevalence is indicated.
Table 1.
Results of federated queries of simulated hospital networks
| Upper bound, no partitions | Upper bound with partitions | Estimate using sampling | Lower bounds with partitions | Lower bounds, no partitions | Patient code exchanges | Standard error of estimate | |
|---|---|---|---|---|---|---|---|
| Number of hospitals | |||||||
| 1 | 10000.0 | 10000.0 | 10000.0 | 10000.0 | 10000.0 | 0.0 | 0.00 |
| 2 | 10204.1 | 10204.1 | 10000.0 | 9109.4 | 5140.6 | 20.0 | 29.37 |
| 3 | 10396.1 | 10396.1 | 9980.6 | 8360.5 | 3524.0 | 120.0 | 25.63 |
| 4 | 10583.6 | 10583.6 | 9958.2 | 7728.5 | 2696.7 | 480.0 | 36.58 |
| 5 | 10798.4 | 10798.4 | 9995.8 | 7218.6 | 2208.3 | 1493.1 | 18.78 |
| 6 | 10993.8 | 10993.8 | 9987.4 | 6795.2 | 1886.0 | 3520.6 | 10.85 |
| 7 | 11198.0 | 11198.0 | 10003.1 | 6459.1 | 1655.9 | 6586.9 | 17.16 |
| 8 | 11421.8 | 11421.8 | 10011.0 | 6267.3 | 1473.0 | 10321.8 | 13.43 |
| 9 | 11604.0 | 11604.0 | 10010.3 | 6125.0 | 1343.2 | 14782.3 | 5.69 |
| 10 | 11798.4 | 11798.3 | 10001.7 | 6116.0 | 1231.2 | 19338.2 | 3.76 |
| Patient overlap | |||||||
| 0 | 10000.0 | 10000.0 | 10000.0 | 10000.0 | 2051.7 | 0.0 | 0.00 |
| 0.1 | 10410.0 | 10410.0 | 9987.5 | 8487.7 | 2132.4 | 938.5 | 13.01 |
| 0.2 | 10803.7 | 10803.7 | 10005.2 | 7218.8 | 2211.6 | 1500.7 | 15.93 |
| 0.3 | 11218.0 | 11218.0 | 10015.5 | 6173.4 | 2295.2 | 1600.0 | 24.69 |
| 0.4 | 11615.6 | 11615.6 | 10037.6 | 5370.4 | 2383.5 | 1600.0 | 23.64 |
| 0.5 | 11999.9 | 11999.9 | 9994.1 | 4712.9 | 2460.1 | 1600.0 | 29.93 |
| 0.6 | 12411.7 | 12411.7 | 10031.7 | 4191.3 | 2546.6 | 1600.0 | 48.51 |
| 0.7 | 12817.5 | 12817.5 | 10047.7 | 3786.5 | 2619.0 | 1600.0 | 62.22 |
| 0.8 | 13218.7 | 13218.7 | 9964.4 | 3437.0 | 2689.7 | 1593.4 | 69.76 |
| 0.9 | 13600.6 | 13600.6 | 10007.4 | 3152.6 | 2788.8 | 1438.5 | 109.42 |
| 1 | 13999.7 | 13999.7 | 10005.3 | 2857.3 | 2857.3 | 200.0 | 242.34 |
| Concept prevalence | |||||||
| 1.00E-05 | 10.8 | 10.8 | 10.0 | 9.8 | 3.7 | 8.3 | 0.00 |
| 0.0001 | 109.2 | 109.2 | 100.0 | 86.0 | 26.7 | 93.1 | 0.00 |
| 0.001 | 1080.4 | 1080.4 | 1002.2 | 756.6 | 234.1 | 685.0 | 0.98 |
| 0.01 | 10812.4 | 10812.4 | 10016.0 | 7205.1 | 2218.2 | 1502.5 | 30.74 |
| 0.1 | 108008.9 | 108008.9 | 100105.1 | 70973.0 | 21757.0 | 1600.0 | 180.61 |
| 0.2 | 215995.9 | 215995.9 | 200228.2 | 141765.1 | 43429.0 | 1600.0 | 437.96 |
| 0.3 | 323976.4 | 323976.4 | 299408.1 | 212319.8 | 65081.9 | 1600.0 | 860.11 |
| 0.5 | 540102.6 | 540102.6 | 500994.3 | 353751.6 | 108317.2 | 1600.0 | 931.12 |
| 0.75 | 810014.8 | 809921.4 | 750978.3 | 530188.1 | 162422.4 | 1600.0 | 1056.48 |
| 1 | 1080088.3 | 1000000.0 | 987211.5 | 706634.7 | 216375.9 | 1600.0 | 1623.11 |
| Data fact overlap | |||||||
| 0 | 10000.0 | 10000.0 | 10000.0 | 6877.6 | 2054.4 | 1443.7 | 0.00 |
| 0.1 | 10797.5 | 10797.5 | 9992.7 | 7209.1 | 2208.8 | 1481.7 | 16.55 |
| 0.2 | 11606.8 | 11606.8 | 10000.2 | 7531.0 | 2383.0 | 1493.4 | 41.70 |
| 0.3 | 12403.7 | 12403.7 | 10050.9 | 7853.6 | 2542.8 | 1526.0 | 32.37 |
| 0.4 | 13182.4 | 13182.4 | 10003.8 | 8158.2 | 2696.1 | 1565.2 | 27.12 |
| 0.5 | 14059.4 | 14059.4 | 10051.7 | 8509.4 | 2866.8 | 1568.8 | 39.38 |
| 0.6 | 14775.1 | 14775.1 | 9956.7 | 8800.9 | 3007.7 | 1569.6 | 24.99 |
| 0.7 | 15568.8 | 15568.8 | 9994.0 | 9122.6 | 3175.7 | 1573.2 | 31.30 |
| 0.8 | 16363.4 | 16363.4 | 9997.6 | 9422.6 | 3328.9 | 1579.6 | 36.47 |
| 0.9 | 17262.9 | 17262.9 | 9991.0 | 9738.8 | 3504.7 | 1600.0 | 14.61 |
| 1 | 18015.8 | 18015.8 | 10000.0 | 10000.0 | 3677.1 | 1594.0 | 0.00 |
| Sample size | |||||||
| 1 | 10813.9 | 10813.9 | 9959.0 | 7190.4 | 2210.4 | 160.0 | 67.53 |
| 2 | 10793.6 | 10793.6 | 9983.0 | 7207.6 | 2221.4 | 316.8 | 29.20 |
| 3 | 10799.6 | 10799.6 | 10004.5 | 7211.7 | 2206.7 | 473.2 | 49.73 |
| 5 | 10792.7 | 10792.7 | 10019.9 | 7210.1 | 2221.5 | 788.4 | 21.68 |
| 10 | 10799.7 | 10799.7 | 10033.0 | 7201.4 | 2208.3 | 1482.8 | 15.42 |
| 20 | 10794.9 | 10794.9 | 9983.9 | 7209.7 | 2206.6 | 2643.1 | 14.37 |
| 50 | 10803.6 | 10803.6 | 10002.2 | 7206.1 | 2210.5 | 5085.5 | 5.50 |
| 100 | 10804.5 | 10804.5 | 10004.7 | 7194.4 | 2212.1 | 6791.9 | 3.73 |
| 500 | 10796.1 | 10796.1 | 10000.0 | 7212.4 | 2209.8 | 9274.4 | 0.00 |
| 1000000 | 10797.2 | 10797.2 | 10000.0 | 7198.9 | 2208.8 | 9286.0 | 0.00 |
Each value represents the mean of 10 simulations.
Figure 1.
Simulated hospital networks, varied by (A) number of hospitals, (B) patient overlap, (C) concept prevalence, and (D) data fact overlap. Vertical bars on the estimate curves represent the 95% CIs. The CIs on the other curves are negligible in size. This figure is only reproduced in colour in the online version.
Without partitioning, as the number of hospitals increases, the upper bound increases and the lower bound decreases. In other words, there becomes greater uncertainty of the actual number of patients. As either the patient overlap or data fact overlap increase, the lower bound increases. However, the upper bound also increases proportionally at approximately the same rate, resulting in a greater range between the two bounds. Changing the concept prevalence has little effect on the bounds relative to the actual number of patients.
In all simulations, partitioning has no effect on the upper bounds. However, the lower bounds are at least as high with partitioning as without, and in most cases several times higher. The greatest benefits of partitioning occur with larger numbers of hospitals and less patient overlap between hospitals. Without partitioning, the lower bound is simply the largest count from any single hospital. The potential of using SHRINE to demonstrate that there enough patients in a multi-institution study to conduct a trial is completely lost. All that can be determined is which one location would be best for a single hospital study. In the extreme case where there is no patient overlap, and patients are evenly distributed across all h hospitals, the lower bound underestimates the number of patients by a factor of h. In contrast, when there is no patient overlap, partitioning yields the exact number of matching patients.
With a 20% patient overlap, which is closer to a real-world scenario, the lower bound with five hospitals was 2210.4 (95% CI 2202.2 to 2218.6) without partitioning and 7190.4 (95% CI 7167.4 to 7213.4) with partitioning, a 3.25-fold increase. The upper bound was 10 813.9 (95% CI 10 796.7 to 10 831.1) in both cases.
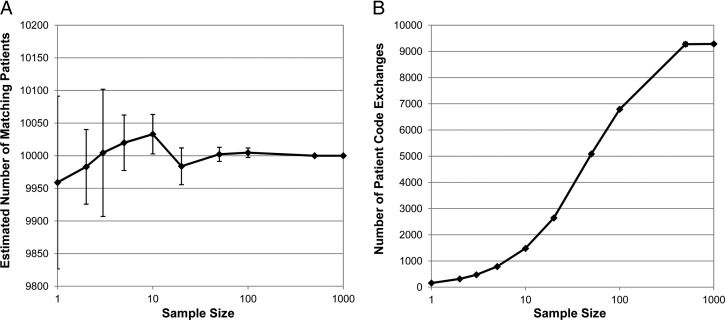
Note that with partitioning, the actual number of matching patients, 10 000, is still 39.1% higher than the lower bound. Figure 2 shows that exchanging individual patient codes between hospitals creates a much better estimate. Even with a sample size of just one patient per hospital pair per partition, which corresponds to 160 total patient code exchanges, the actual number of patients was within the 95% CI of the average estimate of 9959.0 (95% CI 9826.7 to 10 091.4), and the SD of the 10 estimates (ie, the accuracy) was only 2.1% of the actual number of patients. Increasing the sample size narrows the CI and improves the accuracy. The sinusoidal shape in figure 2B is due to the fact that at some point the sample size becomes larger than the number of patients in the partition. Figure 1 shows that a sample size of 10 (up to 1600 patient code exchanges) resulted in a near exact estimate in every combination of simulation parameters tested.
Figure 2.
Estimating the actual number of matching patients in a five-hospital network using sampling. The actual number of patients is 10 000. Vertical bars represent the 95% CIs. (A) The estimated number of patients using different sample sizes. (B) The total number of patient code exchanges.
The prevalence of the medical concept being queried and the data fact overlap have a relatively minor effect on the ratio between the lower bounds with or without partitioning. Though, as the data fact overlap approaches 1, the lower bound with partitioning approaches the actual number of matching patients. When the data fact overlap is 0, the upper bound both with and without partitioning is the same as the actual number of matching patients.
Discussion
When deciding between a central database and a federated architecture for systems like SHRINE that return aggregate counts, issues such as patient privacy, security, scalability, and performance come to mind. However, what is often overlooked is the fact that the sum of the counts returned by each hospital in a federated search does not necessarily equal the count that would be returned by a central database. If the overlap in the patient populations of the hospitals is large, then users can easily be misled by the results if they naively assume they can simply add the individual hospital counts. Therefore, when creating a federated search tool that returns aggregate counts, the bounds should always be presented, so that users can understand how to interpret the results.
Unfortunately, the bounds might not be very helpful in estimating the actual number of patients that match a query. The purpose of creating SHRINE was to provide investigators with a larger population of patients to study. However, adding more hospitals to a network could have no effect on the lower bounds, while at the same time increasing the uncertainty of query results. If investigators assume that there is no patient overlap among the hospitals, then the upper bound can be treated as the actual number of matching patients. This might be valid in certain cases; however, in others, such as the SHRINE network we created in Boston, where patients are routinely seen by more than one hospital, that is a much more difficult assumption to make.
Partitioning can increase the lower bounds of queries by several fold, making SHRINE a much more useful tool. Except for the initial process of identifying which patients receive care from multiple hospitals, partitioning does not change the basic technical architecture of SHRINE, where hospitals retain control over their local databases and only have to return aggregate counts. It might also be relatively easy to implement and have little impact on performance. This is because, to date, the local nodes in SHRINE networks have been implemented using an open source software platform, Informatics for Integrating Biology and the Bedside (i2b2).18 The i2b2 software already includes the ability to partition query results by demographic variables such as gender and race. i2b2 can consider the code corresponding to the list of hospitals that treat a patient as just another demographic variable.
Sampling dramatically reduces the uncertainty of federated queries, and in most cases it gives a result almost the same as the actual number of patients that match Type 1 queries. This is true even when the number of patient code exchanges is small. However, it makes the SHRINE architecture much more complicated, and it requires hospitals to share clinical data about individual patients. Privacy and policy concerns can be partially reduced by the fact that data about individual patients only have to be shared between hospitals that both treat the patient. However, some institutions might consider that simply revealing to one hospital that its patients are also treated at another hospital is an unconsented information disclosure.
Once each hospital has determined which of its patients match a Type 1 query, partitioning and sampling is just one way of estimating the total number of distinct patients (ie, the cardinality) in these lists. Several probabilistic algorithms, such as probabilistic counting, LogLog counting, and adaptive counting, have been described in the literature and are frequently used in applications such as database query optimization and network traffic analysis.19–21 In this scenario, instead of sharing individual patient codes, these algorithms would share m new codes from each hospital. These new codes are generated from hashes of the patient codes such that the probability of a code having a particular value can be used to estimate the number of distinct patients. The average of the m estimates gives an overall estimate with an accuracy on the order of 1/sqrt(m).
Probabilistic counting, which produces one of the best estimates, has a 9.7% accuracy for m=64.19 With five hospitals, this requires sharing 320 values. In contrast, partitioning with sampling achieved 2.1% accuracy using only 160 patient code exchanges (plus the overhead of each hospital sharing the aggregate counts of the partitions). However, there are differences in the amount of processing and bandwidth required by each algorithm. Therefore, depending on the actual network, a probabilistic algorithm might perform better than partitioning and sampling for a desired accuracy. Probabilistic algorithms also have the benefit in that the shared codes are each derived from information about multiple patients. So, the chance that one hospital can learn something about a particular patient from another hospital is very small. This might address privacy and policy concerns about sampling individual patient codes and disclosing at which hospitals patients receive care.
A limitation in the simulations we ran is that the hospitals were the same size, with patients and data facts randomly distributed among them. This is the worst-case scenario for lower bound calculations since one hospital's counts do not dominate the others. Also, investigators often have a sense of what the approximate patient overlap is between hospitals and can use this to gauge where the actual number of patients lies between the lower and upper bounds.
In this study, patients were partitioned according to the combinations of hospitals where they receive care. While less ideal, there are other ways to partition patients that do not require exchanging data about individual patients. Consider a two-hospital network, where H1 treats children and H2 treats mostly adult patients. The knowledge that the hospitals treat different age groups can be used to form a type of partition. For example, we previously saw that if H1 has 1000 patients with diabetes and H2 has 800, then the total number of patients with diabetes has bounds of (1000, 1800) without partitioning. We can divide this into the sum of two separate queries: (1) patients with diabetes who are younger than 18 years; and (2) patients with diabetes who are 18 years old or older (including patients who began at H1 that have now transitioned to H2). If H1 returned 600 and 400 for the two queries, respectively, and H2 returned 100 and 700, then the bounds are (600, 700) and (700, 1100) for the individual queries and (1300, 1800) for their sum.
Conclusion
SHRINE and other federated query tools have potential to expand greatly the number of patients available for clinical research. However, uncertainty in the overlap of patient populations across hospitals limits the effectiveness of these tools. In this study we presented a technique that reduces this uncertainty while retaining an aggregate federated architecture.
Acknowledgments
Isaac S Kohane provided helpful guidance on how to frame this study.
Footnotes
Contributors: GW conceived the study, performed the experiments, and wrote the manuscript.
Funding: This work was conducted with support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award 8UL1TR000170-05, and financial contributions from Harvard University and its affiliated academic health care centers) and Informatics for Integrating Biology and the Bedside (i2b2, National Institutes of Health Office of the Director, National Library of Medicine and the National Institute of General Medical Sciences Award 2U54LM008748).
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: The data generated by the simulations are presented in table 1.
References
- 1.Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc 2009;16:624–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holzbach AM, Chueh H, Porter AJ, et al. A query engine for distributed medical databases. Medinfo 2004 2004:1519CD [Google Scholar]
- 3.Namini AH, Berkowicz DA, Kohane IS, et al. A submission model for use in the indexing, searching, and retrieval of distributed pathology case and tissue specimens. Stud Health Technol Inform 2004;107(Pt 2):1264–7 [PubMed] [Google Scholar]
- 4.Drake TA, Braun J, Marchevsky A, et al. A system for sharing routine surgical pathology specimens across institutions: the Shared Pathology Informatics Network. Hum Pathol 2007;38:1212–25 [DOI] [PubMed] [Google Scholar]
- 5.McMurry AJ, Gilbert CA, Reis BY, et al. Distributed architecture for public health, research, and clinical care. J Am Med Inform Assoc 2007;14:527–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keator D, Grethe J, Marcus D, et al. A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN). IEEE Trans Inf Technol Biomed 2008;12:162–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oster S, Langella S, Hastings S, et al. caGrid 1.0: an enterprise Grid infrastructure for biomedical research. J Am Med Inform Assoc 2008;15:138–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson N, Abend A, Mandel A, et al. Implementation of a deidentified federated data network for population-based cohort discovery. J Am Med Inform Assoc 2012;19:e60–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patten IS, Rana S, Shahul S, et al. Cardiac angiogenic imbalance leads to peripartum cardiomyopathy. Nature 2012;485:333–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohane IS, McMurry A, Weber G, et al. The co-morbidity burden of children and young adults with autism spectrum disorders. PLoS One 2012;7:e33224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandl KD, Kohane IS. Escaping the EHR trap—the future of health IT. N Engl J Med 2012;366:2240–2 [DOI] [PubMed] [Google Scholar]
- 12.Newcombe HB, Kennedy JM, Axford SJ, et al. Automatic linkage of vital records. Science 1959;130:954–9 [DOI] [PubMed] [Google Scholar]
- 13.Fellegi IP, Sunter AB. A theory for record linkage. J Am Stat Assoc 1969;64:1183–210 [Google Scholar]
- 14.Bell RB, Kessey J, Richards T. The urge to merge: linking vital statistics records and medicaid claims. Med Care 1994;32:1004–18 [PubMed] [Google Scholar]
- 15.Trompa M, Ravelli AC, Bonsel GJ, et al. Results from simulated data sets: probabilistic record linkage outperforms deterministic record linkage. J Clin Epidemiol 2001;64:565–72 [DOI] [PubMed] [Google Scholar]
- 16.Grannis SJ, Overhage JM, McDonald CJ. Analysis of identifier performance using a deterministic linkage algorithm. Proc AMIA Symp 2002:305–9 [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy SN, Chueh HC. A security architecture for query tools used to access large biomedical databases. Proc AMIA Symp 2002:552–6 [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010;17:124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flajolet P, Martin GN. Probabilistic counting algorithms for data base applications. J Comp Syst Sci 1985;31:182–209 [Google Scholar]
- 20.Durand M, Flajolet P. Loglog counting of large cardinalities. 11th Annual European Symposium on Algorithms 2003 [Google Scholar]
- 21.Cai M, Pan J, Kwok Y, et al. Fast and accurate traffic matrix measurement using adaptive cardinality counting. Proceedings of the 2005 ACM SIGCOMM Workshop on Mining Network Data; 2005 [Google Scholar]