SUMMARY
A major hurdle in using complex systems for drug screening is the difficulty of defining the mechanistic targets of small molecules. The zebrafish provides an excellent model system for juxtaposing developmental phenotypes with mechanism discovery using organism genetics. We carried out a phenotype-based screen of uncharacterized small molecules in zebrafish that produced a variety of chemically-induced phenotypes with potential genetic parallels. Specifically, kalihinol F caused an undulated notochord, defects in pigment formation, hematopoiesis and neural development. These phenotypes were strikingly similar to the zebrafish mutant, calamity, an established model of copper deficiency. Further studies into the mechanism of action of kalihinol F revealed a novel copper chelating activity. Our data support a novel mechanism of action for kalihinol F and the utility of zebrafish as an effective system for identifying new therapeutics and target pathways.
Keywords: zebrafish, chemical screen, kalihinol F, copper, notochord
INTRODUCTION
A major challenge in the drug discovery process often centers on assigning in vivo activity to candidate compounds. As a potential solution, zebrafish have proven useful for determining both genetic and molecular mechanisms and may offer a powerful tool for understanding drug mechanisms (Kaufman et al., 2009; Taylor et al., 2010; Zon and Peterson, 2005). Small embryo size and high fecundity of adult females make zebrafish amenable to moderate throughput screening and enable screens that directly affect a specific phenotype. Rapid zebrafish development that occurs ex-utero, combined with optical transparency of the embryos, allow for convenient observation of morphological changes under a dissecting microscope without sacrificing the embryo (Kimmel et al., 1995). Small molecule screening in a whole organism also provides a physiological context that is lacking in cell-based and in vitro assays, and affords preliminary toxicity, tissue specificity and pharmacokinetic profiles that greatly facilitate lead compound identification and optimization. Most importantly, the zebrafish is a vertebrate system that shares a high degree of genetic and physiological similarity to humans. Numerous zebrafish genetic mutants have been established that faithfully phenocopy human disease mechanisms and zebrafish-based chemical genetic screens are currently underway to identify compounds that modify disease phenotypes, an approach that promotes development of novel biological tools and therapeutics (Ingham, 2009; Kaufman et al., 2009; Lieschke and Currie, 2007; MacRae and Peterson, 2003; Shin and Fishman, 2002; Taylor et al., 2010; Zon and Peterson, 2005).
Previous efforts have clearly demonstrated the utility of zebrafish in identifying compounds that affect specific developmental phenotypes. In 2000, Peterson et al. carried out one of the earliest zebrafish-based chemical screens and discovered small molecules that perturb normal development of the central nervous system, cardiovascular system, pigmentation and ear (Peterson et al., 2000). Since then, numerous screens have revealed chemical modifiers of zebrafish embryonic development (Kaufman et al., 2009; Taylor et al., 2010). However, another approach may exist to use zebrafish to define mechanisms of uncharacterized compounds and compound collections. For example, the kalihinol family of compounds are isonitrile diterpenoids originally isolated from the marine sponge Acanthella sp. (Patra et al., 1984). These compounds are characterized by a biflorane carbon skeleton and show a variety of biological activities that include antibacterial, antifungal, antiparasitic, antifouling and cytotoxicity against tumor cells (Alvi et al., 1991; Chang et al., 1984; Fusetani et al., 1990; Hirota et al., 1996; Miyaoka et al., 1998; Omar et al., 1988; Sun et al., 2009; Wolf and Schmitz, 1998). Although specific kalihinols, such as kalihinol F, have proposed mechanisms in bacteria and starfish, it is not clear how kalihinols might function within a vertebrate system both from physiologic and mechanistic point of view (Bugni et al., 2004; Ohta et al., 2003).
In this study, we performed a zebrafish-based phenotypic screen of 954 diverse compounds and juxtaposed the observed chemically-induced phenotypes with known genetic mutations. This analysis led us to focus on kalihinol F which produced a wavy notochord, loss of pigmentation, as well as hematologic and neurologic abnormalities. The kalihinol F-induced phenotypes were highly similar to those reported for the zebrafish mutant calamity (cal), which shows evidence of copper deficiency due to a mutation in the copper transporter, atp7a (Mendelsohn et al., 2006). Consistent with reduced copper levels, kalihinol F-induced phenotypes were prevented with exogenous copper. Further studies demonstrate that kalihinol F chelates copper, an activity shared by other kalihinol analogs. Overall, our findings support a novel mechanism of action for kalihinol F and demonstrate the use of zebrafish as an effective system for integrating biological activity with mechanism of action.
RESULTS
A zebrafish-based phenotypic screen identifies small molecule inducing a copper-deficient phenotype
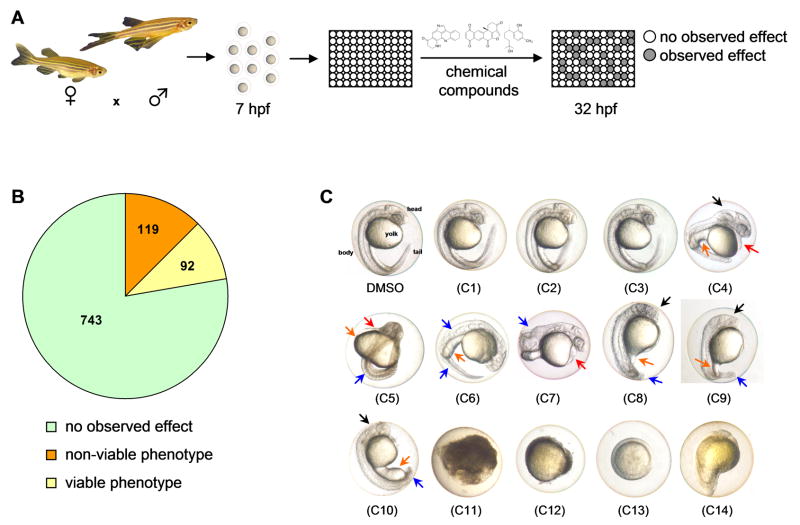
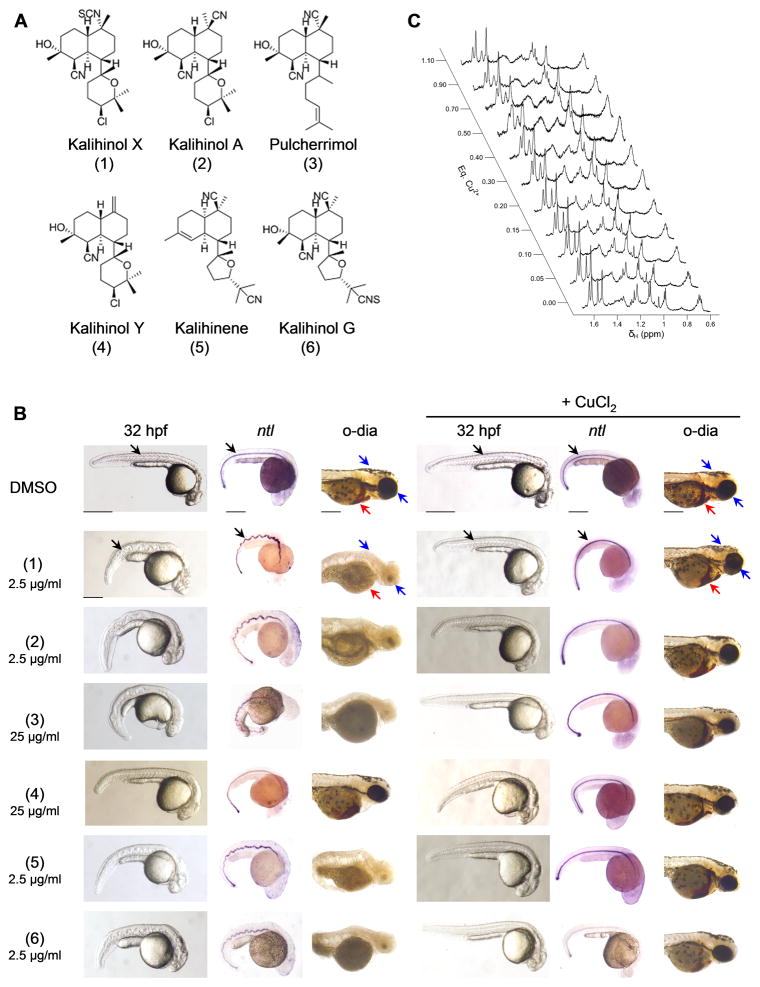
We utilized wild type zebrafish embryos to screen for bioactive small molecules. For our phenotypic assay, we arrayed 7 hpf embryos in 96-well plates at 1 embryo per well, treated with test compounds and visually evaluated developmental phenotypes at 32 hpf (Figure 1a). A total of 954 compounds from different chemical libraries were used in the screen and included synthetic drug-like compounds, FDA-approved drugs and natural products (Figure 1b). No discernible effect was observed for majority of the compounds, resulting in normal-appearing embryos with well-defined body structures at 32 hpf (Figure 1, b–c, DMSO, c1–c3). Of the active compounds, 119 caused overt lethality (Figure 1, b–c, c11–c14) while 92 had viable phenotypes that showed varying developmental defects (Figure 1, b–c, c4–c10). A systematic search of phenotypes elicited by our compound collection against known zebrafish genetic mutants resulted in an interesting match with the zebrafish mutant calamity (cal). The calamity mutant carries a mutation in the ubiquitous copper transporter gene atp7a and displays copper deficiency-induced developmental defects (Mendelsohn et al., 2006). Kalihinol F-treated embryos caused defects consistent with copper deficiency (Figure 2, b–d).
Figure 1.
Zebrafish-based chemical screen leads to identification of interesting phenotypes. a. Schematic of the zebrafish phenotype screen. Wild type embryos were arrayed in 96-well plates and treated with 10 μM or 25 μg/ml of test compounds at 7 hpf. Phenotypes were scored at 32 hpf. b. Pie chart showing phenotype distribution from the zebrafish screen. c. At 32 hpf, zebrafish embryos have a defined head, body and tail, with a prominent yolk (C, DMSO). Normal-looking embryos were observed for 743 compounds (C1–C3), 119 were toxic to embryos (C11–14), and 92 had viable phenotypes (C4–C10) that include edema (red arrow), underdeveloped head (black arrow), yolk (orange arrow) and tail/body (blue arrow) malformations.
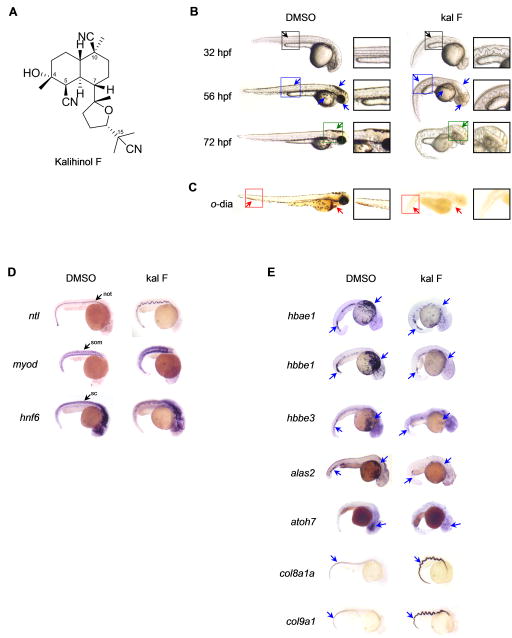
Figure 2.
Embryos treated with kalihinol F show a phenotype consistent with copper deficiency. a. Chemical structure of kalihinol F. b. Exposure of zebrafish embryos to 2.5 μg/ml of kalihinol F (kal F) resulted in a wavy notochord (black arrow, D, ntl), loss of pigmentation (blue arrow) and enlarged hindbrain vesicle (green arrow) as compared to DMSO-treated control embryos. Boxed figures are enlargements of highlighted portions on whole embryos. c. o-dianisidine staining at 72 hpf revealed loss of hematopoiesis (red arrows) in treated embryos versus control. d. In situ hybridization on 32 hpf embryos for ntl, myod and hnf6. not, notochord; som, somites; sc, spinal cord. e. In situ hybridization for hbae1, hbbe1, hbbe3, alas2, atoh7, col8a1a, col9a1 confirms gene expression analysis by RNA sequencing that is consistent with copper-deficient phenotype of kalihinol F. See also Tables S1, S2.
Kalihinol F (Figure 2a) is a marine natural product that has been reported to have antibiotic and cytotoxic activities (Alvi et al., 1991; Bugni et al., 2004; Chang et al., 1984; Fusetani et al., 1990; Hirota et al., 1996; Miyaoka et al., 1998; Ohta et al., 2003; Omar et al., 1988; Sun et al., 2009; Wolf and Schmitz, 1998). It has not, however, been associated with copper homeostasis. At 32 hpf, embryos treated with 2.5 μg/ml kalihinol F had an undulated notochord, as confirmed by in situ staining for ntl, and a complete absence of pigmentation (Figure 2, b–d). In situ staining for myoD and hnf6 also revealed short, deformed somites and wavy spinal cord, respectively (Figure 2d). By 56 hpf, loss of pigmentation was more evident. The embryos also appeared curved and had a slow heart beat with no circulation (Figure 2b). The phenotype became progressively worse by 72 hpf, as evidenced by an enlarged hindbrain vesicle, edema, severe body malformation and crinkly tail (Figure 2b). Loss of blood was confirmed by o-dianisidine staining (Figure 2c).
To confirm the bioactivity of kalihinol F at the molecular level, we looked at the gene expression profile of kalihinol F-treated embryos by RNA sequencing. We found that the differentially expressed genes in treated embryos, some of which we confirmed by in situ hybridization, reflected the morphological defects that we have previously associated with kalihinol F and copper deficiency (Table 1, Figure 2e, Tables S1–2). Downregulation was observed for hemoglobin genes such as hbae1, hbbe2, hbae3, hbbe1, hbaa1, hbbe3 that are involved in oxygen transport, heme binding, and formation of hemoglobin complex. Several genes that are expressed in the brain, eye and blood were also downregulated. Kalihinol F-treated embryos exhibited increased expression of inflammatory and immune response genes, as well as col8a1a and col9a1, genes that are both essential for normal notochord and cartilage development and consistent with previous studies reporting elevated collagen levels during copper deficiency (Gansner et al., 2007; Tilton et al., 2006).
Table 1.
GO analysis of the differentially regulated genes in kalihinol F-treated embryos.
| GO Category | Total Genes | Changed Genes | Enrichment | FDR |
|---|---|---|---|---|
| Hemoglobin complex | 15 | 8 | 51.6 | 0 |
| Oxygen transport | 27 | 8 | 28.7 | 0 |
| Oxygen binding | 40 | 9 | 21.8 | 0 |
| Heme binding | 156 | 13 | 8.1 | 0 |
| Extracellular region | 1430 | 47 | 3.2 | 0 |
| Response to mineralocorticoid stimulus | 31 | 5 | 15.6 | 0.001 |
| Inflammatory response | 254 | 12 | 4.6 | 0.002 |
| Response to chemical stimulus | 2073 | 40 | 1.9 | 0.006 |
| Organic ether metabolic process | 96 | 7 | 7.1 | 0.007 |
| Notochord development | 45 | 5 | 10.7 | 0.007 |
| Fibronectin binding | 11 | 3 | 26.4 | 0.011 |
| Immune response | 548 | 16 | 2.8 | 0.011 |
| Response to progesterone stimulus | 31 | 4 | 12.5 | 0.018 |
| Ossification | 165 | 8 | 4.7 | 0.020 |
| Regulation of angiogenesis | 91 | 6 | 6.4 | 0.020 |
| Lipoprotein metabolic process | 96 | 6 | 6.0 | 0.025 |
| Eukaryotic cell surface binding | 15 | 3 | 19.3 | 0.025 |
| Triglyceride metabolic process | 66 | 5 | 7.3 | 0.031 |
| Vesicle lumen | 38 | 4 | 10.2 | 0.032 |
| Protein activation cascade | 40 | 4 | 9.7 | 0.035 |
| Cartilage development | 151 | 7 | 4.5 | 0.043 |
| Protein maturation | 111 | 6 | 5.2 | 0.043 |
| Response to other organism | 401 | 12 | 2.9 | 0.044 |
Exogenous copper prevents kalihinol F copper-deficient phenotype
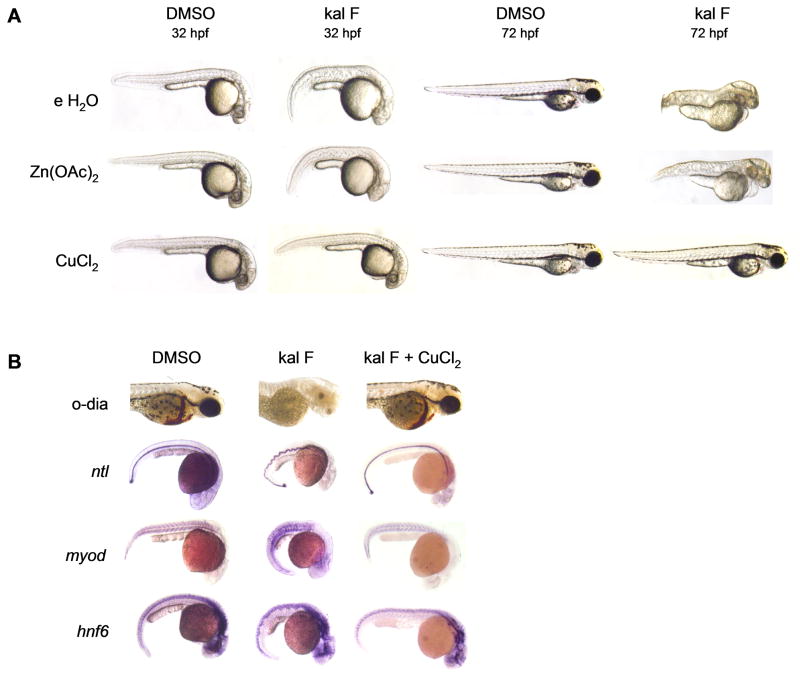
Since kalihinol F-treated embryos phenocopy cal, we hypothesized that kalihinol F might be inducing copper deficiency either by targeting atp7a, resulting in impaired cellular uptake of copper or by reducing overall levels of bioavailable copper for normal growth and development through copper chelation. To distinguish between the two, treated embryos were given exogenous copper, which will reverse copper chelation but will not overcome defective copper absorption due to inactivated atp7a, as previously shown (Mendelsohn et al., 2006). Exposure of treated embryos to 10 μM CuCl2 led to a complete prevention of the kalihinol F phenotype (93%, n=120). The notochord appeared normal, as did the somites and spinal cord (Figure 3, a–b). Normal pigmentation returned and healthy blood circulation was observed (Figure 3, ab). To evaluate specificity for copper rescue, embryos were also given zinc and calcium, both +2 metals. Neither zinc (100%, n=82) nor calcium (100%, n=27) were capable of reverting any of the kalihinol F-induced embryo defects (Figure 3, a–b, Figure S1).
Figure 3.
Kalihinol F phenotype is prevented by addition of copper. a. 5 hpf embryos were treated with either DMSO or 2.5 μg/ml kal F then rescued at 7 hpf with 10 μM CuCl2, Zn(OAc)2 and embryo water (e H2O). Only copper-treated embryos showed normal pigmentation, notochord and hindbrain comparable to control embryos (bottom row, second and fourth figures from left). b. Normal hematopoiesis and rescue of notochord, somites, spinal cord with copper were confirmed by o-dianisidine staining and in situ for ntl, myod, hnf6. See also Figure S1.
Kalihinol F chelates copper
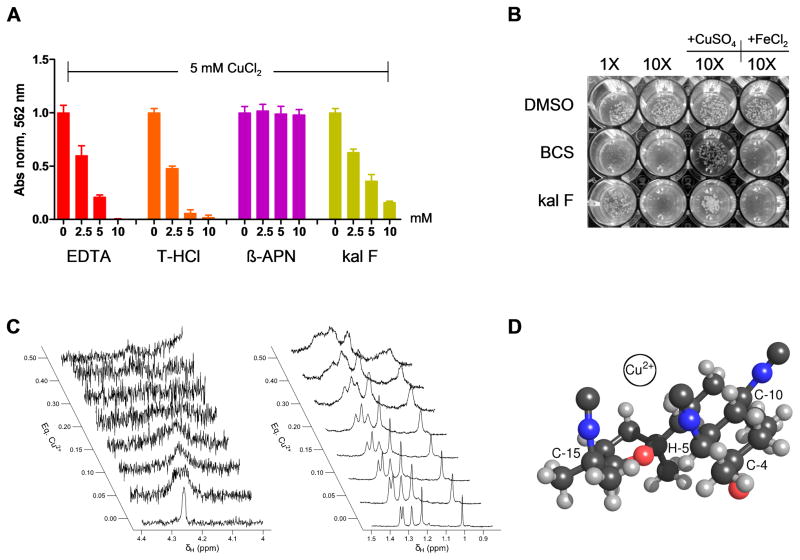
Based on the phenotype and copper prevention experiments, our data suggest that kalihinol F might act as a copper chelator. To investigate this further, we performed a modified BCA assay to assess copper chelating ability of kalihinol F in comparison with several controls. A constant concentration of CuCl2 was mixed with various concentrations of each of kalihinol F, β-aminopropionitrile (β-APN), and the positive controls EDTA and trientine HCl (T-HCl), a copper chelator used in the clinic to treat Wilson disease (Ding et al., 2011). As expected, a decrease in absorbance due to free copper was observed with increasing concentrations of EDTA and T-HCl (Figure 4a). At 2-fold molar excess, copper was almost completely chelated. A similar behavior was displayed by kalihinol F; there was a reduction in absorbance that was concentration-dependent, although not as steep as was seen with EDTA and T-HCl. The lysyl oxidase inhibitor β-APN did not exhibit any copper chelating activity (Figure 4a).
Figure 4.
Kalihinol F is a copper chelator. a. BCA assay was performed on 5 mM CuCl2 supplemented with EDTA, T-HCl, β-APN or kal F, in increasing molar excess. Copper chelation was determined by measuring absorbance at 562 nm. Graph shown above is representative of multiple experiments. b. Wild type W303 yeast cells were grown on YP-agar plate containing respiration-selective media treated with DMSO, BCS or kal F. Growth inhibition due to limiting copper levels was evaluated visually. Addition of copper, but not iron, was able to rescue yeast growth (lanes 3,4). Picture shown above is representative of multiple experiments. c. Effects of addition of Cu2+ to the 1H NMR linewidths of H-5 (left panel) and the methyl group signals (right panel) of kalihinol F. d. Proposed binding site of copper based on previously determined x-ray structure of kalihinol F (Patra et al., 1984). Note that the isonitrile groups attached to C-5 and C-15 are oriented towards the same face of the molecule and potentially forms a bidentate coordination site for Cu2+ as depicted. Note also the proximity of H-5 and the furan ether group. See also Figure S3.
To further validate the mode of action of kalihinol F, we turned to yeast, whose growth in respiratory-selective media is dependent on copper, a cofactor for Cox1 and Cox2 which are essential for respiration (Weintraub and Wharton, 1981). Wild type yeast cells grown in glycerol-lactate media supplemented with either kalihinol F or with the known copper chelator, bathocuproine disulphonate (BCS), showed a dose-dependent growth inhibition that was rescued with exogenous copper, but not iron, which is consistent with the dependency of yeast respiration on copper (Figure 4b).
Because of its paramagnetic nature, copper (II) causes broadening of resonance signals in NMR spectroscopy as a result of distance-dependent paramagnetic relaxation enhancement (PRE) of nuclear spins (Bloembergen, 1957; Solomon, 1955). To confirm the interactions between kalihinol F and copper by NMR, a titration experiment was performed to measure the effects of addition of CuCl2 on the spectral line widths of kalihinol F in DMSO-d6. Kalihinol F was present at 11 mM and NMR spectra were collected at the titration end points 0.00, 0.05, 0.10, 0.15, 0.20, 0.30, 0.40, and 0.50 mole equivalents of CuCl2 in D2O. There was a difference among the signals in the degree of line broadening that occurred in response to copper addition. Some signals exhibited a more rapid onset and more extensive degree of broadening as the titration progressed. The signal for H-5, in particular, was strongly broadened at 0.05 eq Cu (II) and had essentially merged with the baseline by 0.30 eq Cu (II) (Figures 2a, 4c). In addition, the center frequency for H-5 gradually moved upfield as the titration progressed. Before addition of Cu (II), δH-5 occurred at 4.26 ppm. The frequency shifted by 0.03 ppm upfield for each addition of 0.05 eq Cu (II). The remaining signals also exhibited increased broadening as the titration progressed, but did so to a much lesser extent than H-5 (Figure 4c). The remaining signals also displayed smaller shifts in frequency than H-5. Overall these results suggest that kalihinol F does bind to Cu (II) in DMSO and that the binding site is likely near H-5.
A novel bioactivity for kalihinol family of compounds
Kalihinols belong to a large family of multifunctional diterpenes that exhibit a wide array of biological properties (Alvi et al., 1991; Bugni et al., 2004; Chang et al., 1984; Fusetani et al., 1990; Hirota et al., 1996; Miyaoka et al., 1998; Ohta et al., 2003; Omar et al., 1988; Sun et al., 2009; Wolf and Schmitz, 1998). To investigate whether the copper chelating activity is shared by other kalihinol analogs, 7 hpf embryos were treated with 6 additional kalihinol compounds with DMSO as control (Figure 5a). Kalihinol X (100%, n=50), kalihinol A (99%, n=75), kalihene (95%, n=141) and kalihinol G (100%, n=149) exhibited a similar copper-deficient phenotype at 2.5 μg/ml (Figure 5b). Pulcherrimol (95%, n=107) only showed activity at a higher concentration of 25 μg/ml and this was reflected in the NMR analysis of the interaction between pulcherrimol and copper (Figure 5, b–c). When a second titration was carried out on a sample containing pulcherrimol, the onset of line broadening was less extensive for pulcherrimol compared to kalihinol F, which suggests a weaker affinity of pulcherrimol for Cu (II) (Figures 4c, 5c). Kalihinol Y (100%, n=48), was not active at all (Figure 5b). As previously observed with kalihinol F, the copper-deficient phenotype induced by the kalihinol analogs were prevented by treatment with copper (Figure 5b).
Figure 5.
Other members of the kalihinol family also show copper chelating activity. a. Chemical structure of kalihinol analogs. b. 7 hpf embryos were treated with either 25 μg/ml or 2.5 μg/ml of kalihinol analogs and DMSO vehicle control (first, second, third columns). For phenotype rescue experiments, 5 hpf embryos were treated with kalihinols and given CuCl2 at 7 hpf (fourth, fifth, sixth columns). Embryos were monitored for wavy notochord (black arrow), loss of pigmentation (blue arrow) and hematopoeisis (red arrow). c. Effects of addition of Cu2+ to the 1H NMR linewidths of the methyl signal region of pulcherrimol.
Kalihinol F mitigates toxicity associated with overexposure to copper
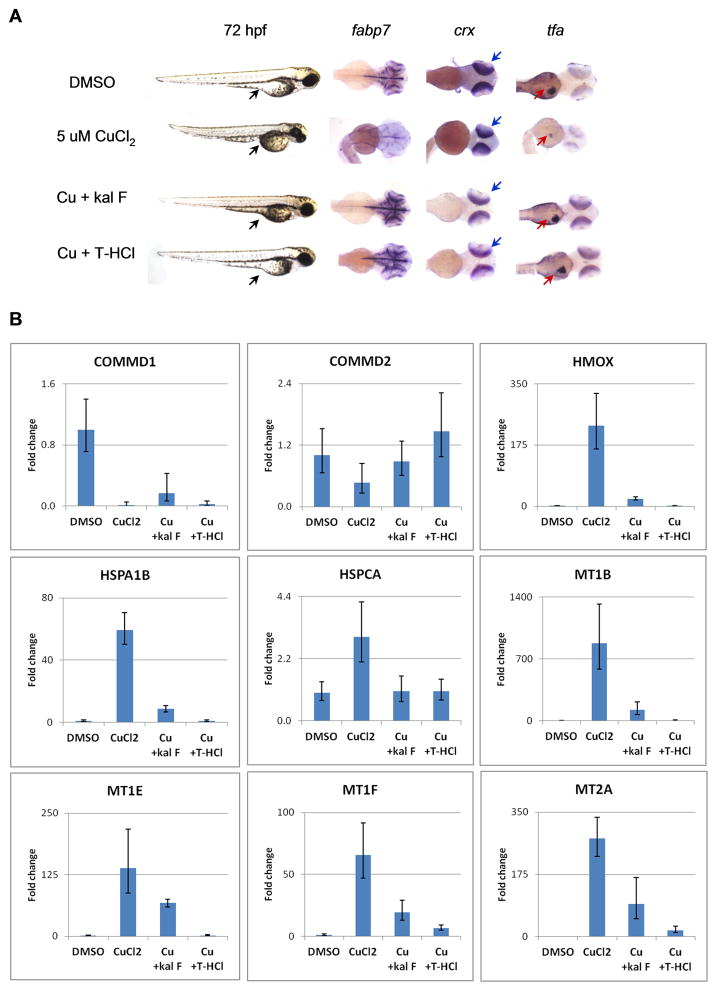
Copper chelation therapy has long been used to treat Wilson disease (WD), a disorder characterized by hepatic accumulation of copper due to mutations in ATP7b, a liver-specific copper transporter (Huster, 2010). To determine if kalihinol F can prevent harmful effects of excess copper, 5 hpf embryos were given 5 μM CuCl2 and were further treated with 5 μg/ml kalihinol F or 10 μM T-HCl. At 72 hpf, embryos exposed to copper remained in their chorion. They appeared shorter and exhibited growth defects that include a pinched yolk extension (96%, n=142) and a smaller head, as confirmed by in situ for fabp7 (95%, n=48, Figure 6a). In situ hybridization for crx and tfa also revealed smaller eyes (100%, n=52) and liver (91%, n=74), respectively (Figure 6a). Treatment with kalihinol F reversed the developmental defects associated with copper toxicity. The rescued embryos were indistinguishable from control embryos, with normal yolk extension (100%, n=102) and well-developed head (100%, n=42), eyes (100%, n=50) and liver (91%, n=55, Figure 6a). Similar results were observed for T-HCl (Figure 6a).
Figure 6.
Kalihinol F reverses adverse effects of copper toxicity. a. 5 hpf embryos were treated with 5 μM CuCl2 and rescued at 7 hpf with either 5 μg/ml kalihinol F or 10 μM T-HCl. Excessive copper resulted in a smaller embryo with pinched yolk extension (lane 1, black arrow), small head (lane 2) and eyes (lane 3), and impaired liver development (lane 4). In situ hybridization for fabp7, crx (blue arrow) and tfa (red arrow) revealed normal head development, eye size and proper liver formation, respectively, for embryos rescued with kalihinol F or T-HCl b. Human HepG2 cells were treated with 80 μM copper and rescued with either 30 μg/ml kalihinol F or 160 μM T-HCl. Quantitative RT-PCR showed gene expression changes induced by copper were reversed by either kalihinol F (Cu + kal F) or T-HCl (Cu + T-HCl). Graph shown above is representative of at least 2 experiments. See also Figure S2, Table S3.
To determine if kalihinol F can overcome toxic effects of copper in liver cells, human hepatocarcinoma cells (HepG2) were treated with 80 μM CuCl2 and then rescued with 30 μg/ml kalihinol F or 160 μM T-HCl. Quantitative RT-PCR was done to evaluate gene expression changes associated with chemical treatments over 24 h. Consistent with previous studies by Muller et al., HepG2 cells incubated with copper showed reduced expression of COMMD1 and COMMD2 while metallothioneins such as MT1B, MT1E, MT1F and MT2A, along with HMOX and heat shock proteins, HSPA1B and HSPCA, were significantly upregulated (Figure 6b) (Muller et al., 2007). Further treatment with either kalihinol F or T-HCl reversed these trends, although the effect seen with T-HCl is more pronounced compared with kalihinol F (Figure 6b). This could be due to less amount of kalihinol F (30 μg/ml ~ 78 μM) given to cells because of limited supply of the compound. There also was toxicity in HepG2 cells exposed to copper, as evidenced by floating debris, which was alleviated in rescued cells (Figure S2).
DISCUSSION
Defining the bioactivity and mechanism of action of small molecules is often a challenge in the development of potential therapeutics. We have demonstrated that chemical screening in zebrafish, coupled with genetic analyses, allows for evaluation of the biological effects of active compounds with better insight into their molecular mechanism. Our data revealed kalihinol F as a novel copper chelator, which evoked developmental abnormalities similar to the known copper transport mutant, calamity (cal) (Mendelsohn et al., 2006).
Prior work on kalihinols has been limited to understanding their bioactivity in non-vertebrate systems and to date, has not identified a specific molecular mechanism (Alvi et al., 1991; Bugni et al., 2004; Chang et al., 1984; Fusetani et al., 1990; Hirota et al., 1996; Miyaoka et al., 1998; Ohta et al., 2003; Omar et al., 1988; Sun et al., 2009; Wolf and Schmitz, 1998). Consistent with the phenotype observed for kalihinol F-treated embryos, our BCA assay data indicate that kalihinol F can sequester copper, although not as well as EDTA or T-HCl, a commonly used drug in copper chelation therapy against Wilson disease (Figure 4a) (Ding et al., 2011). Kalihinol F exhibited the same activity in yeast, where we exploited the dependency of yeast cells on copper for growth under respiratory-selective media (Figure 4b) (Weintraub and Wharton, 1981). Utilizing NMR to detect copper binding with kalihinol F, we found that it displayed a significant affinity for Cu (II) but did not exhibit the tightness of binding seen with β-penicillamine, another clinically used copper chelator (Figure 4c, Figure S3) (Ding et al., 2011). Although the affinity of kalihinol F for Cu (II) is weaker, it is predominantly hydrophobic and may offer the advantage of being able to penetrate hydrophobic media and potentially to transport Cu (II) passively through barriers such as cell membranes. Interestingly, both T-HCl and β-penicillamine did not result in any phenotype when given to zebrafish embryos (data not shown).
Based on the previously published x-ray structure of kalihinol F, we propose that copper sits in a binding pocket in the vicinity of H-5, formed by the isonitrile groups on C-5 and C-15 (Figure 4d) (Patra et al., 1984). Both H-5 and the tetrahydrofuran ring are located equatorially on their respective rings and are in close proximity to each other. Moreover, the isonitrile groups on C-5 and C-15 are both located on the same face of the ring assembly. The lone-pair electrons on the carbon atoms of these groups are strong candidates for being involved in coordination to Cu (II), suggesting that it might be possible for them to form a bidentate binding site for Cu (II). It is interesting to note, however, that the inactive analogue, kalihinol Y, lacks the C15 isonitrile group, which suggests that the C5 isonitrile moiety is critical for copper binding (Figure 5a, b). This is consistent with the effect of copper on H5, as seen in our NMR data (Figure 4c). The other isonitrile on C-10 is located equatorially and is, therefore, not capable of intramolecularly coordinating a Cu (II) atom at the proposed binding site (Figure 4). In the x-ray structure of kalihinol F, the isonitrile groups at C-5 and C-15 are roughly parallel to one another and 4.1 Å apart, which would not be a favorable geometry for binding to the same Cu (II) atom. However, a slight rotation around the C-7/C-11 bond and/or the C-14/C-15 bond could yield a more favorable binding site. Such a rotation could also change the magnetic environment near H-5, accounting for the strong perturbation in its chemical shift and its line width.
The differences in the bioactivity of the kalihinol compounds that we additionally screened revealed functionalities that may be important for copper chelation (Figure 5). In comparing the structural differences among the kalihinol analogs, our data indicate the significance of the substituted tetrahydropyranyl/tetrahydrofuranyl moiety and the cyano functional group at C-11 and C-10, respectively. Further studies are needed to examine how the cyano group at C-10 contributes to the overall stability of the copper-kalihinol complex, despite its distance from the proposed binding site on the molecule.
Copper is an essential mineral for normal growth and development of living organisms. It is an important co-factor for a number of metabolic enzymes involved in cellular respiration, energy metabolism, collagen cross-linking, antioxidation, and pigment formation, among others (Tisato et al., 2009). Copper homeostasis is tightly regulated, as evidenced by human ailments resulting from unbalanced copper levels (Tisato et al., 2009). Wilson disease (WD), characterized by copper accumulation in the liver and marked by neurodegenerative symptoms and hepatotoxicity, is a genetic disease caused by mutations in ATP7B, an ATPase copper-transporting enzyme that is primarily responsible for copper elimination in the liver (Huster, 2010). Copper chelation therapy has been a successful treatment strategy for WD, however, the spectrum of chelators available are limited and harbor some toxicities (Ding et al., 2011; Huster, 2010; Tisato et al., 2009; Tumer and Moller, 2010). The ability of kalihinol F to reverse copper toxicity in zebrafish embryos and human HepG2 cells to a similar extent as T-HCl, suggests that kalihinols may have clinical relevance as copper chelators (Figures 6 a, b).
The copper-chelating activity exhibited by kalihinols is consistent with a large body of literature reporting isonitrile/isocyanide compounds interacting with copper and cuproenzymes (Liu and Reiser, 2011; Reedy et al., 1995; Rhames et al., 2001). One of the most recent is a report by Zhang et al. where they show that an isocyanide anti-fouling compound, Isocyanide 1, caused a wavy notochord in zebrafish embryos and phenocopied copper deficiency (Zhang et al., 2012). The discovery of kalihinols as copper chelators might aid in identifying potential binding partners responsible for the wide array of biological activity reported for these compounds.
In conclusion, we have used zebrafish to rapidly assign a novel activity and mechanism of action to kalihinol F. Interestingly, a combined zebrafish-yeast chemical genetic screen undertaken by Ishizaki et al. has identified 45 compounds affecting copper metabolism in zebrafish embryos that included reported copper chelators and the known MEK inhibitor, U0126, revealing a novel copper-dependent target pathways for U0126 and further supporting the utility of the zebrafish in elucidating mechanism of action of small molecules (Ishizaki et al., 2010). The kalihinol family of compounds signifies a new class of copper chelators that may be used as a biological tool to better elucidate the molecular mechanisms underlying copper homeostasis. Kalihinols may represent a novel copper chelating pharmacophore. However, in order to effectively use kalihinols as copper chelators, further studies are required to fully characterize its binding affinity to copper. Kalihinol F may not complex copper as strongly as clinically used copper chelators but preliminary SAR analysis presented in this study can facilitate in developing more potent, kalihinol-based copper chelators that are already membrane-permeable and bioavailable.
SIGNIFICANCE
Elucidating the mechanistic action of small molecules is frequently restricted by the complexities of vertebrate model systems. By using a novel approach of juxtaposing chemically-induced phenotypes from our zebrafish screen to known genetic zebrafish mutations, we identified kalihinol F and the kalihinol family as a new class of copper chelators. Our findings may lead to the development of more potent copper chelators that may prove useful in the clinic. We have also shown the utility of zebrafish as an effective system for rapidly assigning mechanism of action to small molecules which may be used to more efficiently identify novel therapeutics and target pathways.
EXPERIMENTAL PROCEDURES
Zebrafish maintenance
Wild-type Danio rerio (zebrafish) were maintained as previously described (Westerfield, 1995). Fertilized embryos were collected following natural spawnings either in 1× E3 medium (286 mg/L NaCl, 13 mg/L KCl, 48 mg/L CaCl2·2H2O, 40 mg/L MgSO4, 0.01% methylene blue) or 2× PTU (1× E3 medium, 30.4 mg/L phenylthiourea) and allowed to develop at 28.5 °C.
Zebrafish phenotype screen and additional drug treatments
Embryos were periodically checked for death and developmental delay before treatment. At 7 hours post-fertilization (hpf), embryos were arrayed in 96-well plates at 1 embryo/well. Sample compounds were then added to the desired concentration, with dimethyl sulfoxide (DMSO) as control. DMSO was kept at 0.5% of the total assay volume.
Embryos were examined visually for viable and non-viable phenotypes with a dissecting microscope at 24 hours post treatment (hpt). Embryos were photographed live with an Olympus DP71 digital camera. All experiments were repeated at least twice, in duplicate.
Compounds used in the screen were prepared in DMSO either as 10 mM or 5 mg/ml stocks. Further dilutions were carried out in DMSO. The Spectrum Collection library was purchased from MicroSource Discovery Systems (Gaylordsville, CT). The DIVERSet library was purchased from ChemBridge (San Diego, CA). The chemistry and marine natural product libraries are proprietary to the University of Utah.
For copper prevention experiments, embryos were treated with kalihinol compounds at 5 hpf. Copper chloride (CuCl2·2H2O), calcium chloride (CaCl2) or zinc acetate (ZnOAc2) was added 2 h later at 5 or 10 μM final concentration, with embryo water as control. Phenotype suppression was evaluated 24 h after treatment with CuCl2.
For kalihinol rescue experiments, embryos were treated with 5 uM CuCl2 at 5 hpf. Trientine·2HCl (T-HCl) or kalihinol F was added 2 h later at 10 μM or 5 μg/ml final concentration, with DMSO as control. Phenotype rescue was evaluated 72 h after treatment with copper chelators.
In situ hybridization
In situ hybridizations were performed as previously described using digoxigenin-labeled riboprobes for ntl (no tail), myod (myogenic differentiation 1), hnf6 (hepatocyte nuclear factor 6), hbae1 (hemoglobin alpha embryonic-1), hbbe1 (hemoglobin beta embryonic-2), hbbe3 (hemoglobin beta embryonic-3), alas2 (δ-aminolevulinate synthetase 2), atoh7 (atonal homolog 7), col8a1a (collagen type VIII, alpha 1a), col9a1 (collagen type IX, alpha 1), fabp7a (fatty acid binding protein 7), crx (cone-rod homoebox) and tfa (transferrin-a) (Nadauld et al., 2004).
o-Dianisidine staining
o-Dianisidine staining was performed as previously described (Mendelsohn et al., 2006). The embryos were then fixed with 8% paraformaldehyde/2× sucrose buffer at room temperature for 2 h or overnight at 4 °C.
RNA sequencing and data analyses
RNA was isolated from embryos treated with kalihinol F or DMSO at 32 hpf using the RNeasy kit (Qiagen). Quadruplicate biological replica samples were prepared and sequenced using Illumina HiSeq 2000 (Illumina). Raw data were aligned using Novoalign (Novocraft) against the Zv9 genome build containing known and theoretical splice junctions from Ensembl transcript annotation. After converting splice junction matches to genomic coordinates, the OverDispersedRegionScanSeqs (Useq) was used to identify differentially expressed genes using an estimated FDR of 10% and twofold difference. GO term analysis was performed using GoMiner (discover.nci.nih.gov/gominer).
BCA assay
Bicinchoninic acid (BCA) assay was performed according to the manufacturer’s instructions with minor modifications (Pierce, Thermo Scientific). EDTA, T-HCl, β-aminopropionitrile (β-APN) or kalihinol F was added to CuCl2 in 0, 0.5, 1, 2 molar ratios. The resulting solutions were incubated at room temperature for 30 min to allow copper chelation to proceed and then incubated at 37 °C for 15 min after addition of BCA and bovine serum albumin (BSA). Samples were read on the spectrophotometer at 562 nm.
NMR analysis
NMR experiments were carried out on a Varian INOVA 500 MHz spectrometer with a 3 mm Nalorac MDBG probe. Kalihinol F and pulcherrimol were each dissolved in DMSO-d6 (240 μL, 11 mM) and were titrated using 26 mM CuCl2 in D2O. β-penicillamine was dissolved in DMSO-d6 (240 μL, 18 mM) and was titrated using 48 mM CuCl2 in D2O. 1H NMR spectra were recorded after each titration step with mixing at a sample temperature of 298 K. For each spectrum, 16384 data points were acquired (acquisition time 2.05 s) with a spectral width of 8000 Hz. Spectra were Fourier-transformed directly with no window functions applied.
Yeast culture and drug treatments
Wild type W303 yeast cells were cultured in liquid yeast peptone (YP)-glucose at 30°C overnight. Approximately 400 cells were spotted per well in 48-well YP-agar plate prepared with 2%-glycerol-2% lactate as carbon source and supplemented with kalihinol F, bathocuproine disulphonate (BCS) or DMSO as control. The culture plate was incubated at 30°C and yeast cell growth inhibition due to chelation of available copper in the media was monitored visually for 2 days. Final drug concentration was either 0.5 or 5 μg/ml for kalihinol F and 0.1 or 1 mM for BCS.
For growth rescue experiments, YP-agar plates were additionally treated with either 50 uM or equimolar copper sulfate (CuSO4) or iron chloride (FeCl2) for kalihinol F- and BCS-containing wells, respectively.
Cell culture and drug treatments
Human hepatocellular carcinoma (HepG2) was obtained from ATCC and was cultured according to the manufacturer’s protocols. Cells were plated in 6-well plates, seeded at 500,000 cells/well on day 1 then treated with 80 μM CuCl2 or DMSO the following day. The cells were then rescued with 30 μg/ml kalihinol F or 160 μM T-HCl 1 h later and incubated at 37°C for 24 h.
Quantitative RT-PCR
RNA from cell lysates was isolated using the RNeasy kit (Qiagen). cDNA was synthesized from 4 μg of total RNA using Superscript III (Invitrogen). When possible, intron-spanning primers were designed using the Universal ProbeLibrary Assay Design Center (Roche Applied Science). A complete list of primer sets is provided in Table S3.
PCR master mix was prepared with the LightCycler® 480 Probes Master kit and Universal ProbeLibrary probes according to the manufacturer’s protocols (Roche Applied Science). PCR was performed in quadruplicate using the LightCycler® 480 System (Roche Applied Science) with 45 cycles of amplification and annealing temperature of 60°C. Fold change in gene expression was measured by normalizing against 18S rRNA and comparing compound-treated samples with DMSO-treated control.
Supplementary Material
HIGHLIGHTS.
Screened compound libraries using zebrafish to identify modulators of embryogenesis
Reported novel bioactivity for kalihinol F as inducing a copper-deficient phenotype
Revealed MOA of kalihinol F by juxtaposing phenotype with zebrafish genetic mutants
Identified kalihinol family of compounds as a new class of copper chelators
Acknowledgments
We wish to thank the following: R. Looper and M. Sigman for providing the University of Utah Chemistry Department chemical library, D. Nix and S. Hammoud for helping in analyzing the RNA sequencing data, and HSC core facilities at the University of Utah. This work was supported by NIH T32 Postdoctoral Training Grant (R.G.C.D., D.R.W), NIH CA36622 (C.M.I), NCI 5PO1CA073992-14 (D.A.J.), NCI 5RO1CA116468-08 (D.A.J.) and the Huntsman Cancer Foundation (D.A.J.). Funding for the NMR spectrometer was provided by NSF grant DBI-0002806 and NIH grants RR06262 and RR14768.
The project described was supported by Award Number P30CA042014 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
The authors declare no competing financial interests.
ITS and DAJ conceived the study design. ITS, EJM, RMV, RGCD and KE performed the experiments. ITS, EJM, RMV, RGCD, DRW, CMI and DAJ analyzed the data. ITS, RMV, RGCD and DAJ wrote the paper. All the authors have read, revised, and approved the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvi KA, Tenenbaum L, Crews P. Anthelmintic polyfunctional nitrogen-containing terpenoids from marine sponges. J Nat Prod. 1991;54:71–78. doi: 10.1021/np50073a002. [DOI] [PubMed] [Google Scholar]
- Bloembergen N. Proton relaxation times in paramagnetic solutions. J Chem Phys. 1957;27:572–573. [Google Scholar]
- Bugni TS, Singh MP, Chen L, Arias DA, Harper MK, Greenstein M, Maiese WM, Concepción GP, Mangalindan GC, Ireland CM. Kalihinols from two Acanthella cavernosa sponges: Inhibitors of bacterial folate biosynthesis. Tetrahedron. 2004;60:6981–6988. [Google Scholar]
- Chang CWJ, Patra A, Roll DM, Scheuer PJ, Matsumoto GK, Clardy J. Kalihinol-A, a highly functionalized diisocyano diterpenoid antibiotic from a sponge. J Am Chem Soc. 1984;106:4644–4646. [Google Scholar]
- Ding X, Xie H, Kang YJ. The significance of copper chelators in clinical and experimental application. J Nutr Biochem. 2011;22:301–310. doi: 10.1016/j.jnutbio.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Fusetani N, Yasumuro K, Kawai H, Natori T, Brinen L, Clardy J. Kalihinene and isokalihinol B, cytotoxic diterpene isonitriles from the marine sponge Acanthella klethra. Tetrahedron Lett. 1990;31:3599–3602. [Google Scholar]
- Gansner JM, Mendelsohn BA, Hultman KA, Johnson SL, Gitlin JD. Essential role of lysyl oxidases in notochord development. Dev Biol. 2007;307:202–213. doi: 10.1016/j.ydbio.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota H, Tomono Y, Fusetani N. Terpenoids with antifouling activity against barnacle larvae from the marine sponge Acanthella cavernosa. Tetrahedron. 1996;52:2359–2368. [Google Scholar]
- Huster D. Wilson disease. Best Pract Res Cl Ga. 2010;24:531–539. doi: 10.1016/j.bpg.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Ingham PW. The power of the zebrafish for disease analysis. Hum Mol Genet. 2009;18:R107–R112. doi: 10.1093/hmg/ddp091. [DOI] [PubMed] [Google Scholar]
- Ishizaki H, Spitzer M, Wildenhain J, Anastasaki C, Zeng Z, Dolma S, Shaw M, Madsen E, Gitlin J, Marais R, et al. Combined zebrafish-yeast chemical-genetic screens reveal gene-copper-nutrition interactions that modulate melanocyte pigmentation. Dis Models Mech. 2010;3:639–651. doi: 10.1242/dmm.005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman CK, White RM, Zon L. Chemical genetic screening in the zebrafish embryo. Nat Protoc. 2009;4:1422–1432. doi: 10.1038/nprot.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dynam. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. Animal models of human disease: Zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- Liu M, Reiser O. A copper(I) isonitrile complex as a heterogeneous catalyst for azide alkyne cycloaddition in water. Org Lett. 2011;13:1102–1105. doi: 10.1021/ol103134c. [DOI] [PubMed] [Google Scholar]
- MacRae CA, Peterson RT. Zebrafish-based small molecule discovery. Chem Biol. 2003;10:901–908. doi: 10.1016/j.chembiol.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Mendelsohn BA, Yin C, Johnson SL, Wilm TP, Solnica-Krezel L, Gitlin JD. Atp7a determines a hierarchy of copper metabolism essential for notochord development. Cell Metab. 2006;4:155–162. doi: 10.1016/j.cmet.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Miyaoka H, Shimomura M, Kimura H, Yamada Y, Kim HS, Yusuke W. Antimalarial activity of kalihinol A and new relative diterpenoids from the Okinawan sponge, Acanthella sp. Tetrahedron. 1998;54:13467–13474. [Google Scholar]
- Muller P, Bakel H, Sluis B, Holstege F, Wijmenga C, Klomp LJ. Gene expression profiling of liver cells after copper overload in vivo and in vitro reveals new copper-regulated genes. J Biol Inorg Chem. 2007;12:495–507. doi: 10.1007/s00775-006-0201-y. [DOI] [PubMed] [Google Scholar]
- Nadauld LD, Sandoval IT, Chidester S, Yost HJ, Jones DA. Adenomatous polyposis coli control of retinoic acid biosynthesis is critical for zebrafish intestinal development and differentiation. J Biol Chem. 2004;279:51581–51589. doi: 10.1074/jbc.M408830200. [DOI] [PubMed] [Google Scholar]
- Ohta E, Ohta S, Hongo T, Hamaguchi Y, Andoh T, Shioda M, Ikegami S. Inhibition of chromosome separation in fertilized starfish eggs by kalihinol F, a topoisomerase I inhibitor obtained from a marine sponge. Biosci Biotech Bioch. 2003;67:2365–2372. doi: 10.1271/bbb.67.2365. [DOI] [PubMed] [Google Scholar]
- Omar S, Albert C, Fanni T, Crews P. Polyfunctional diterpene isonitriles from marine sponge Acanthella carvenosa. J Org Chem. 1988;53:5971–5972. [Google Scholar]
- Patra A, Chang CWJ, Scheuer PJ, Van Duyne GD, Matsumoto GK, Clardy J. An unprecedented triisocyano diterpenoid antibiotic from a sponge. J Am Chem Soc. 1984;106:7981–7983. [Google Scholar]
- Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. P Natl Acad Sci USA. 2000;97:12965–12969. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reedy BJ, Murthy NN, Karlin KD, Blackburn NJ. Isocyanides as ligand-directed indicators of cu(I) coordination in copper proteins. Probing the inequivalence of the cu(I) centers in reduced dopamine-beta-monooxygenase. J Am Chem Soc. 1995;117:9826–9831. [Google Scholar]
- Rhames F, Murthy N, Karlin K, Blackburn N. Isocyanide binding to the copper(I) centers of the catalytic core of peptidylglycine monooxygenase (PHMcc) J Biol Inorg Chem. 2001;6:567–577. doi: 10.1007/s007750100233. [DOI] [PubMed] [Google Scholar]
- Shin JT, Fishman MC. FROM ZEBRAFISH TO HUMAN: Modular Medical Models. Annu Rev Genomics Hum Genet. 2002;3:311–340. doi: 10.1146/annurev.genom.3.031402.131506. [DOI] [PubMed] [Google Scholar]
- Solomon I. Relaxation processes in a system of two spins. Phys Rev. 1955;99:559. [Google Scholar]
- Sun JZ, Chen KS, Yao L-g, Liu HL, Guo YW. A new kalihinol diterpene from the Hainan sponge Acanthella sp. Arch Pharmacal Res. 2009;32:1581–1584. doi: 10.1007/s12272-009-2110-4. [DOI] [PubMed] [Google Scholar]
- Taylor KL, Grant NJ, Temperley ND, Patton EE. Small molecule screening in zebrafish: An in vivo approach to identifying new chemical tools and drug leads. Cell Commun Signal. 2010:8. doi: 10.1186/1478-811X-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilton F, La Du JK, Vue M, Alzarban N, Tanguay RL. Dithiocarbamates have a common toxic effect on zebrafish body axis formation. Toxicol Appl Pharm. 2006;216:55–68. doi: 10.1016/j.taap.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Tisato F, Marzano C, Porchia M, Pellei M, Santini C. Copper in diseases and treatments, and copper-based anticancer strategies. Med Res Rev. 2009;30:708–749. doi: 10.1002/med.20174. [DOI] [PubMed] [Google Scholar]
- Tumer Z, Moller LB. Menkes disease. Eur J Hum Genet. 2010;18:511–518. doi: 10.1038/ejhg.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub ST, Wharton DC. The effects of copper depletion on cytochrome c oxidase. J Biol Chem. 1981;256:1669–1676. [PubMed] [Google Scholar]
- Westerfield M. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 3. Vol. 385. Eugene, OR: University of Oregon Press; 1995. The Zebrafish Book. [Google Scholar]
- Wolf D, Schmitz FJ. New diterpene isonitriles from the sponge Phakellia pulcherrima. J Nat Prod. 1998;61:1524–1527. doi: 10.1021/np980240g. [DOI] [PubMed] [Google Scholar]
- Zhang YF, Kitano Y, Nogata Y, Zhang Y, Qian PY. The mode of action of isocyanide in three aquatic organisms, Balanus amphitrite, Bugula neritina and Danio rerio. PLoS ONE. 2012;7:e45442. doi: 10.1371/journal.pone.0045442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.