Abstract
Clinically oriented interface terminologies support interactions between humans and computer programs that accept structured entry of healthcare information. This manuscript describes efforts over the past decade to introduce an interface terminology called CHISL (Categorical Health Information Structured Lexicon) into clinical practice as part of a computer-based documentation application at Vanderbilt University Medical Center. Vanderbilt supports a spectrum of electronic documentation modalities, ranging from transcribed dictation, to a partial template of free-form notes, to strict, structured data capture. Vanderbilt encourages clinicians to use what they perceive as the most appropriate form of clinical note entry for each given clinical situation. In this setting, CHISL occupies an important niche in clinical documentation. This manuscript reports challenges developers faced in deploying CHISL, and discusses observations about its usage, but does not review other relevant work in the field.
Keywords: Interrface Terminology, Clinical Terminology, Computer-based Documentation, Support, U.S. Gov't, P.H.S.;, Medical Informatics Applications;, User-Computer Interface;
Background
To standardize and support structured clinical documentation across their institution as one of several options for electronic patient documentation, the authors have since 2000 developed, iteratively expanded, and evaluated a new interface terminology called CHISL: Categorical Health Information Structured Lexicon.1 The CHISL terminology derives from prior work on the INTERNIST-1 and Quick Medical Reference (QMR) diagnostic expert systems’ lexicon during 1973–88 (note: the QMR lexicon extended the INTERNIST-1 lexicon without substantial changes to its basic format) developed by Myers, Miller, and colleagues at the University of Pittsburgh.2 3 Through a 1981–3 National Library of Medicine-sponsored research project, Miller developed a structured ‘deep’ representation format for the original precoordinated text strings comprising the INTERNIST-1 and QMR lexicons.4 Later, Miller and Masarie and collaborators (including David Evans of Carnegie Mellon University) further elaborated this deep finding representation scheme during 1988–92, in the early phases of the US National Library of Medicine Unified Medical Language System project. As part of a collaboration with Anne-Marie Rassinoux and colleagues from Geneva, Switzerland, the group further demonstrated that combining superficial and deep representation schemes better enabled automated parsing of texts than did either approach alone.5–8 Miller recently published a comprehensive history of the INTERNIST-1 and QMR projects that explained the origins of their related terminologies.2
This manuscript describes how developers extended the INTERNIST-1/QMR lexicon to create CHISL with respect to published interface terminology desiderata, and reviews experience with its implementation and broad usage at our institution.9 10 To meet the desiderata, CHISL attempts (1) to have adequate concept and term coverage for a number of clinical domains where it is used for structured clinical documentation; (2) to balance precoordination and the need for postcoordination (ie, ‘compositional balance’); (3) to define clinically useful relationships through use of assertional knowledge; (4) to map to SNOMED CT (Systematized Nomenclature Of Medicine—Clinical Terms), a standard terminology with a formal semantic structure; (5) to include mechanisms that support human readability; and (6) to be application independent, so that different computer-based documentation systems could implement CHISL. The fourth attribute, CHISL's mapping to SNOMED CT terminology has been covered in detail elsewhere,11 12 and is not discussed in this manuscript.
Approach
CHISL concept and term coverage
Interface terminologies should include the major concepts and terms used in the domain they cover.9 10 The CHISL developers created concepts and terms in response to user-identified clinical documentation needs, as described below. The initial seed for creating CHISL was the existing INTERNIST-1/QMR lexicon. To create the INTERNIST-1/QMR terminology, Dr Jack Myers and colleagues reviewed published biomedical research on over 650 disorders in internal medicine beginning in 1972, and created terms that were precisely worded, explicit, diagnostically discriminating, and clinically meaningful.2 3 The project's knowledge base about each disease entailed careful review of 50–250 peer-reviewed articles that described how patients present at time of diagnosis with the disorder. The resultant INTERNIST-1/QMR lexicon consisted of a carefully constructed, precoordinated terminology of 4500 manifestations of disease, including past history, symptoms, signs, and findings from laboratory and imaging tests.2
Because CHISL was designed to support structured clinical documentation across the clinical institution, its content required broader coverage of clinical concepts than the internal medicine-oriented INTERNIST-1/QMR lexicon. Use of CHISL initially targeted specific multispecialty clinics, including general internal medicine, adult subspecialty cardiology, general neurology, general pediatrics, gynecology, subspecialty pulmonary medicine, urology, subspecialty gastroenterology, and emergency medicine. CHISL development involved an iterative collaboration among healthcare providers who were potential CHISL users, clinical informaticians who additionally had formal training in a clinical field (eg, nursing, internal medicine), and software developers. This team determined, for each clinical subdomain, what CHISL's specific scope and content would comprise—for example, necessary contexts, concepts, qualifiers, synonyms, and relevant sanctioning relationships. Three information sources drove the CHISL customization process: stated requirements from participating healthcare providers, review of clinical notes the providers had generated during pre-CHISL patient encounters, and post facto consolidation of end-users’ suggestions for new terms that they registered during use of CHISL in clinics. Through this process, developers expanded the number of available concepts and modifiers, and increased the number of synonyms for each concept.
CHISL compositional balance
To enhance usability, interface terminologies should model concepts at a level of detail that minimizes both the need to search through long lists of highly detailed precoordinated concepts, and the need to take steps to add detail to relatively general concepts through postcoordination.9 10 Interface terminologies striving for compositional balance should attempt to model concepts as general as possible while still retaining clinical meaning, and then should link to relevant modifiers to minimize the effort required to perform postcoordination. This is in contrast to large-scale terminologies that attempt to represent every possible entity within a medical domain at every level of detail.
The approach Miller took during a 1981–83 National Institutes of health (NIH) grant project to restructure precoordinated INTERNIST-1 findings served as a model for bringing these attributes to CHISL. In this project, as in CHISL, Miller extended INTERNIST-1 precoordinated finding names to include a ‘deeper’ multi-axial frame-based representation for the involved concept.4 13 The ‘deep’ terminology representation involved two related concept classes, one called ‘generic finding frames’ and one called ‘instantiated frames’. The generic finding frames represented relatively general concepts, and specified rules and patterns for how additional detail could be specified for a concept. Instantiated frames were more specific concepts that postcoordinated relevant axes from the generic finding frames with specific values to represent the full meaning of the original precoordinated INTERNIST-1 finding.
Generic finding frame instantiation involved selecting from the generic finding frame's allowed qualifier axes, which consisted of pick lists, called ‘item lists’ (ie, sanctioned list of modifiers, or value sets). In the generic frame system, each finding frame had a specific required element (ie, the name of an item list) that indicated how one could describe the status of the finding in a patient. The frame developer also had to specify which item in the status-related item list corresponded to the default status of the finding in a ‘normal’ (healthy) patient. For example, the status descriptor for the generic frame ‘jaundice’ was the item list ‘presence-or-absence’, which contained elements ‘present’ and ‘absent’. The ‘normal’ status of jaundice was designated as ‘absent’. The item lists limited (ie, sanctioned) qualifiers available to users to those that were clinically relevant to the generic finding defined by the frame. For example, associated with the generic frame finding ‘chest pain’ is an item list covering ‘chest pain quality’ that includes the modifiers ‘dull aching’, ‘sharp’, ‘crushing’, and ‘burning’.7
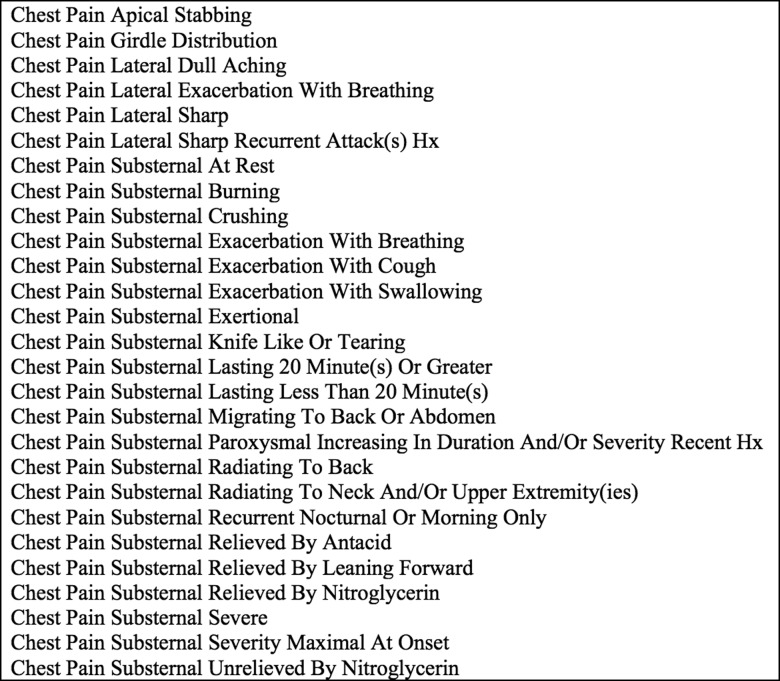
Instantiated finding frames could thus represent the actual findings reported in the literature to occur in patients with any of the 650 disorders in the INTERNIST-1/QMR knowledge base.6 8 Figure 1 illustrates how the generic finding frame ‘Chest Pain’ could represent dozens of INTERNIST-1/QMR finding names through creation of instantiated frames. In a similar manner, CHISL's relatively generic finding frames are linked to item lists containing modifiers and status values. Healthcare providers using CHISL can instantiate finding frames with greater detail and clinical statuses in real time when using it to document patient cases.
Figure 1.
Examples of precoordinated concepts (called instantiated findings) for ‘Chest Pain’ from INTERNIST-1/Quick Medical Reference. Hx, history.
Assertional knowledge-defined relationships
We have previously described assertional knowledge in a terminology as the non-definitional information that provides clinically oriented context and meaning.9 Examples of assertional knowledge include whether a concept is normally absent or present in the population being represented by the terminology and the set of qualifiers and modifiers common to each concept. The use of sanctioned item lists in the INTERNIST-1 frames provided a framework for including clinically oriented assertional knowledge in the terminology. Operationally, item lists can be constructed to include only those qualifiers, modifiers, and associated concepts that are assertionally relevant to the parent concept.
Team members adopted these approaches when constructing CHISL. CHISL concepts are modeled to be relatively general, to retain clinical meaning, and to associate with sanctioned lists of qualifiers and modifiers. An example of the CHISL concept for ‘Chest Pain’ and some of the associated item lists is presented in figure 2.
Figure 2.
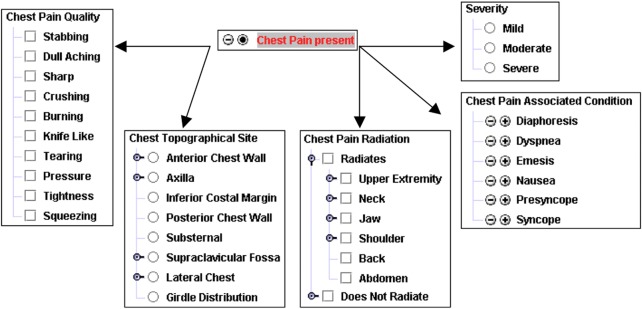
A model for CHISL (Categorical Health Information Structured Lexicon) concept. The concept ‘Chest Pain History or Symptom’ is modeled in CHISL with a preferred synonym ‘Chest Pain’. Chest Pain may be set to absent or present (with ‘absent’ the explicitly defined normal status) and can be modified, including severity, chest pain quality, chest pain radiation, chest pain-associated conditions, among others not listed here. The arrows represent references from the concept to its modifiers. This figure is only reproduced in colour in the online version.
CHISL support for human readability
While a major goal of structured clinical documentation is to create a database of standardized clinical data, the primary goal of clinical documentation, in general, is to create human-readable notes that healthcare providers can reference when caring for patients. Structured data entered into a computer using an interface terminology may not be easily readable in its native form. Interface terminologies can incorporate several methods to enhance readability of any data that they encode. The two main approaches include applying a rich, clinically oriented set of synonyms, and incorporating metadata into the terminology to support computer generation of a naturalistic text presentation.
CHISL leverages both of these approaches. First, and as above, we developed CHISL to have an adequate coverage of the concepts and common colloquial terms (ie, as synonyms) used in standard clinical discourse and documentation. Second, every CHISL concept, modifier, and list includes attributes specifying the part of speech it takes, and whether any special grammatical rules apply. Using these attributes, a computer application can display CHISL content in any of various human user-friendly ways. In a relatively straightforward example, the CHISL concept for ‘headache’ carries a ‘part of speech’ attribute with a value of ‘noun’. The modifier, ‘frontal’ has the part of speech value of ‘adjective’. Applying basic English grammar rules, a documentation system using CHISL would be able to weave the sentence, ‘frontal headache is present’ after a user sets the status for the concept ‘headache’ to present and then modifies it with the qualifier ‘frontal’ from the item list of ‘head pain locations’. A different documentation system could use these inputs to construct sentences that are either more elegant or synoptic, such as: ‘the patient has a frontal headache’ or ‘frontal headache—present’, respectively. This type of natural language generation system is similar to a simple augmented transition network, such as previously described in other structured documentation and expert systems.14 15
CHISL's application independence
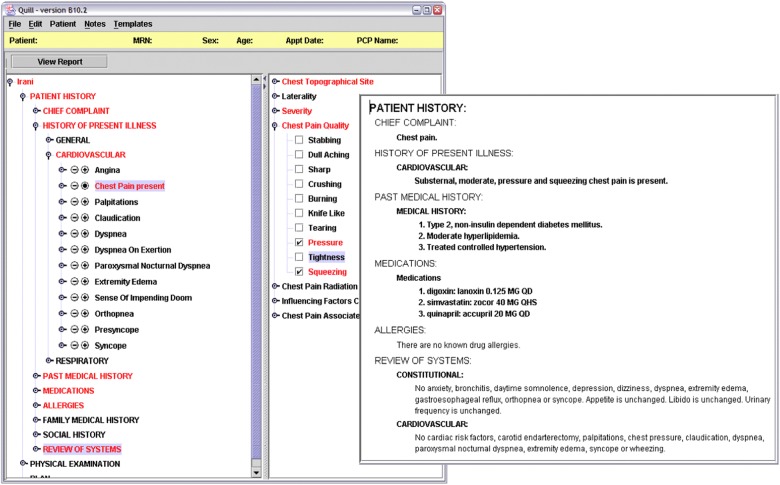
Since 2002, CHISL has been rolled out in a number of different clinical settings and as part of various structured computer-based documentation systems throughout our institution. The primary software using CHISL is a locally developed structured documentation system called Quill.16–18 Quill allows users to document clinical encounters using flexible templates consisting of aggregated CHISL findings and their associated item lists of qualifiers. Quill leverages CHISL attributes, specifying the finding's normal status (eg, that ‘chest pain’ is normally absent in a general population of healthy adults), what appropriate synonyms findings have, and how to generate narrative text from concepts and qualifiers selected as part of documenting clinical care. Quill has been implemented in cardiology, neurology, thoracic surgery, and HIV/AIDS specialty clinics, and in several inpatient services. Figure 3 displays an image of a Quill note and its associated autogenerated narrative text note. In addition, CHISL has been implemented in a static web form used to triage patients being evaluated in the emergency department since May 2005, in a surgical intensive care unit registry tool since February 2006, and in support of several web-based patient intake forms and computer-based documentation systems used across the institution's outpatient sites since October 2011.
Figure 3.
Structured entry tool screen snapshot. The figure above is an image of two views of the user interface for a template created to evaluate a patient with chest pain. The left window shows the interface designed for user input and the right window (inset) shows the narrative report generated by the user's input. This figure is only reproduced in colour in the online version.
Observations
Between the start of CHISL development and the end of 2011, developers have created a total of 3801 findings, 2225 item lists, and 3973 qualifiers aggregated in the item lists. In the interval from 2002 through September 2010, CHISL supported structured clinical documentation in Quill, emergency department triage, and the surgical intensive care unit. Quill users have saved a total of 175 296 notes. In addition, since May 17, 2005 a total of 528 455 emergency department triage forms and 5999 surgical intensive care unit registry forms have been completed. As we have published elsewhere, CHISL use supported 4.3% of all outpatient clinical documentation from 2004 to 2008 at our medical center, increasing from 22 548 clinical notes in 2004 to 150 476 in 2008.18 This increase is, in part, because Quill was implemented in a rolling fashion one clinic at a time.
Discussion
The CHISL terminology described in this manuscript has been developed and iteratively refined to serve as an interface terminology to support and standardize clinical documentation at the authors’ institution. CHISL has been developed to reflect previously published interface terminology desiderata. In particular, CHISL developers partnered with clinical domain experts to ensure adequate contexts, concept, modifier, and term coverage for those clinical areas in which it is used. Developers applied methods successfully used to support the INTERNIST-1 and QMR diagnostic expert systems’ lexicons to achieve compositional balance and to define clinically useful relationships between concepts and modifiers through use of assertional knowledge. In addition, developers structured CHISL and its metadata to allow extensions that can support human readable representation when used in certain clinical software, and to be usable in numerous computer-based documentation systems. Since its initial deployment, CHISL has supported clinical documentation in a growing number of settings throughout the clinical enterprise.
One of the most notable challenges CHISL developers have encountered relates to support for prose output. The current fixed CHISL data model allows for individual concepts and modifiers to encode only single prose styles or parts of speech. The data model does not accommodate cases where concepts or modifiers may have different parts of speech depending on the context in which they are used. For example, the modifying concepts represented by ‘mild’, ‘moderate’, and ‘severe’ would be used as adjectives when documenting chest pain (eg, to achieve the output, ‘severe chest pain is present’), but as adverbs when documenting a change to muscle bulk (eg, ‘muscle bulk is severely decreased’). Given the static data model, developers worked around this gap, creating duplicate versions of the modifiers with each having different parts of speech assigned for prose output. This approach has the advantage of allowing multiple types of nuanced prose output, but the disadvantage of promoting potentially unrecognized ambiguity. However, duplicate concepts or modifiers can be mapped to each other or to an external standard, as necessary, to diminish ambiguity.
In an alternative method, developers created scripts that would process the raw prose output and create complex sentences in using computer-based documentation systems. This approach has the advantage of being customizable to the level of the individual user's documentation styles, but the disadvantage of being labor intensive to build out in each using documentation system as new content comes available.
Ultimately, an updated interface terminology data model that accommodates varied prose outputs based on context, or linking to an external prose-generation service would eliminate the need for workarounds that risk introducing ambiguity, and would allow prose output to be standardized across the computer-based documentation systems used. Additionally, implementing and upper layer ontology on top of CHISL's data model may allow for mapping metadata to decrease ambiguity.
A second challenge observed by the developers is the time- and labor-intensive work required to acquire knowledge from clinical domain experts, and to model it as CHISL frames. The authors speculate that natural language processing tools coupled to improved data mining algorithms might increase the efficiency of modeling clinical domains with interface terminologies. For instance, natural language processing tools could screen clinical documents from an electronic medical records system or data warehouse for frequently occurring medical concepts, modifiers, terms, and relationships, and could determine patterns that occur within given healthcare providers or clinical domains. Identified items that occur frequently could then be refined either through human review or through machine learning-driven techniques that can isolate relevant content missing from the existing terminology.
Footnotes
Funding: This work has also been supported by grants from the National Library of Medicine (K22LM8576 and R01LM009591; STR).
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Rosenbloom ST, Brown SH, Froehling D, et al. Using SNOMED CT to represent two interface terminologies. J Am Med Inform Assoc 2009;16:81–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller RA. A history of the INTERNIST-1 and Quick Medical Reference (QMR) computer-assisted diagnosis projects, with lessons learned. Yearb Med Inform 2010:121–36 [PubMed] [Google Scholar]
- 3.Miller RA, Pople HE, Jr., Myers JD. Internist-1, an experimental computer-based diagnostic consultant for general internal medicine. N Engl J Med 1982;307:468–76 [DOI] [PubMed] [Google Scholar]
- 4.Parker RC, Miller RA. Creation of realistic appearing simulated patient cases using the INTERNIST-1/QMR knowledge base and interrelationship properties of manifestations. Methods Inf Med 1989;28:346–51 [PubMed] [Google Scholar]
- 5.Rassinoux AM, Miller RA, Baud RH, et al. Modeling principles for QMR medical findings. Proc AMIA Annu Fall Symp 1996:264–8 [PMC free article] [PubMed] [Google Scholar]
- 6.Rassinoux AM, Miller RA, Baud RH, et al. Compositional and enumerative designs for medical language representation. Proc AMIA Annu Fall Symp 1997:620–4 [PMC free article] [PubMed] [Google Scholar]
- 7.Rassinoux AM, Miller RA, Baud RH, et al. Modeling just the important and relevant concepts in medicine for medical language understanding: a survey of the issues. In: Proceedings of the IMIA WG6 Working Conference, Jacksonville, FL, 1997 [Google Scholar]
- 8.Rassinoux AM, Miller RA, Baud RH, et al. Modeling concepts in medicine for medical language understanding. Methods Inf Med 1998;37:361–72 [PubMed] [Google Scholar]
- 9.Rosenbloom ST, Miller RA, Johnson KB, et al. Interface terminologies: facilitating direct entry of clinical data into electronic health record systems. J Am Med Inform Assoc 2006;13:277–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbloom ST, Miller RA, Johnson KB, et al. A model for evaluating interface terminologies. J Am Med Inform Assoc 2008;15:65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wade G, Rosenbloom ST. Experiences mapping a legacy interface terminology to SNOMED CT. BMC Med Inform Decis Mak 2008;8(Suppl 1):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wade G, Rosenbloom ST. The impact of SNOMED CT revisions on a mapped interface terminology: terminology development and implementation issues. J Biomed Inform 2009;42:490–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller R, Masarie FE, Myers JD. Quick medical reference (QMR) for diagnostic assistance. MD Comput 1986;3:34–48 [PubMed] [Google Scholar]
- 14.Shortliffe EH, Davis R, Axline SG, et al. Computer-based consultations in clinical therapeutics: explanation and rule acquisition capabilities of the MYCIN system. Comput Biomed Res 1975;8:303–20 [DOI] [PubMed] [Google Scholar]
- 15.Miller PL. Critiquing anesthetic management: the ‘ATTENDING’ computer system. Anesthesiology 1983;58:362–9 [DOI] [PubMed] [Google Scholar]
- 16.Shultz EK, Rosenbloom ST, Kiepek WT, et al. Theater Style Demonstration—Quill: a novel approach to structured reporting. Proc AMIA Annu Fall Symp 2003:1074. [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenbloom ST, Kiepek W, Belletti J, et al. Generating complex clinical documents using structured entry and reporting. Stud Health Technol Inform 2004;107:683–7 [PubMed] [Google Scholar]
- 18.Rosenbloom ST, Stead WW, Denny JC, et al. Generating clinical notes for electronic health record systems. Appl Clin Inform 2010;1:232–43 [DOI] [PMC free article] [PubMed] [Google Scholar]