Abstract
Objectives
To present a system that uses knowledge stored in a medical ontology to automate the development of diagnostic decision support systems. To illustrate its function through an example focused on the development of a tool for diagnosing pneumonia.
Materials and methods
We developed a system that automates the creation of diagnostic decision-support applications. It relies on a medical ontology to direct the acquisition of clinic data from a clinical data warehouse and uses an automated analytic system to apply a sequence of machine learning algorithms that create applications for diagnostic screening. We refer to this system as the ontology-driven diagnostic modeling system (ODMS). We tested this system using samples of patient data collected in Salt Lake City emergency rooms and stored in Intermountain Healthcare’s enterprise data warehouse.
Results
The system was used in the preliminary development steps of a tool to identify patients with pneumonia in the emergency department. This tool was compared with a manually created diagnostic tool derived from a curated dataset. The manually created tool is currently in clinical use. The automatically created tool had an area under the receiver operating characteristic curve of 0.920 (95% CI 0.916 to 0.924), compared with 0.944 (95% CI 0.942 to 0.947) for the manually created tool.
Discussion
Initial testing of the ODMS demonstrates promising accuracy for the highly automated results and illustrates the route to model improvement.
Conclusions
The use of medical knowledge, embedded in ontologies, to direct the initial development of diagnostic computing systems appears feasible.
Keywords: Ontology, Diagnostic System, Data Mining, Pneumonia
Introduction
Modern medicine has attained a degree of complexity that limits human ability to deliver care consistently and effectively. At the same time, the growth of electronic health records (EHRs) has allowed clinicians increased access to the large amounts of data collected during the care of each patient. This combination has resulted in a degree of information overload that challenges the clinician's ability to focus on relevant information, to align this information with standards of clinical practice, and to use this combination of clinical data and medical knowledge to deliver care reflecting the best available medical evidence. The result echoes the observation of David Eddy1 in 1990, ‘… all confirm what would be expected from common sense: the complexity of modern medicine exceeds the inherent limitations of the unaided human mind.’
A side effect of the population of EHRs with patient data is the creation of large data warehouses containing accumulated clinical data that represent the care of hundreds of thousands of patients. Analysis of these massive data collections can provide insight into the character of disease and can indicate which among the available diagnostic and therapeutic approaches is most likely to yield desired outcomes. Moreover, this is exactly the information needed to support the creation of computer-based clinical decision support (CDS) tools, which can change the information dynamic at the bedside.
In order to use this information to develop CDS applications, an organization must marshal the resources necessary to extract data from a data warehouse, analyze it, and construct from it decision support tools that can contribute to care. This typically requires the collaboration of clinicians, database analysts, statisticians/data miners, and software developers. This large resource commitment is a key impediment to the broad use of the data stored in clinical data warehouses to develop decision support applications.
In this paper, we describe a data analysis environment that is part of an effort to address these challenges. This system is designed to demonstrate ways in which ontologies coupled with specialized programs for data analysis can reduce the resource requirements needed to develop diagnostic CDS applications. We called this environment ‘the ontology-driven diagnostic modeling system’ (ODMS). Below, we provide a brief description of the ODMS. We then give an example of its intended use through the description of a pilot project in which the system is used to develop a diagnostic model for screening emergency department patients and identify those patients with pneumonia. We compare the system produced with an older manually created system that is currently in production.
Materials and methods
The ODMS is an experimental system whose goal is to create an environment that combines a medical ontology with an enterprise data warehouse (EDW) to support the development of diagnostic modules for use in screening for disease and other clinical conditions. The ODMS has five key components:
An ontology designed to represent the class hierarchies for essential medical concepts (diseases/conditions, therapies, clinical observations, outcomes, and procedures) and to capture the relationships between these medical concepts.
Tools for extracting collections of concepts from the ontologies that are relevant to a specific clinical question.
A mechanism for retrieving the data represented by these concepts from a data warehouse. While many of these data are stored in the EDW as structured and coded data objects, some of the data are present only in free-text, clinical documents. For these, we demonstrate the use of natural language processing (NLP) tools to extract the necessary data.
An ‘analytic workbench’ which processes the data extracted from the EDW and constructs diagnostic screening models.
The EDW itself. To better support research and quality initiatives, we are developing a specialized abstract of the EDW designed to support queries focused on generally recognized classes of medical data and expressed using standard medical terminologies such as the International Classification of Diseases (ICD-9), LOINC, Snomed, etc. The ODMS takes advantage of this system, ‘the analytic health repository’ (AHR).
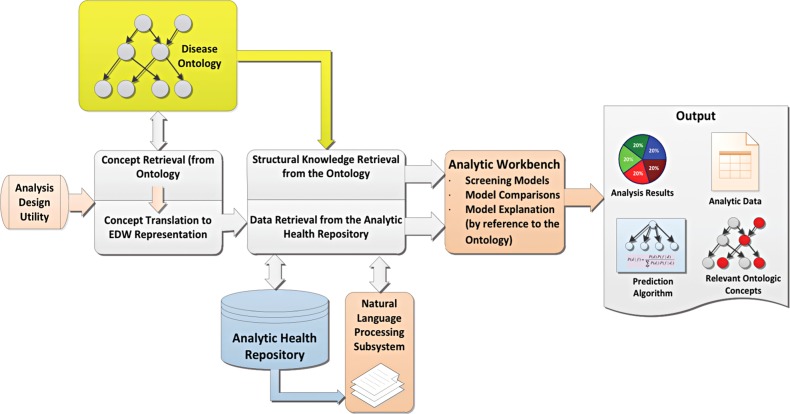
Figure 1 provides an overview of the ODMS. Below we describe these components in the context of the development of a diagnostic screening system for pneumonia.
Figure 1.
Overview of ontology-driven diagnostic modeling system (ODMS). The system can respond to queries that reference concepts in the ontology. A typical query might reference the disease concept ‘pneumonia’ from within the disease ontology and then indicate that the ODMS was to build one or more models designed to diagnose it. EDW, enterprise data warehouse.
Developing a model to diagnose pneumonia
The behavior of the ODMS is driven by a clinically focused disease ontology. This ontology has as its goal a model that captures (1) the relationships among diseases, (2) the relationships between diseases and relevant observations, (3) the relationships between diseases and typical therapeutic interventions, and (4) the relationships between diseases and anticipated outcomes.
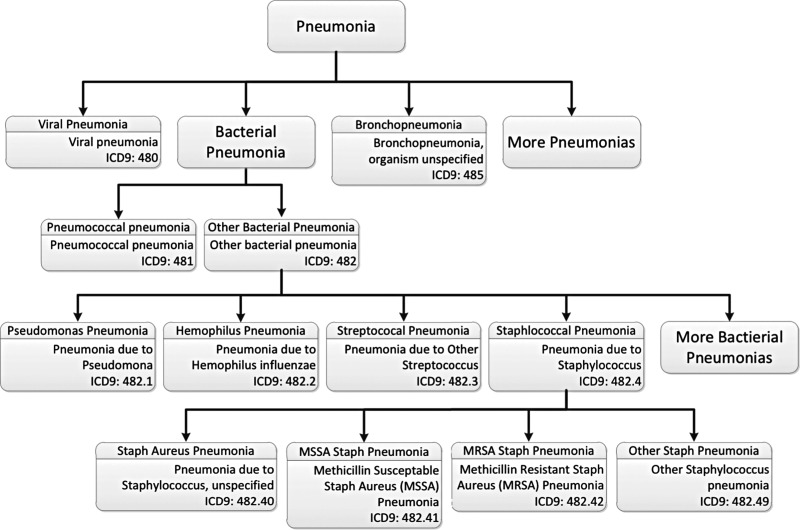
In order to provide value to researchers and clinicians, the ontology is enriched with links from the concepts embedded in it to data stored in Intermountain Healthcare’s EDW. In the case of pneumonia, the necessary information includes the knowledge necessary to identify patients with pneumonia, the knowledge necessary to choose those clinical data elements that would be useful in diagnosing pneumonia, and the knowledge necessary to find these data in the EDW. Identifying a collection of patients with pneumonia is accomplished using the ICD-9 codes assigned to each patient after discharge. Figure 2 displays a fragment of the disease class hierarchy maintained within the ODMS. For this component of the ontology, we have chosen to largely mirror the structure of ICD-9. Under the general heading of pneumonias, one finds different types of pneumonia with increasing specificity at lower levels in the hierarchy. Concepts that match ICD-9 concepts are labeled with appropriate ICD-9 codes.
Figure 2.
A fragment of the disease class hierarchy from the ontology-driven diagnostic modeling system ontology. The system can traverse the ontology to collect the different kinds of pneumonia and can use the storage information (in this case International Classification of Diseases (ICD) codes) to develop structured query language queries designed to identify the subset of the population whose discharge diagnosis was one of the pneumonias.
As a result of this organization, the system can use taxonomic explosion to help design the queries necessary to find groups of patients with pneumonia. In order to search for these patients, the system will traverse the hierarchy and bind all of the relevant ICD codes into a query (expressed in structured query language (SQL)), which will be run against a target population and return a list of patients whose discharge diagnoses include one of the aggregated codes. Other members of the target population are collected into a list of ‘non-pneumonia’ patients.
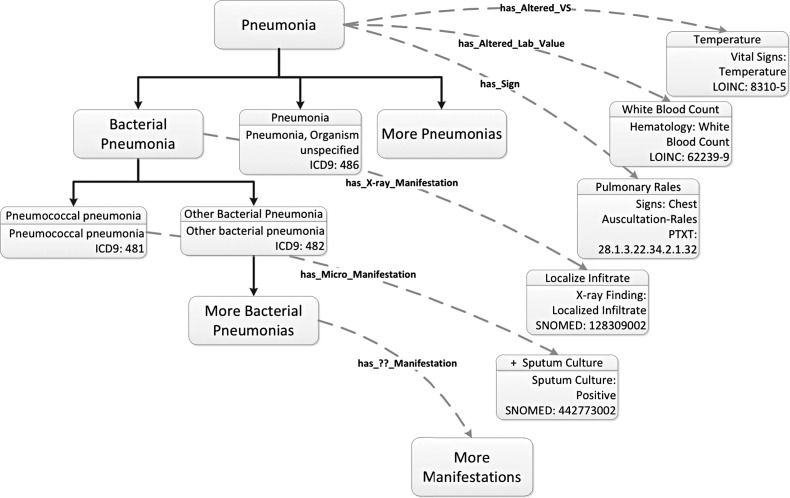
The result is a group of disease and control patients (ie, patients with and without pneumonia) whose stored, clinical data can be used to construct a diagnostic system. The next step is to extract these data. This activity is supervised by the ontology. In addition to the class hierarchy of diseases described above, the system contains class hierarchies for other medical concepts including laboratory results, vital signs, and x-ray results. Properties defined within the ontology connect diseases to relevant concepts in these hierarchies (figure 3).
Figure 3.
Connections within the ontology-driven diagnostic modeling system (ODMS) ontology. Oncologic properties are used to connect different diseases to relevant disease manifestations. These manifestations are inspected by the ODMS and used to develop search strategies to extract relevant clinical data from the enterprise data warehouse for analysis. Note that relevant concepts are identified using International Classification of Diseases (ICD), LONIC, SNOMED, and a local coding system called PTXT. VS, vital signs.
These connections can be exploited to generate queries that bring back relevant clinical findings. Again, taxonomic explosion can be used within the targeted findings hierarchies to build collections of potentially useful clinical data. Once these queries are constructed, they are used to extract data from the EDW/AHR for both patients with and without pneumonia. This dataset provides the substrate used by the analytic workbench to generate diagnostic decision support systems.
Invoking the system
The process of building a diagnostic model begins with the definition of the patient subpopulations from which the model will be derived. They are derived from a larger population representing those patients who are the target population for the model. In the case of our example—the diagnosis of pneumonia—the population of interest is patients admitted to the emergency departments in two hospitals in Salt Lake City. We represent these ‘populations of interest’ in a class hierarchy of encounter types, which allows us to specify increasingly restrictive encounter criteria as we move down the tree.
To identify criteria to separate patients into positive and negative disease subgroups, we can select a single node in the disease hierarchy and let taxonomic explosion identify the collection of concepts from which to build the query (see figure 2) or we can create a composite concept, a ‘Pneumonia-Definition,’ which provides a logical combination of concepts from which to construct the query. An example would be ‘hasPrimaryDischargeDiagnosis some ((Influenza or Pneumonia) and (not (Influenza_with_Other_Manifestations)))’ (displayed in the syntax of Protégé, a system for authoring ontologies2).
Extracting the data
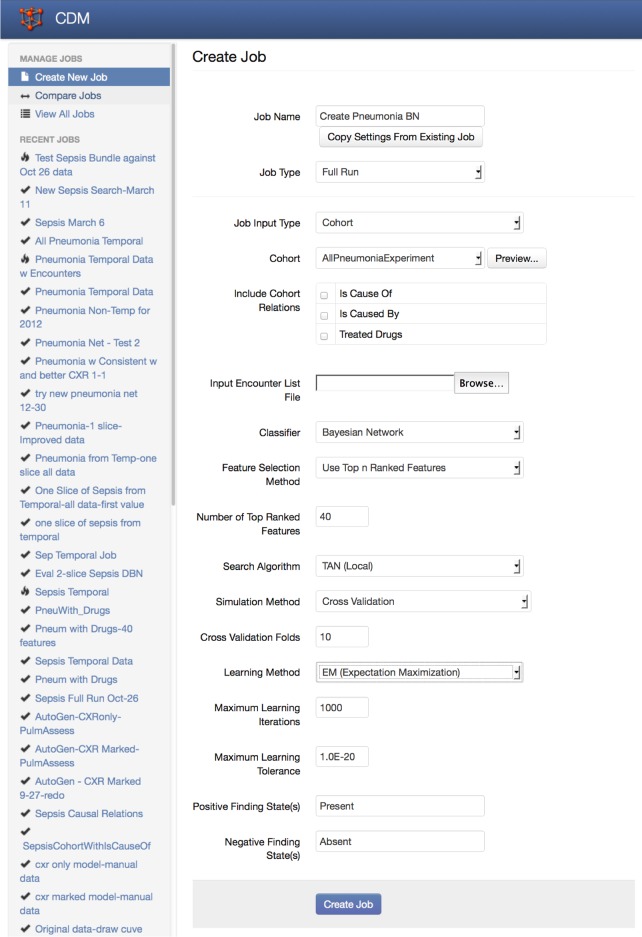
Once we have defined the parent population and the criteria for identifying diseased and non-diseased subpopulations, the ODMS carries out its analysis. It queries the AHR to identify patient cohorts with and without the defined diseases. It then traverses relevant ontological properties to identify those findings that are associated with these diseases. The definitions for these findings are used to construct queries that return the necessary data. These data are organized into a standard format (Weka's attribute-relation file format (ARFF))3 for input into the component of the ODMS called the analytic workbench. Here it undergoes a semiautomated analysis to generate preliminary results. This semiautomated analysis is triggered by a user through interaction with an initial form that allows the user to alter the default configuration of the analytic workbench (figure 4). The typical analysis for the system seeks to create a Bayesian network classifier using:
A supervised discretization algorithm for continuous variables4
A simple feature-selection algorithm that automatically chooses the 15 variables with the highest χ2 value
An algorithm to infer Bayesian network structure from data (tree-augmented naïve Bayes (TAN)5)
Estimation of Bayesian network parameters using expectation maximization6
Iterative analysis using 10-fold cross-validation to effectively measure system accuracy.
Figure 4.
Initial screen for setting up analyses in the analytic workbench. The setup screen begins by displaying default settings and allows the user to modify the settings. Shown are the typical settings for the initial development of the pneumonia and Bayesian network described here.
This process generates a set of preliminary results, which can be inspected by the user to determine whether further analysis will be profitable. These preliminary results consist of:
The list of concepts chosen from the ontology for inclusion in the query.
The raw data file extracted by the system.
The diagnostic model generated.
Accuracy statistics such as sensitivity, specificity, positive and negative predictive value, and the area under the receiver operating characteristics (ROC) curve
Various graphical representations of the results.
A key challenge in modeling diagnoses such as pneumonia is a requirement for data not typically found in structured form in the EHR or EDW. These are the data that are captured as narrative text. They include the many medical documents generated through dictation and transcription, by the clinician typing into the EHR or, more recently, through speech recognition tools that convert dictated information directly into medical reports. For pneumonia, a diagnostic system would be incomplete without the results of the radiographic examinations of the chest. These results are available only as dictated radiology reports.
To accommodate this type of data, we have integrated an NLP component into the ODMS. The concepts necessary to trigger data extraction from free-text reports are included in the ontology. When these are referenced in the definition of disease findings, the ODMS invokes the NLP component to acquire the essential information.
The NLP component used by the analytic workbench for pneumonia was developed as a part of the earlier project whose goal was to predict pneumonia in the emergency department7. This simple system consists of a random forest classifier8 trained to classify individual sentences within a chest x-ray report. A heuristic aggregates results across the sentences within the report and returns simple output indicating the presence or absence of support for pneumonia within the report. The system was originally tested on 4009 chest x-ray reports that had been adjudicated by a group of physicians. It demonstrated a sensitivity of 0.95 and a specificity of 0.74.
Analyzing the data
Within the ODMS, the analytic workbench is the component that receives data retrieved from the EDW/AHR and constructs relevant diagnostic screening models. Its output includes a listing of pertinent clinical concepts extracted from the ontology, the raw dataset retrieved from the data warehouse, a prediction algorithm for the diagnostic screening model, and an analysis of the model's accuracy.
The analytic workbench is intended to support the group of analytic activities required to produce and explore different preliminary diagnostic models. It is designed to consume the dataset extracted defined within the ontology. An ARFF file containing relevant data elements for this diagnostic problem is created, and the workbench uses this data file to construct and test diagnostic models for the target disease.
Our approach to the construction of the analytic workbench has been to combine components from various, readily available, modeling systems whose functions can be integrated through their application programming interfaces (APIs). The prototype workbench includes tools from Weka 3.6.59 10 (a general-purpose, open-source, data-mining toolkit) and Netica11 (a commercially available, Bayesian network authoring and execution application). Future versions will include components from R12 (an open-source, statistical package). We intend to take advantage of the high-quality graphical output provided by this package as well as some of its statistical features, notably its built-in functions for estimating area under the ROC curve and CIs.
The current version of the analytic workbench is focused on the development of Bayesian network13–15 models displaying varying degrees of sophistication. Other algorithms will be added as their utility becomes apparent. We have chosen Bayesian networks as the initial modeling paradigm for the analytic workbench based on two factors: previous experience with pneumonia screening and the similarity of Bayesian networks to the structural components in ontologies. In the late 1990s, a pneumonia diagnostic system was developed and deployed at LDS Hospital in Salt Lake City using Bayesian networks.16–18 This system proved surprisingly accurate. However, its development was labor-intensive. Variable selection, discretization of continuous variables, development of the network structure, and analysis of system accuracy required large amounts of manual effort.
A key goal for the initial version of the analytic workbench is to determine whether modern tools for developing Bayesian networks will noticeably reduce the effort required. In the interval since the first pneumonia screening system, a variety of tools to assist with the sequence of steps required for Bayesian model construction have become available. These new tools exist in the form of open-source and commercial data mining and statistical analysis software such as those referenced above. The products selected have APIs that allow them to be embedded in systems such as the analytic workbench. A goal of the work described here is to test our ability to configure components from different toolsets in various potentially advantageous ways.
The second reason for the choice of Bayesian networks is the similarity in certain structural characteristics between these networks and ontologies. Bayesian networks are built around directed links reflecting mathematical relationships between variables. Ontologies are built around directed links representing semantic relationships between concepts. These relationships have attracted the attention of researchers who are interested in extending ontologies to reason in areas of uncertainty.19–21 We hope in future experiments to determine the value of translating the ontological connections into the directed links necessary for the Bayesian models.
Returning the results of the analysis
In most cases, we expect the ODMS to be used as an automated way to generate an exploratory analysis of a disease or diseases. The user interface allows specification of a number of parameters that modify the analytic process. However, while the system supports a number of different analytic paths, we anticipate that its primary use will be to generate an initial analysis and then to provide these results and the raw data collected by the system to the user for further evaluation. Users are expected to define the diagnostic conditions that they wish to examine, accept (or potentially modify) the default settings provided for the analysis, and use the results as a starting point for their own further analysis of the dataset provided.
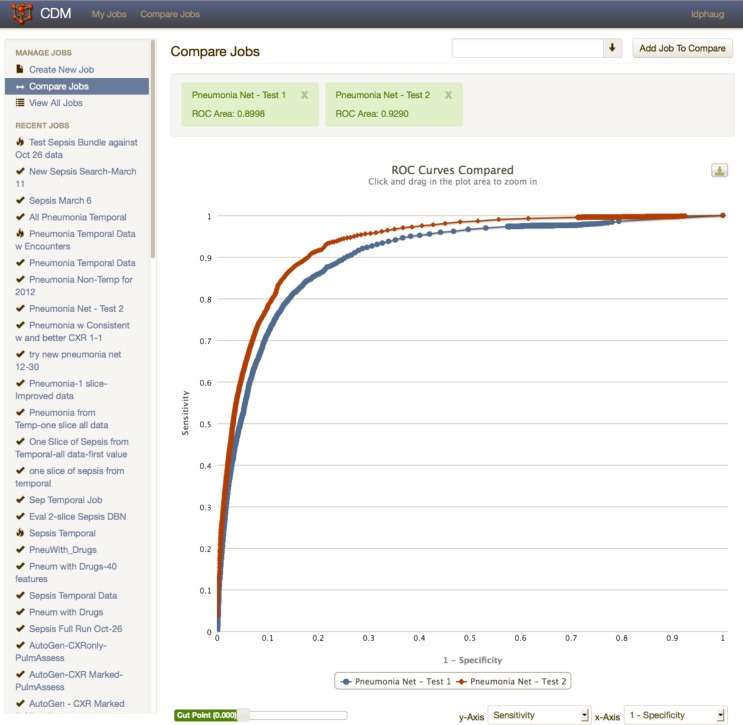
Because the goal of the system is to provide an initial look at the accuracy of diagnostic predictive models derived from clinical data, several charts useful for visualizing these models are immediately available. Figure 5 demonstrates one of these, the classic ROC curve used to explore the performance of predictive models across a range of thresholds for the output of the model. If the researcher has used the system to generate more than one predictive model, the resulting ROC curves can be displayed together for comparison. The workbench also calculates an area under the ROC curve for each model.
Figure 5.
Graphics produced by the analytic workbench. Receiver operating characteristics (ROC) curves for two different models produced during the study of a pneumonia diagnostic model are displayed. The area under the ROC curve is a good overall measure of the accuracy of a diagnostic predictive model.
Finally, the ODMS returns the model itself to the researcher. The researcher can then choose to return to the system for further analysis, take the data provided and continue the analysis on a different platform, or integrate the diagnostic model provided into a clinical or research application.
Sample evaluation
As an initial demonstration of the ODMS, we chose to use it to reproduce a tool for pneumonia diagnosis that had originally been created manually for use in the emergency department.6 This manually constructed, pneumonia diagnostic system was developed by researchers over an 8-week period through a process that involved manual identification of candidate data necessary for diagnosis, manual construction of SQL queries to extract these data, manual review and cleansing of the research dataset, use of Netica to develop a group of diagnostic Bayesian networks, and manual comparison of these Bayesian models to determine the diagnostic network with the best overall performance.
The process began with construction of a dataset consisting of 91 candidate clinical features for a curated set of 2413 positive pneumonia cases and 46 036 negative pneumonia cases seen in emergency department patient encounters at LDS Hospital and Intermountain Medical Center in Salt Lake City, Utah between 1 January 2008 and 1 January 2011. Since the diagnostic gold standard for pneumonia is a chest radiograph compatible with pneumonia, patient encounters without a chest radiograph were excluded from this cohort. Pneumonia was defined by ICD-9 codes as compatible with a primary discharge diagnosis of pneumonia (480–487.1).
Features were defined iteratively through discussion with clinicians and analysis of data available in the EDW. Table 1 contains a list of the features selected during this process. Data corresponding to these features were collected from the EDW using manually constructed SQL queries. These data were collected into tabular structures and submitted to Weka for supervised feature selection using Fayyad and Irani's minimum description length method.4 Subsequently, the feature set was submitted to Netica to develop a Bayesian network structure using the TAN5 mechanism. This Bayesian network was then trained using Netica's expectation maximization algorithms.6 Accuracy statistics are reported in table 2.
Table 1.
Features selected for the manually developed predictor and the ODMS-generated predictor
| Manually developed predictor features | ODMS-generated predictor features |
|---|---|
| Demographics | Demographics |
| Age value | Vital signs |
| Vital signs | Temperature |
| Diastolic BP | Heart rate |
| Mean pressure | Respiratory rate |
| Heart rate | Laboratory data |
| Respiratory rate | Anion gap |
| Systolic BP | BUN |
| Temperature | Chloride |
| Laboratory data | Spo2 |
| BUN | Fio2 percent |
| Chloride | Sodium |
| Creatinine | WBC |
| Sodium | Chest x-ray results |
| Spo2 | Single lobe infiltrate |
| WBC | Multi lobar infiltrates |
| Chest x-ray results | Nursing assessment |
| NLP finding | Abdomen not distended |
| Nursing assessment | Abnormal abdominal exam |
| Abnormal breath sounds | Abnormal breath sounds |
| Absent breath sounds | Alert and oriented ×3 |
| Absent cough | Distended abdomen |
| Clear breath sounds | Dull aching pain |
| Coarse breath sounds | Dyspneic |
| Cough clears secretions | Firm abdomen |
| Cough doesn't clear secretions | Frequent cough |
| Crackles | Incisional pain |
| Decreased breath sounds | Moderate cough |
| Fine crackles | No abnormal cough |
| Frequent cough | No tenderness on palpation |
| Infrequent cough | Non-productive cough |
| Moderate cough | Not oriented to place |
| Non-productive cough | Not oriented to time |
| Abnormal breath sounds on expiration | Not oriented ×3 |
| Abnormal breath sounds on inspiration | Oriented ×3 |
| Productive cough | Pleuritic pain |
| Rales breath sounds | Productive cough |
| Rhonchi breath sounds | Rales breath sounds |
| Stridor breath sounds | Sharp stabbing pain |
| Strong cough | Soft abdomen |
| Tubular breath sounds | Strong cough |
| Upper airway congestion | Tender abdomen |
| Weak cough | Throbbing pain |
| Wheezes | Wheezes |
| ED chief complaint | ED chief complaint |
BP, blood pressure; BUN, blood urea nitrogen; ED, emergency department; Fio2, fractional inspired oxygen; NLP, natural language processing; ODMS, ontology-driven diagnostic modeling system; Spo2, saturation of peripheral oxygen; WBC, white blood cells.
Table 2.
Accuracy of the models developed manually and by the ODMS
| Area under ROC curve | ||
|---|---|---|
| Manually developed predictor | ODMS-generated predictor | |
| Manually curated data | 0.944 (95% CI 0.942 to 0.947) | 0.881 (95% CI 0.875 to 0.887) |
| ODMS-generated data | 0.756 (95% CI 0.747 to 0.766) | 0.920 (95% CI 0.916 to 0.924) |
ODMS, ontology-driven diagnostic modeling system; ROC, receiver operating characteristics.
The model manually derived from this dataset was compared with a model automatically derived by the ODMS. For this experiment, we triggered the system using the ontology-based definition of pneumonia described above. The ODMS automatically extracted the data consistent with its ontological model, executed a feature-selection algorithm, and constructed and tested the diagnostic model. The process required approximately 22 h to extract the data and approximately 24 h to analyze it and provide the appropriate model. No user interaction was required during this process.
We chose to compare the two models by using accuracy statistics generated through testing each model with its own dataset and then with the dataset produced during the construction of the alternate model. With this as the goal, we produced accuracy statistics for the original manually developed predictor using the manually curated data by applying 10-fold cross-validation. In this procedure, different 10% subsamples of the data are set aside, a model is generated using the remaining 90%, and the 10% test dataset is used to evaluate this model. Similarly, we tested the ODMS-generated predictor against the ODMS-generated data that were created during predictor development, again applying 10-fold cross-validation. In each case bootstrapping was used to determine CIs.22
We then tested each of these diagnostic predictors using data generated during the alternate system's construction. The key accuracy statistic used was the area under the ROC curve. The results are described below.
Results
As mentioned above, the manual process produced a dataset of 91 candidate clinical features for a curated set of 2413 positive pneumonia cases and 46 036 negative pneumonia cases. Continuous data were discretized as necessary, and, after manual review, 40 features were selected for inclusion in the diagnostic model (table 1).
The ODMS automatically produced a set of 101 candidate clinical features for an automatically extracted set of 4240 positive pneumonia cases and 310 235 negative pneumonia cases. In the system's initialization screen, we specified that the feature-selection step should restrict the number of features used in the model to 40 (table 1), the number used in the manually developed predictor.
We tested each diagnostic model with its own dataset using 10-fold cross-validation in which the model's parameters were retrained during each fold cycle. We then took each diagnostic model (using its overall parameterization) and tested it with the dataset generated during the creation of the other model. The test statistic used was the area under the ROC curve and the associated 95% CIs. Table 2 shows the resulting values.
Perhaps the most interesting statistic in this table is the accuracy of the pneumonia diagnostic system developed automatically by the ODMS. This predictor represents a highly automated, preliminary modeling effort. However, when applied to the manually curated data, accuracy fell off significantly. This is not necessarily surprising. The manually developed predictor was created using a restricted subset of the entire emergency department patient population. We trained only with those patients who had a radiograph of the chest reported as a part of their visit. This predictor has proven reasonable accuracy in that patient subgroup, but fails to accommodate the different data patterns seen in the much larger population of patients who do not have a chest x-ray examination.
To illustrate a possible next step that a modeler might take, we retrained the ODMS-generated predictor with a dataset constrained to more closely match the manually curated data. To accomplish this, we retrieved the ODMS-generated data from the ODMS and removed patients without x-ray examination of the chest. The resulting dataset contained 2899 positive pneumonia cases and 78 798 negative pneumonia cases. We returned this modified dataset to the ODMS and requested a new model.
The system then produced a model specific to emergency department patients with chest imaging examinations. In a 10-fold cross-validation evaluation against the modified dataset, it produced an area under the ROC curve of 0. 902 (95% CI 0.897 to 0.908). When tested directly against the manually curated data, the area under the ROC curve was modestly improved at 0.899 (95% CI 0.893 to 0.905).
This simple example illustrates the anticipated use of the ODMS. A model developer will request an initial model and then engage in a series of interactions with the ODMS to improve the operating characteristics of this predictive model for a population of interest.
Discussion
The example above illustrates the workings of the ODMS. The results, although preliminary, are encouraging. Although not reported here, we have seen similar results in a system for early diagnosis of sepsis, which is used in several of our institutions.
Note that the statistics above should not be interpreted as a formal comparative evaluation. There is substantial overlap in patients represented in the manually curated data and the ODMS-generated data. However, the two datasets used were somewhat different. The manually curated data were not only limited to a set of patients who had a radiographic examination of the chest, but the chest x-rays of a significant subset of these patients were reviewed by a group of pulmonologists to confirm the presence of pneumonia. The categories to which they were assigned were altered if the reviewer disagreed.
These interventions could be expected to produce a less ‘noisy’ dataset, with fewer patients misidentified. It reduced the reliance on ICD coding, a process that is known to introduce error into research datasets.23 The ODMS-generated data were not similarly reviewed. Not only did it include patients both with and without radiographic chest examination, but none of the chest radiographs were reviewed to confirm the diagnosis. The results of the simple manipulation of the ODMS-generated data by restricting them to patients with chest radiographs demonstrated the types of interventions that modelers will use to refine a diagnostic system if the initial, automatically generated results are promising.
Conclusion
The ODMS that we have described above is being used to develop a group of diagnostic models that are destined to play a part in healthcare in our facilities. Future success is dependent on extending and enriching the clinical ontology that we are developing. Our ability to drive these analyses using this resource is dependent on its completeness and on our ability to link it effectively to data sources in the EDW.
Development of the ontology has, so far, largely been a manual process (although informed by terminologies such as ICD-9); however, we are actively exploring approaches to importing or automatically inferring key concept classes. Existing structured terminologies, including Snomed-CT, RxNorm, and others, hold promise as sources of large collections of relevant concepts.
We also foresee adding to the available analytic tools in the analytic workbench. A variety of predictive modeling tools is available and provides attractive alternatives to the Bayesian models that have been our focus so far. In addition, we are committed to providing better tools for data visualization and for visualizing the results of the analyses produced by the system. Our goal is to make the system accessible to a range of clinical researchers who need assistance in integrating a knowledge of medicine with data-storage practices used in large EDWs.
The system we have described is focused on the discovery of predictive diagnostic models. However, altering it to support research into questions of comparative effectiveness is entirely feasible. The medical knowledge captured in the ontology could be used to identify relevant study populations, therapeutic alternatives worthy of study, predisposing clinical factors, and significant medical outcomes. This promises to be a fruitful area for future research.
Finally, the ontology itself may someday be able to play a more direct role in clinical care. Its use in research will test and refine the relationships between diseases and supporting clinical findings, diseases and therapies, and diseases and outcomes. This information may find use at the bedside by supporting novel tools for viewing the course and status of a patient’s illness. We would hope that efforts to extend and validate this resource will provide value for research now and someday for the direct delivery of clinical care.
Footnotes
Contributors: All of the authors contributed to the ideas behind this paper. ND provided the sample problem (pneumonia) and initial system against which the ODMS-develop system was compared. JPF, XW, KM, and ME made particular contributions to the execution and evaluation of the project.
Funding: Supported by Grant LM010482 from the National Library of Medicine.
Competing interests: None.
Ethics approval: Intermountain Healthcare Institutional Review Board.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: We would be pleased to share the research ontology. An effort is being made to construct it from generally available components with few licensing restrictions.
References
- 1.Eddy DM. Clinical decision making. JAMA 1990;263:1265–75 [DOI] [PubMed] [Google Scholar]
- 2.Protégé-OWL: http://protege.stanford.edu/overview/protege-owl.html (accessed 21 Dec 2012).
- 3.ARFF: http://weka.wikispaces.com/ARFF (accessed 21 Dec 2012).
- 4.Fayyad UM, Irani KB. Multi-interval discretization of continuous valued attributes for classification learning. Proceedings of the Thirteenth International Joint Conference on Artificial Intelligence (IJCAI-93); Chamberry, France. 1993:1022–7 [Google Scholar]
- 5. Friedman N, Geiger D, Goldszmidt M. Bayesian Network Classifiers. Machine Learning 1997;29:131–63.
- 6.http://en.wikipedia.org/wiki/Expectation_maximization
- 7.Dean NC, Jones BE, Ferraro JP, et al. Performance and utilization of an emergency department electronic screening tool for pneumonia. JAMA Intern Med. Published Online first 18 Mar 2013. 10.1001/jamainternmed.2013.3299 [DOI] [PubMed] [Google Scholar]
- 8.http://en.wikipedia.org/wiki/Random_forest (accessed 21 Dec 2012).
- 9.Hall M, Frank E, Holmes G, et al. The WEKA data mining software: an update. ACM SIGKDD Explorations Newsletter 2009;11:10–8 [Google Scholar]
- 10.Weka: http://www.cs.waikato.ac.nz/ml/weka/ (accessed 21 Dec 2012).
- 11.Netica Software. Norsys Software Corporation; [March 16, 2011]. http://www.norsys.com (accessed 21 Dec 2012).
- 12.R: http://www.r-project.org/ (accessed 21 Dec 2012).
- 13.Pearl J. Probabilistic reasoning in intelligent systems. San Francisco: Morgan-Kaufmann, 1988 [Google Scholar]
- 14.Montironi R, Whimster WF, Collan Y, et al. How to develop and use a Bayesian Belief Network. J Clin Pathol 1996;49:194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SM, Abbott PA. Bayesian networks for knowledge discovery in large datasets: basics for nurse researchers. J Biomed Inform 2003;36:389–99 Review [DOI] [PubMed] [Google Scholar]
- 16.Aronsky D, Haug PJ. Diagnosing community-acquired pneumonia with a Bayesian network. Proceedings AMIA Symposiuml Lake Buena Vista, Florida, USA; 1998:632–6 [PMC free article] [PubMed] [Google Scholar]
- 17.Aronsky D, Chan KJ, Haug PJ. Evaluation of a computerized diagnostic decision support system for patients with pneumonia: study design considerations. J Am Med Inform Assoc 2001;8:473–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aronsky D, Haug PJ. Automatic identification of patients eligible for a pneumonia guideline. Proceedings AMIA Symposium Los Angeles, California, USA; 2000:12–16 [PMC free article] [PubMed] [Google Scholar]
- 19.Ding Z, Peng Y. A probabilistic extension to ontology language OWL. Proceedings of the 37th Hawaii International Conference on System Sciences (HICSS-37), Big Island, Hawaii, USA; 2004 [Google Scholar]
- 20.Ding Z, Peng Y, Pan R, et al. A Bayesian methodology towards automatic ontology mapping. Proceedings of the AAAI-05 C&O Workshop on Contexts and Ontologies: Theory, Practice and Applications, Pittsburg, Pennsylvania; 2005 [Google Scholar]
- 21.Costa PCG, Laskey KB, Alghamdi G. Bayesian ontologies in AI systems. Proceedings of the Fourth Bayesian Modeling Applications Workshop: Bayesian Models meet Cognition, held at the Twenty Second Conference on Uncertainty in Artificial Intelligence (UAI 2006), Cambridge, MA, Arlington: AUAI Press, 2006 [Google Scholar]
- 22.Efron B, Tibshirani RJ. An introduction to the bootstrap. Boca Raton: CRC Press LLC, 1998 [Google Scholar]
- 23.Aronsky D, Haug PJ, Lagor C, et al. Accuracy of administrative data for identifying patients with pneumonia. Am J Med Qual 2005;20:319–28 [DOI] [PubMed] [Google Scholar]