Abstract
Objective
Incorporating accurate life expectancy predictions into clinical decision making could improve quality and decrease costs, but few providers do this. We sought to use predictive data mining and high dimensional analytics of electronic health record (EHR) data to develop a highly accurate and clinically actionable 5 year life expectancy index.
Materials and methods
We developed the index using EHR data for 7463 patients ≥50 years old with ≥1 visit(s) in 2003 to a large, academic, multispecialty group practice. We extracted 980 attributes from the EHRs of the practices and affiliated hospitals. Correlation feature selection with greedy stepwise search was used to find the attribute subset with best average merit. Rotation forest ensembling with alternating decision tree as underlying classifier was used to predict 5 year mortality. Model performance was compared with the modified Charlson Comorbidity Index and the Walter life expectancy method.
Results
Within 5 years of the last visit in 2003, 838 (11%) patients had died. The final model included 24 attributes: two demographic (age, sex), 10 comorbidity (eg, cardiovascular disease), one vital sign (mean diastolic blood pressure), two medications (loop diuretic use, digoxin use), six laboratory (eg, mean albumin), and three healthcare utilization (eg, the number of hospitalizations 1 year prior to the last visit in 2003). The index showed very good discrimination (c-statistic 0.86) and outperformed comparators.
Conclusions
The EHR based index successfully distinguished adults ≥50 years old with life expectancy >5 years from those with life expectancy ≤5 years. This information could be used clinically to optimize preventive service use (eg, cancer screening in the elderly).
Background and significance
Accurate life expectancy prediction is essential for clinical decision making—it helps physicians weigh the benefits and risks of alternative care strategies and identify the best option for each patient. Failure to consider life expectancy leads to poor quality care and wastes healthcare resources. For example, patients with life expectancy <5 years often receive cancer screening even though its potential harms outweigh any benefits in this population.1–6
Although incorporating accurate life expectancy predictions into clinical decision making could improve quality and decrease costs, few clinicians actually do this—perhaps because existing life expectancy indices are inaccurate and/or burdensome. Indices can be inaccurate because they use imperfect claims data.7 More accurate indices often include additional clinical information (eg, functional status) but its collection is burdensome—providers do not routinely assess things like functional status.8 9
Using comprehensive electronic health record (EHR) data for life expectancy prediction could address the limitations of existing indices. The EHR contains rich clinical data traditionally absent from claims (eg, vital signs, laboratory results) that could improve accuracy without increasing provider burden.10–13 However, analyzing the large amount of information within a comprehensive EHR is challenging.
Predictive data mining and high dimensional analytics can generate actionable insights based on massive and high dimensional data, such as that within a comprehensive EHR. For example, many companies (eg, Amazon, Netflix, Google) use predictive mining and analytics to generate individualized recommendations and personalized news on a massive scale—improving both sales and customer satisfaction.14–18 In healthcare, predictive data mining has been explored as a means to improve treatment of infections and cancer, identify adverse drug events, measure quality of asthma care, and predict cancer outcomes.19–24
Objective
Our goals were to: (1) present a set of approaches for predictive mining and analysis of high dimensional EHR data, (2) develop a highly accurate non-burdensome 5 year life expectancy index for outpatients aged 50 years and older, and (3) compare the new index with other better known prognostic indices (a modified Charlson Comorbidity Index25 and a modified Walter Life Expectancy Index26).
Methods
Patient population
EHR data were extracted for patients ≥50 years old with ≥1 visit(s) to the Northwestern Medical Faculty Foundation (NMFF) during 2003. NMFF is an urban, academic, multispecialty group practice with EpicCare EHR. Many NMFF patients receive hospital care at Northwestern Memorial Hospital, an urban academic hospital with Cerner EHR.
Ascertainment of 5 year survival
Outcome was death within 5 years of the last outpatient encounter in 2003 (ie, the index visit). This outcome was selected because decisions about preventive service use (eg, cancer screening, aggressive glucose control) should include consideration of 5 year life expectancy.3 6 26 Vital status was determined using the National Center for Health Statistics National Death Index (NDI) for the years 2003–2008. All patients were linked to the NDI using extracted EHR data. The probabilistic scoring approach with NDI recommended cut-off points was used to identify true matches.27
Predictive attributes
We extracted 980 distinct predictive attributes for 7463 patients. These attributes included all a priori plausible predictors of mortality available within the EHR, including sociodemographic data, comorbidities, vital signs, laboratory results, medications, and healthcare utilization (see online supplementary appendix).
Sociodemographic data
We extracted 11 sociodemographic attributes from Epic: age, sex, marital status, race/ethnicity (white, black, Hispanic, Asian, other, declined, or unknown), and socioeconomic status (zip code matched Agency for Healthcare Research and Quality Index of Socioeconomic Status and its components using 1990 census data).28 To protect patient privacy, all patients ≥90 years old (n=53) were considered to be 90.
Comorbidities
We extracted 117 comorbidity attributes from Epic. International Classification of Diseases-9 (ICD-9) codes, current procedural terminology codes, or substance use statuses were grouped to reflect specific comorbidity attributes (see online supplementary etable 1). Codes were extracted from encounter diagnoses in the year prior to the index visit, and the past medical history, past surgical history, social history, and problem list as of the index visit. Comorbidity attributes included individual diagnoses (eg, coronary artery disease, cerebrovascular disease, peripheral arterial disease (PAD)), groups of related diagnoses (eg, any cardiovascular disease included coronary artery disease, cerebrovascular disease, or PAD), and a count of the comorbidities identified. An additional 26 attributes were counts of encounters in the year prior to the index visit for which the primary diagnosis was a comorbidity for which frequent exacerbations predict life expectancy (eg, heart failure) or for which identification of an active (ie, non-historical) diagnosis might be important (eg, cancer).
Vital signs
We extracted 20 vital sign attributes from Epic including the mean, SD, median, high, and low heart rate, systolic blood pressure, diastolic blood pressure, and pulse pressure recorded in the year prior to the index visit.
Medications
We extracted 664 possible medication attributes from Epic. Medications were classified into Veterans Administration Classes using National Drug Classification Codes.29 Codes were extracted from the medication list as of the index visit (binary and count attributes for each medication class) or from medications ordered in the year prior to the index visit (count attributes). Additional medication attributes included counts of antihypertensive medications, diabetic medications, and antiplatelet/anticoagulant medications and a count of total medications prescribed (see online supplementary etable 2).
Laboratory results
We extracted 120 laboratory attributes from Epic, including mean, median, SD, high, and low for 24 laboratory tests (eg, creatinine, albumin) recorded in the year prior to the index visit (see online supplementary etable 3).
Healthcare utilization
We extracted 44 healthcare utilization attributes from Cerner and six from Epic. Utilization attributes extracted from Cerner included discharge status (eg, to home, skilled nursing facility) and counts of hospital admissions, emergency department visits, and home health referrals either ≤1 or 1–2 years prior to the index visit. Utilization attributes extracted from Epic included counts of visits to a primary care provider, any general medicine provider, and any NMFF provider either ≤1 or 1–2 years prior to the index visit.
Feature selection
Feature selection aims to reduce the number of attributes while retaining the predictive power of the original attribute set. We analyzed our entire data set using Correlation Feature Selection (CFS) to identify a subset of features highly correlated with the outcome (dichotomous 5-year mortality) and weakly correlated amongst themselves.30 CFS was used in conjunction with a greedy stepwise search to find subsets with best average merit (see online supplementary eMethods). CFS identified a subset of 52 features, which was manually reviewed to eliminate: (1) 12 features with low face validity (eg, milk of magnesia use highly predictive of mortality—the two patients who received it both died), (2) 5 redundant features (eg, PAD already included in ‘any cardiovascular disease’), and (3) 3 features with potentially problematic reliability (eg, very low/high vital signs more susceptible to random error because of manual data entry). Manual reduction reduced the subset to 32 features. CFS was again applied to identify a subset of 23 features, to which sex was added for a final set of 24 features. Their relative predictive power was assessed using the information gain metric, which evaluates the worth of an attribute by measuring the information gain with respect to the outcome status.
Comparison prognostic indices
We calculated a modified Charlson Comorbidity Index (CCI).7 We extracted demographic data and ICD-9 codes from Epic past medical history, past surgical history, and problem list as of the index visit and encounter diagnosis 1 year prior to the index visit to calculate an outpatient CCI adjusted for age, sex, and race/ethnicity (white, black, Hispanic, Asian, other, declined, or unknown). Although this index typically applies only to hospitalized patients, investigators have previously used outpatient Charlson listed diagnoses and inpatient diagnoses to compute the score.25
We also calculated predicted life expectancy using a modified Walter method.26 We used comorbid diagnosis counts as a surrogate for provider classification into mortality risk groups (ie, highest quartile of comorbid diagnoses is equivalent to the sick group, lowest quartile is equivalent to the healthy group).31 Life expectancy was calculated for each group using age–sex matched life tables from 2003—the sick group is likely to live only as long as 25% of their age–sex matched cohort, the healthy group is likely to live as long as 75% of their age–sex matched cohort, and the intermediate group is likely to live as long as 50% of their age–sex matched cohort.
Statistical analysis
We used the rotation forest ensembling technique with alternating decision tree as the underlying classifier to predict 5 year mortality. The rotation forest ensembling technique is presented her because it was superior to models generated using other techniques (eg, logistic regression, support vector machines, neural networks, naïve Bayes, random forest, and Bayesian networks) (see online supplementary eMethods) Tenfold cross validation was used to evaluate the model in order to ensure that the model was tested on data that it had not seen while training, thus minimizing the chance of over fitting (see online supplementary eMethods ).
The discriminatory power of the predictive models was assessed using c statistics and binary classification metrics: sensitivity (recall), specificity, positive predictive value (precision), negative predictive value, the percentage of correct predictions, and the F measure (the harmonic mean of precision and recall). We used risk categories of <50% and ≥50% because this is equivalent to a median life expectancy of 5 years—a life expectancy at which point consideration of the benefits and risks of continued cancer screening is particularly important. Reclassification tables and the net reclassification improvement was used to compare the Ensemble Index to the comparison indices.32
To analyze calibration, the mean predicted and the observed risk of death within 5 years were compared across deciles of predicted risk. The Hosmer–Lemeshow χ2 test was used to determine if the difference between predicted and actual risks was statistically significant. Because our large sample had a relatively low incidence of death, we also compared predicted and observed risk of death within 5 years across risk deciles (<10%, 10%≤×<20%, 20%≤×<30%, etc). Statistical analysis was performed using R V.2.11.1, WEKA V.3.6.3, ROC Web-calculator,33 and STATA/SE V.10.1. All predictive modeling was done using WEKA implementations of various techniques with default parameters, unless otherwise stated. This study was approved by the institutional review board at Northwestern University.
Results
Patient characteristics
We identified 7463 patients aged 50 years or greater with one or more visits to NMFF in 2003. Selected characteristics are displayed in table 1. Mean age of the participants was 62 years. Forty per cent were men and 51% were white. The most common diagnoses were hypertension (52%), any cardiovascular disease (17%), diabetes (17%), and cancer (15%).
Table 1.
Selected characteristics of the study patients at the index visit in 2003
| Full cohort (n=7463) | Dead at 5 years (n=838) | Alive at 5 years (n=6625) | |
|---|---|---|---|
| Demographics | |||
| Age (years) (mean±SD) | 62±10 | 70±11 | 61±9 |
| Male sex (n (%)) | 2993 (40) | 397 (47) | 2596 (39) |
| Race/ethnicity (n (%)) | |||
| White | 3838 (51) | 411 (49) | 3427 (52) |
| Black | 1772 (24) | 273 (33) | 1499 (23) |
| Hispanic | 359 (5) | 40 (5) | 319 (5) |
| Asian | 230 (3) | 14 (2) | 216 (3) |
| Other | 548 (7) | 65 (8) | 483 (7) |
| Unknown/declined | 716 (1) | 35 (4) | 681(10) |
| Diagnoses (n (%)) | |||
| Any vascular disease | 1233 (17) | 926 (14) | 307 (37) |
| Heart failure | 352 (5) | 140 (17) | 212 (3) |
| Hypertension | 3880 (52) | 534 (64) | 3346 (51) |
| Tobacco use | 841 (11) | 141 (17) | 700 (11) |
| Chronic kidney disease | 248 (3) | 106 (13) | 142 (2) |
| Diabetes mellitus | 1281 (17) | 254 (30) | 1027 (16) |
| Dementia | 1128 (2) | 58 (7) | 60 (1) |
| HIV | 15 (<1) | 4 (<1) | 11 (<1) |
| Anemia | 633 (8) | 167 (20) | 466 (7) |
| Any cancer | 1133 (15) | 244 (29) | 889 (13) |
| Any liver disease | 181 (2) | 43 (5) | 138 (2) |
| Comorbidity count (mean±SD) | 2.5±2.0 | 4.1±2.3 | 2.3±1.8 |
| Vital signs (mean±SD) | |||
| Systolic blood pressure (mm Hg) | 131±16 | 134±19 | 131±16 |
| Diastolic blood pressure (mm Hg) | 79±9 | 76±10 | 80±9 |
| Laboratory results | |||
| Albumin (g/dl) | 3.7±0.4 | 3.3±0.5 | 3.8±0.4 |
| Creatinine (mg/dl) | 1.1±0.8 | 1.5±1.6 | 1.0±0.6 |
| Outpatient medications (n (%)) | |||
| Digoxin prescription | 197 (3) | 81 (10) | 116 (2) |
| Loop diuretic prescription | 576 (8) | 219 (26) | 357 (5) |
| Healthcare utilization (mean±SD) | |||
| Primary care provider visits 0–1 years prior to index visit | 1.2±1.8 | 1.1±2.3 | 1.2±1.7 |
| Hospitalizations 0–1 years prior to index visit | 0.3±1.6 | 1.0±1.6 | 0.2±0.6 |
| Hospitalizations 1–2 years prior to index visit | 0.2±0.7 | 0.7±1.3 | 0.1±0.5 |
Within 5 years of their index visit, 838 (11%) patients died (table 1). These patients were older (mean age 70 vs 61 years), more likely to be black (33% vs 23%), had more comorbid diagnoses (4.1±2.3 vs 2.3±1.8), and were hospitalized more often in the 2 years prior to their index visit. Patients who died had lower albumin (3.3 vs 3.8) and a higher creatinine (1.5 vs 1.0).
Feature selection results
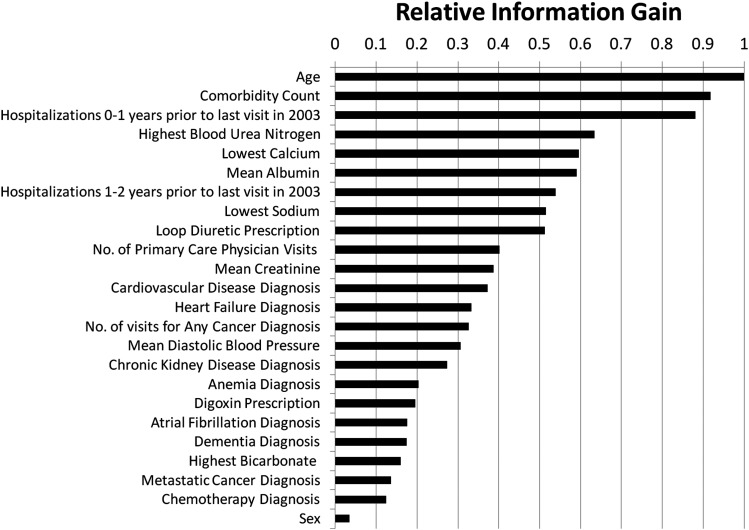
The final model included age, sex, 10 comorbidity attributes (eg, cardiovascular disease, chronic kidney disease), mean diastolic blood pressure, loop diuretic use, digoxin use, six laboratory attributes (eg, mean albumin, mean creatinine), number of visits to primary care provider in the year prior to the index visit, and number of hospitalizations 0–1 and 1–2 years prior to the index visit. Those attributes with the greatest predictive power (information gain) were age, comorbidity count, hospitalizations 1 year prior to the index visit, the highest blood urea nitrogen in the year prior to the index visit, and the lowest calcium in the year prior to the index visit (figure 1).
Figure 1.
Relative information gain of features included in the final Ensemble Index. We used multiple feature selection techniques and manual review to arrive at the final set.
Ensemble Index results
Model discrimination was very good (c statistic 0.86, 95% CI 0.85 to 0.87). Using a predicted 5 year mortality ≥50% as a cut-off, the sensitivity of the Ensemble Index for predicting 5 year mortality was 31%, specificity was 98%, and the F measure was 41% (table 2). The difference between predicted and observed mortality was <3% across all deciles of risk. The Hosmer–Lemeshow statistic was 18.7 (p=0.02) for deciles of risk and 12.2 (p=0.20) for risk deciles (table 3).
Table 2.
Performance of the Ensemble Index, Charlson Comorbidity Index, and Walter life expectancy method for predicting 5 year survival, and reclassifications by the Ensemble Index
| Evaluation metric | Walter life expectancy method | Charlson Comorbidity Index | Ensemble Index |
|---|---|---|---|
| C statistic (95% CI) | 0.78(0.77 to 0.80)* | 0.81 (0.79 to 0.83)* | 0.86 (0.85 to 0.87) |
| Sensitivity (recall) (%) | 22.8 | 13.1 | 30.7 |
| Specificity (%) | 96.8 | 98.5 | 97.7 |
| Positive predictive value (precision) (%) | 47.0 | 52.6 | 63.0 |
| Negative predictive value (%) | 90.8 | 90.0 | 91.8 |
| F measure (%) | 30.7 | 21.0 | 41.3 |
| Correct predictions (%) | 88.4 | 88.9 | 90.2 |
| No of individuals reclassified by the Ensemble Index | |||
| Events moved to life expectancy <5 years | 144 | 181 | Reference |
| Events moved to life expectancy >5 years | 78 | 34 | Reference |
| Non-events moved to life expectancy <5 years | 99 | 110 | Reference |
| Non-events moved to life expectancy >5 years | 163 | 59 | Reference |
| Net reclassification improvement (%) | 8.8† | 16.8† | – |
*p<0.001 for comparison with Ensemble Index; †p<0.001.
Table 3.
Calibration of the Ensemble Index: predicted and observed 5 year mortality by risk groups and each group's contribution to the Hosmer–Lemeshow statistic
| n | n≥75 years old | Observed 5 year mortality (%) | Predicted 5 year mortality (%) | Contribution to Hosmer– Lemeshow statistic | |
|---|---|---|---|---|---|
| Decile of risk* (5 year mortality predicted risk range) | |||||
| 1 (1.1–1.8%) | 741 | 0 | 0.8 | 1.6 | 3.1 |
| 2 (1.9–2.2%) | 723 | 0 | 1.4 | 2.0 | 1.5 |
| 3 (2.3–2.6%) | 781 | 0 | 1.4 | 2.5 | 3.7 |
| 4 (2.7–3.0%) | 790 | 0 | 2.0 | 2.8 | 1.9 |
| 5 (3.1–3.8%) | 738 | 2 | 4.2 | 3.4 | 1.4 |
| 6 (3.9–5.3%) | 733 | 22 | 4.8 | 4.5 | 0.1 |
| 7 (5.4–8.4%) | 719 | 63 | 6.7 | 6.7 | 0.0 |
| 8 (8.5–15.3%) | 749 | 189 | 14.0 | 11.3 | 5.6 |
| 9 (15.4–34.3%) | 741 | 317 | 24.7 | 23.6 | 0.5 |
| 10 (34.4–94.2%) | 748 | 374 | 52.5 | 54.4 | 1.0 |
| Total | 7463 | 967 | 11.2 | 11.3 | 18.7 (p=0.02) |
| 5 year mortality predicted risk deciles | |||||
| <10% | 5471 | 145 | 3.3 | 3.6 | 1.3 |
| 10≤×<20% | 738 | 232 | 18.2 | 14.0 | 9.3 |
| 20≤×<30% | 372 | 165 | 24.2 | 24.4 | 0.0 |
| 30≤×<40% | 264 | 98 | 30.7 | 34.6 | 1.1 |
| 40≤×<50% | 210 | 100 | 44.3 | 44.8 | 0.0 |
| 50≤×<60% | 164 | 75 | 55.5 | 54.8 | 0.0 |
| 60≤×<70% | 121 | 65 | 63.7 | 64.6 | 0.0 |
| 70≤×<80% | 80 | 53 | 68.8 | 74.7 | 0.4 |
| 80≤×<90% | 36 | 28 | 77.8 | 84.4 | 0.1 |
| ≥90% | 7 | 6 | 85.7 | 92.5 | 0.0 |
| Total | 7463 | 967 | 11.2 | 11.3 | 12.2 (p=0.20) |
*Number of subjects in each decile of risk is not equivalent due to ties. Predicted risks were rounded to the first decimal place.
Comparison with other prognostic indices
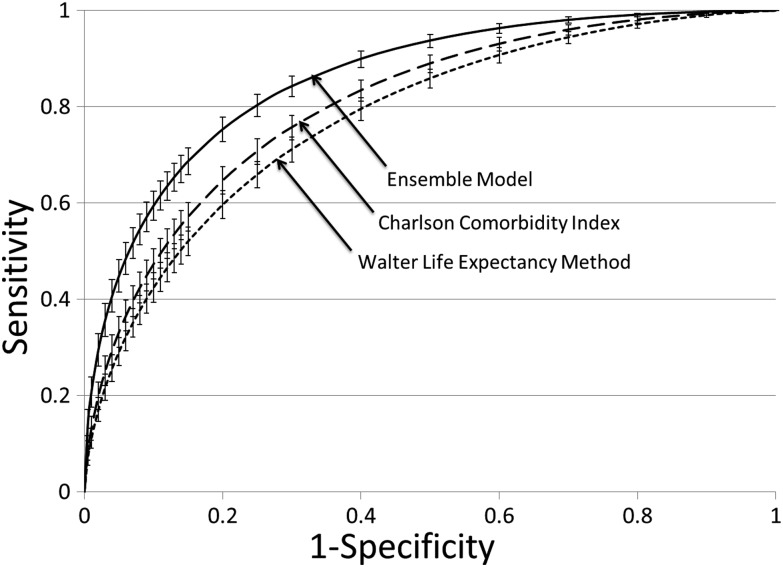
Ensemble Index discrimination was significantly better than both the modified Charlson Index (c statistic 0.81, 9% CI 0.79 to 0.83; p value for comparison <0.001) and the Walter method (c statistic 0.78, 95% CI 0.77 to 0.80; p value for comparison <0.001) (table 2, figure 2). Using predicted 5 year mortality ≥50% as a cut-off, the Ensemble Index outperformed both the Charlson model and the Walter model on all performance measures except specificity (98% Ensemble vs 99% Charlson) (table 2).
Figure 2.
Receiver operating characteristic curves for Ensemble Index, modified Charlson Comorbidity Index, and modified Walter life expectancy method. Our proposed Ensemble Index outperforms the other two indices. Error bars indicate 95% CIs.
Compared with the modified Charlson Index, the Ensemble Index reclassified 181 patients as high risk that ultimately died within 5 years. Compared with the Walter method, the Ensemble Index reclassified 144 patients as high risk that ultimately died within 5 years. Net reclassification improvement for the Ensemble Index over the modified Charlson Index was 16.8% (p<0.001) and over the modified Walter was 8.8% (p<0.001) (table 2).
Discussion
We developed an index that successfully distinguishes between outpatients ≥50 years old with life expectancy<5 years from those with a longer life expectancy. To address the limitations of existing prognostic indices, we used predictive data mining and high dimensional analysis to generate meaningful predictions from the wealth of clinical data in a comprehensive EHR.
Our index is highly discriminative—the c statistic (0.86, 95% CI 0.85 to 0.87) is similar to or better than the best models in the existing literature.8 9 Our index is highly discriminative without being burdensome—using existing EHR data eliminates the need for providers to collect additional information (eg, functional status or activities of daily living). Clinicians should feel comfortable using this highly discriminative, well calibrated, non-burdensome prognostic index in clinical decision making.
Ideally, patients with a life expectancy long enough to benefit from service use should receive the service, while those with limited life expectancies should be spared potentially harmful services that are unlikely to improve outcomes. For example, some cancer screening guidelines recommend against screening patients with a life expectancy <5 years because the potential harms of screening are immediate while the benefits are not realized until 5 years later.3 34 Our index could be used to optimize cancer screening practices—differentiating those patients for whom cancer screening is likely to improve outcomes (life expectancy >5 years) from those in whom cancer screening is unlikely to improve outcomes and may cause harm (life expectancy <5 years). In our study, over half of all patients ≥75 years old had a predicted 5 year mortality <50% and were likely to benefit from continued screening, despite their advanced age. For example, an 81-year-old woman with cardiovascular disease, a mean diastolic blood pressure of 79 mm Hg, unremarkable laboratory studies, and no hospitalizations has a predicted 5 year mortality <10% and is likely to benefit from continued screening despite her advanced age. On the other hand, approximately 5% of patients <75 years old had a predicted 5 year mortality of ≥50% and were likely to be harmed by continued screening. For example, a 63-year-old woman with cardiovascular disease, chronic kidney disease, diastolic blood pressure 75 mm Hg, low albumin, high blood urea nitrogen, on a loop diuretic, and one hospitalization in the year prior to her last visit has a predicted 5 year mortality of 67% and is likely to experience only the harms of continued screening despite her relative youth. Although individual patients and providers may value this predictive information differently, making the information available could facilitate informed decision making and improve quality care.
Our index compares favorably with existing life expectancy indices. In this study, the Ensemble Index outperformed both the modified Charlson Index and the modified Walter life expectancy method. In order to automate the Walter method, we removed provider input. Although this may have marginally worsened its predictive ability, this change is unlikely to explain the poor discrimination of the method relative to the Ensemble Index. Our index is more discriminative and less burdensome than similar indices reported in the literature—Lee et al8 and Schonberg et al9 used survey data (including functional status measures) to predict 4 year (Lee) and 5 year (Schonberg) life expectancy with c statistics of 0.84 and 0.75, respectively.
Limitations
Our index has limitations. First, our index lacks functional status information. While our index's c statistic was similar to that of indices including functional status measures, adding this information would likely have further improved discrimination of the Ensemble Index. It is now possible to efficiently collect and record this information in the EHR using tablet computers.35 Second, our index does not include rare conditions (eg, amyotrophic lateral sclerosis) that influence life expectancy—clinicians must exercise their own judgment when caring for patients with these conditions. Third, the Hosmer–Lemeshow statistic was statistically significant (18.69, p=0.02). Although we believe that calibration and discrimination are equally important for a mortality prediction model such as ours, the significant Hosmer–Lemeshow statistic does not necessarily mean that the index is not useful—even well calibrated models will often have significant Hosmer–Lemeshow statistics when the sample size is large.36 The absence of any systematic variation between predicted and observed risk, and the difference in observed and expected risk of less than 3% across all deciles both suggest that the Ensemble Index was well calibrated. Fourth, the index had low sensitivity (31%). Although this may limit its potential impact, any increases in sensitivity would result in undesirable decreases in specificity. Finally, our index was developed using patient data from a single multispecialty practice and its affiliated hospital. As such, the utility of our index in other settings is unknown—it should be tested in other populations, clinics, and EHRs to evaluate generalizability.
Healthcare organizations should also consider using predictive data mining and high dimensional analytics on their own data—generating life expectancy indices specific to their patient population, provider EHR documentation practices, and available data. As EHR adoption increases, healthcare organizations grow, genetic testing increases, and medical knowledge expands, the availability of highly detailed, patient specific, potentially predictive information will increase. For this information to improve patient care it must be incorporated into clinical decision making, but this likely will be difficult for already burdened providers. Predictive data mining and high dimensional analytics use all available information to provide healthcare organizations with actionable insights that can improve the quality of patient care and decrease costs. Life expectancy indices developed using this methodology are likely to be less expensive than more generalizable indices developed using prospectively collected survey data (eg, Health and Retirement Study). Furthermore, indices developed using data mining and analytics can be automated and their predictions integrated into the EHR—driving clinical decision support algorithms, providing prognostic information at the point of care, or measuring the quality of care.
Conclusion
In summary, we successfully used predictive data mining and high dimensional analysis of EHR data to develop an highly discriminative, non-burdensome, 5 year life expectancy index for outpatients aged 50 years old or older using computer intensive analysis of EHR data. Our index had very good discrimination, was well calibrated, and compared favorably to existing indices. The new index could improve clinical decision making by optimizing use of preventive services like cancer screening—targeting screening to those patients most likely to benefit. Furthermore, similar application of our methodology could use increasingly available EHR data to predict almost anything of interest (eg, readmissions, total costs). These predictive models could ultimately guide interventions (eg, quality measurement, clinical decision support) that improve clinical decision making, improve quality, and decrease costs.
Footnotes
Contributors: All authors have made substantial contributions to the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript. All authors have reviewed the final version of the manuscript as submitted and approved it for publication. JSM and AA had full access to all of the data in this study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Funding: This work is supported in part by the following grants: NSF awards CCF-1029166 and OCI-1144061; DOE awards DE-SC0005340 and DE-SC0007456. JSM's fellowship is funded by AHRQ grant 5T32HS000078-13.
Competing interests: None.
Ethics approval: The study was approved by the institutional review board at Northwestern University.
Provenance and peer review: Not commissioned; externally peer reviewed.
Correction notice: This paper has been corrected since it was published Online First. The funding statement has been updated.
References
- 1.Fisher DA, Galanko J, Dudley TK, et al. Impact of comorbidity on colorectal cancer screening in the veterans healthcare system. Clin Gastroenterol Hepatol 2007;5:991–6 [DOI] [PubMed] [Google Scholar]
- 2.Walter LC, Bertenthal D, Lindquist K, et al. PSA screening among elderly men with limited life expectancies. JAMA 2006;296:2336–42 [DOI] [PubMed] [Google Scholar]
- 3.Walter LC, Lewis CL, Barton MB. Screening for colorectal, breast, and cervical cancer in the elderly: a review of the evidence. Am J Med 2005;118:1078–86 [DOI] [PubMed] [Google Scholar]
- 4.Walter LC, Lindquist K, O'Hare AM, et al. Targeting screening mammography according to life expectancy among women undergoing dialysis. Arch Intern Med 2006;166:1203–8 [DOI] [PubMed] [Google Scholar]
- 5.Walter LC, Lindquist K, Nugent S, et al. Impact of age and comorbidity on colorectal cancer screening among older veterans. Ann Intern Med 2009;150:465–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yourman LC, Lee SJ, Schonberg MA, et al. Prognostic indices for older adults: a systematic review. JAMA 2012;307:182–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deyo RA, Cherkin DC, Ciol MA. Adapting a clinical comorbidity index for use with ICD-9-CM administrative databases. J Clin Epidemiol 1992;45:613–19 [DOI] [PubMed] [Google Scholar]
- 8.Lee SJ, Lindquist K, Segal MR, et al. Development and validation of a prognostic index for 4-year mortality in older adults. JAMA 2006;295:801–8 [DOI] [PubMed] [Google Scholar]
- 9.Schonberg MA, Davis RB, McCarthy EP, et al. Index to predict 5-year mortality of community-dwelling adults aged 65 and older using data from the National Health Interview Survey. J Gen Int Med 2009;24:1115–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devoe JE, Gold R, McIntire P, et al. Electronic health records vs Medicaid claims: completeness of diabetes preventive care data in community health centers. Ann Fam Med 2011;9:351–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayrinen K, Saranto K, Nykanen P. Definition, structure, content, use and impacts of electronic health records: a review of the research literature. Int J Med Inform 2008;77:291–304 [DOI] [PubMed] [Google Scholar]
- 12.Weiner M, Callahan CM, Tierney WM, et al. Using information technology to improve the health care of older adults. Ann Intern Med 2003;139:430–6 [DOI] [PubMed] [Google Scholar]
- 13.Tierney WM, Takesue BY, Vargo DL, et al. Using electronic medical records to predict mortality in primary care patients with heart disease: prognostic power and pathophysiologic implications. J Gen Intern Med 1996;11:83–91 [DOI] [PubMed] [Google Scholar]
- 14.Linden G, Smith B, York J. Amazon.com recommendations: Item-to-item collaborative filtering. IEEE Internet Comput 2003;7:76–80 [Google Scholar]
- 15.Mobasher B. Data mining for web personalization. The adaptive web. Lect Notes Comput Sci 2007;4321:90–135 [Google Scholar]
- 16.Zhou Y, Wilkinson D, Schreiber R, et al. Large-scale parallel collaborative filtering for the Netflix prize. 2008; http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.173.2797 (accessed 19 Jun 2012).
- 17.Heidelberge C. Data mining at Netflix. 2009; http://cahdsu.wordpress.com/2009/08/04/infs-762-data-mining-at-netflix/ (accessed 19 Jun 2012). [Google Scholar]
- 18.Das A, Datar M, Garg A, et al. Google news personalization: Scalable online collaborative filtering. WWW'07: the 16th International Conference on World Wide Web 2007, Banff, Alberta, Canada, May 2007: 271–80
- 19.Hampton T. Data mining approach shows promise in detecting unexpected drug interactions. JAMA 2011;306:144. [DOI] [PubMed] [Google Scholar]
- 20.Jackson HA, Cashy J, Frieder O, et al. Data mining derived treatment algorithms from the electronic medical record improve theoretical empirical therapy for outpatient urinary tract infections. J Urol 2011;186:2257–62 [DOI] [PubMed] [Google Scholar]
- 21.Bereznicki BJ, Peterson GM, Jackson SL, et al. Data-mining of medication records to improve asthma management. Med J Aust 2008;189:21–5 [DOI] [PubMed] [Google Scholar]
- 22.Cakir A, Demirel B. A software tool for determination of breast cancer treatment methods using data mining approach. J Med Syst 2011;35:1503–11 [DOI] [PubMed] [Google Scholar]
- 23.Agrawal A, Misra S, Narayanan R, et al. A lung cancer outcome calculator using ensemble data mining on SEER data. Proceedings of the Tenth International Workshop on Data Mining in Bioinformatics (BIOKDD). San Diego, CA: ACM; 2011 [Google Scholar]
- 24.Yoo I, Alafaireet P, Marinov M, et al. Data mining in healthcare and biomedicine: a survey of the literature. J Med Syst 2012;36:2431–48 [DOI] [PubMed] [Google Scholar]
- 25.Perkins AJ, Kroenke K, Unutzer J, et al. Common comorbidity scales were similar in their ability to predict health care costs and mortality. J Clin Epidemiol 2004;57:1040–8 [DOI] [PubMed] [Google Scholar]
- 26.Walter LC, Covinsky KE. Cancer screening in elderly patients: a framework for individualized decision making. JAMA 2001;285:2750–6 [DOI] [PubMed] [Google Scholar]
- 27.Bilgrad R. National Death Index: User's Guide. In: Services Department of Health and Human Services, Prevention Centers for Disease Control, Statistics. Atlanta, GA 2009
- 28.Bonito AJ, Bann C, Eicheldinger C, et al. Creation of new race-ethnicity codes and socioeconomic status (SES) indicators for Medicare beneficiaries, 2008. http://www.ahrq.gov/qual/medicareindicators/ (accessed 5 Apr 2012). [Google Scholar]
- 29.United States Department of Veterans Affairs National Formulary, 2011. http://www.pbm.va.gov/NationalFormulary.aspx (accessed 5 Apr 2012). [Google Scholar]
- 30.Hall MA. Correlation-based feature selection for machine learning, 1999. http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.37.4643 (accessed 19 Jun 2012). [Google Scholar]
- 31.Centers for Disease Control and Prevention Life Tables. Publications and information products. http://www.cdc.gov/nchs/products/life_tables.htm (accessed 5 Apr 2012). [Google Scholar]
- 32.Pencina MJ, D'Agostino RB, Sr, D'Agostino RB, Jr, et al. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 2008;27:157–72 [DOI] [PubMed] [Google Scholar]
- 33.Eng J. ROC Analysis. 2007. http://www.rad.jhmi.edu/jeng/javarad/roc/JROCFITi.html (accessed 23 May 2012)
- 34.Albert RH, Clark MM. Cancer screening in the older patient. Am Fam Physician 2008;78:1369–74 [PubMed] [Google Scholar]
- 35.Hess R, Santucci A, McTigue K, et al. Patient difficulty using tablet computers to screen in primary care. J Gen Intern Med 2008;23:476–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: the Hosmer-Lemeshow test revisited. Crit Care Med 2007;35:2052–6 [DOI] [PubMed] [Google Scholar]