Abstract
The quality of protein structures determined by nuclear magnetic resonance (NMR) spectroscopy is contingent on the number and quality of experimentally-derived resonance assignments, distance and angular restraints. Two key features of protein NMR data have posed challenges for the routine and automated structure determination of small to medium sized proteins; (1) spectral resolution – especially of crowded nuclear Overhauser effect spectroscopy (NOESY) spectra, and (2) the reliance on a continuous network of weak scalar couplings as part of most common assignment protocols. In order to facilitate NMR structure determination, we developed a semi-automated strategy that utilizes non-uniform sampling (NUS) and multidimensional decomposition (MDD) for optimal data collection and processing of selected, high resolution multidimensional NMR experiments, combined it with an ABACUS protocol for sequential and side chain resonance assignments, and streamlined this procedure to execute structure and refinement calculations in CYANA and CNS, respectively. Two graphical user interfaces (GUIs) were developed to facilitate efficient analysis and compilation of the data and to guide automated structure determination. This integrated method was implemented and refined on over 30 high quality structures of proteins ranging from 5.5 to 16.5 kDa in size.
Keywords: NMR data collection and processing, Chemical shift assignment, Protein structure determination and refinement, Structure validation
Introduction
Multidimensional heteronuclear nuclear magnetic resonance (NMR) spectroscopy is the methodology of choice for the experimental determination of three-dimensional protein structures in solution at atomic resolution, and is an invaluable tool for the biophysical and biochemical characterization of proteins and other biomolecules (Wüthrich 1986; Zuiderweg 2002). These data have been shown to be highly complementary to X-ray crystallography (Christendat et al. 2000; Snyder et al. 2005; Yee et al. 2002; Yee et al. 2005). However, the complexity and diversity of current NMR-based protocols limit its use as a routine strategy to study biological systems (Billeter et al. 2008). For example, the number and type of multidimensional NMR datasets recorded for complete resonance assignment of proteins (< 160 residues) can vary greatly, depending not only on the nature and size of the protein, but also on the data collection strategy and the level of expertise and instrumentation in the laboratory (Billeter et al. 2008; Montelione et al. 2000; Yee et al. 2002). Indeed, it has been argued that if higher levels of conformity and automation are not reached to improve accessibility and guarantee accuracy and quality of the structures obtained, NMR will risk being left behind in the ongoing evolution of Structural Biology (Billeter et al. 2008).
Currently, there are several procedural bottlenecks in obtaining high-resolution structures by NMR. First, the data collection protocol for resonance assignment of proteins centers around the 13C/15N-mediated sequential assignment strategy originally developed 20 years ago by Bax and colleagues (Grzesiek and Bax 1992a, b; Grzesiek and Bax 1993; Ikura et al. 1990a; b; c; Ikura et al. 1991a, b, c; Kay et al. 1990b). A number of methods for semi-automated assignment of backbone and 13Cβ/1Hβ resonances have been developed for this type of assignment strategy (Atreya et al. 2000; Helgstrand et al. 2000; Slupsky et al. 2003; Zimmerman et al. 1997). However, these protocols are highly dependent on uninterrupted sequential scalar connectivities along the protein backbone, especially the less sensitive “out and back”-type of experiments such as the HNCACB (Grzesiek and Bax 1992b), yielding assignments for only main chain atoms. Based on the outcome of these protocols, several important NMR-based computational approaches can provide structural information, such as structural models from CS-Rosetta (Shen et al. 2008; Shen et al. 2009b), dihedral angle restraints predicted by TALOS (Delaglio et al. 1995; Shen et al. 2009a), and residual dipolar couplings for either structure validation (Tjandra et al. 1997), refinement or de novo protein fold determination (Valafar et al. 2004).
In contrast to these computationally-derived models, experimentally-derived high-resolution protein solution structures requires the near-complete assignment of protein side chain resonances, which can often be manually intensive and prone to inaccuracies and subjectivities as the protein spectra become more crowded and complex. Data collection strategies to simplify the manner in which these resonance assignments are obtained have focused on reducing spectral complexity that correlate side chain resonances by reducing the number of correlations in an experiment (Grzesiek and Bax 1993; Ikura et al. 1990a, b, c, Ikura et al. 1991a, b, c; Kay et al. 1990b), or increasing the effective dimensionality of an experiment (Kay et al. 1990a). However, such datasets may lead to significant increases in data collection times with data quality often compromised by sample instability, thereby complicating analysis or limiting the types of samples than can be analyzed. This prompted the development of reduced dimensionality approaches such as G-matrix Fourier transform (GFT)-NMR (Kim and Szyperski 2003) and projection reconstruction methods (Freeman and Kupce 2003) to be used as an alternative to higher (>3)-dimensional FT NMR. However, these experiments are less sensitive than conventional FT three-dimensional experiments, and therefore are limited to proteins with greater solubility.
The development of automated algorithms for NOE-derived protein structure calculations has been an important advance in the field (Güntert 2004; Linge et al. 2003; Rieping et al. 2007; Zheng et al. 2003) as well as more comprehensive approaches to data collection and structure determination, including KUJIBA, PINE-SPARKY, PINE-NMR (Bahrami et al. 2009; Kobayahi et al. 2007; Lee et al. 2009; Wong et al. 2008). These programs all make use of a user-defined resonance assignment list (as discussed above), which is matched with a peak list derived from heteronuclear edited NOESY spectra to assign distance restraints to pairs of protons. Any inaccuracies or incompleteness in the assignment list, and/or mismatches between the scalar derived peak assignments and NOE-derived peak lists (e.g. resulting from minor chemical shift changes due to differences in sample pH or temperature) can lead to inaccuracies in the final protein structure. Thus, early identification of such errors, or ideally, the development of procedures that minimize the occurrence of such errors, is of critical importance to ensure the convergence and accuracy of the final ensemble of structures. For the reasons described above, we sought to develop a comprehensive protocol that avoids some of the vulnerabilities of the “sequential assignment followed by NOE-derived structure” paradigm.
To this end, we describe a method for full 13C/15N/1H protein NMR assignments that integrate four key features to overcome many of the limitations often encountered using these traditional approaches. First, we make use of a minimal self-consistent dataset which fulfils all requirements for both complete resonance assignments and NOE structure determination, enabling complete and consistent data recordings even on delicate samples. Second, non-uniform sampling (NUS) with processing by multidimensional decomposition (MDD) is used to obtain high resolution multidimensional heteronuclear data without lengthening data acquisition times (Barna and Laue 1987; Orekhov et al. 2003). This data collection strategy is particularly useful for peak picking and interpretation of crowded spectra such as 13C-edited TOCSY data for side chain assignments, and 13C- and 15N-edited NOESY spectra for accurate NOE assignments. Third, scalar coupled spin systems are assigned to the protein sequence using HNCA and NOE information in the ABACUS procedure (Lemak et al. 2008), yielding highly accurate backbone and side chain resonance assignments, even for cases in which backbone resonances are missing or for which only partial spin systems are available. Finally, two graphical user interfaces (GUI) have been developed: MDDGUI to facilitate data processing with MDD and a Fragment Monte Carlo (FMC)-GUI to assist in the management of peak lists, execution of the ABACUS protocol, probabilistic evaluation of ABACUS resonance assignments for subsequent structure calculation using programs such as CYANA (Güntert 2004), and refinement with strategies such as restrained molecular dynamics in explicit water (Brünger et al. 1998) or CS-Rosetta (Shen et al. 2008). Importantly, because the resonance assignments and NOE data are part of the same, self-consistent dataset (derived from ABACUS), fewer mismatches exist between NOE distance assignments and the reference chemical shift list, thus making the final convergence of structure calculations more robust and efficient in the derivation of high quality structures for proteins up to at least 150 residues.
The ABACUS dataset
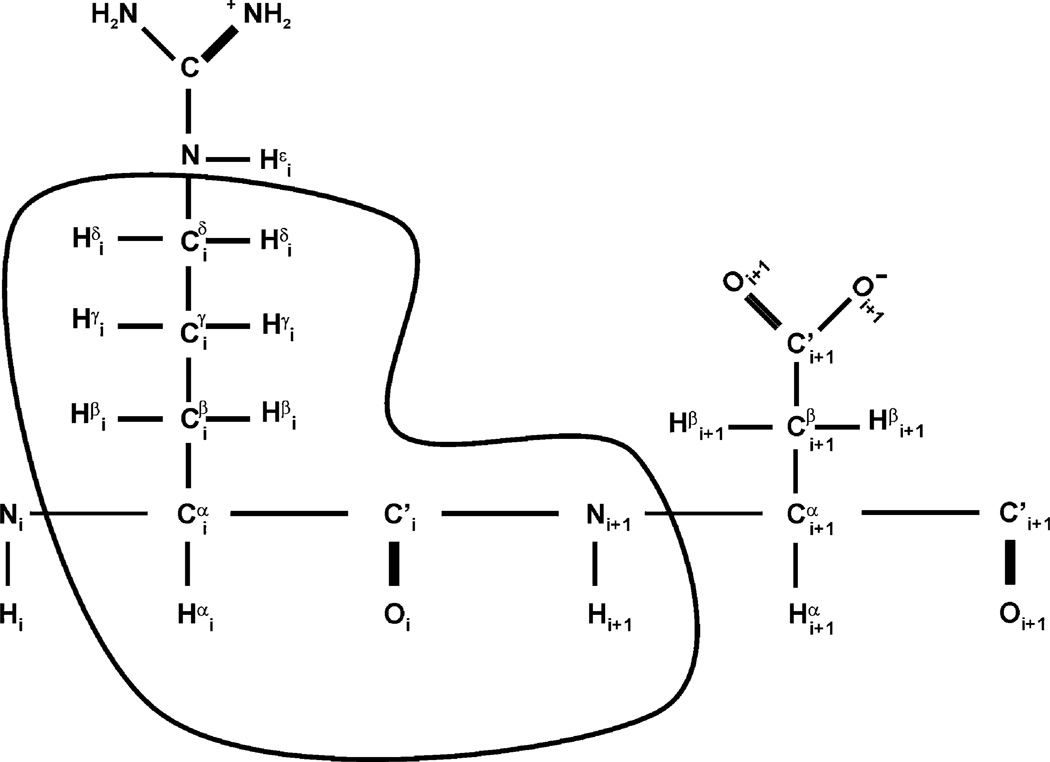
In order to streamline the data collection and analysis process, we sought to define a minimal set of NMR experiments that consistently delivered a sufficient number of correlations both for complete backbone and side chain protein resonance assignments as well as for NOE-based structure determination on a variety of protein samples within a timeframe that does not threaten sample integrity. To this end, we have selected a small set of scalar-coupled NMR experiments that have relatively high sensitivity as compared to the many possible heteronuclear triple resonance experiments, combined with heteronuclear edited NOESY spectra, to make up the ABACUS dataset (Table 1; Bax et al. 1990; Grzesiek and Bax 1992c; Grzesiek and Bax 1993; Kay et al. 1990b; Marion et al. 1989a, b; Muhandiram and Kay 1994; Vuister and Bax 1992). Owing to fewer magnetization transfers and relaxation losses, the CBCA (CO)NH and HNCA, for example, are more robust and of considerable benefit to the ABACUS-based approach to automated assignment as opposed to the less sensitive HNCACB experiment (Grzesiek and Bax 1992b). The same reasoning explains why the spectral features are superior in the HBHA(CBCACO)NH as opposed to the HC(CO)NH experiment (Logan et al. 1992). Importantly, these triple resonance experiments yield a network of scalar couplings that defines a spin system for each amino acid side chain, i: aliphatic H(i), C(i), C’(i), N(i + 1) and HN(i + 1). We refer to this spin system as a Peptide Bond (PB) spin system or a PB fragment (Fig. 1), the basic unit of assignment in the subsequent ABACUS assignment protocol whereupon PB spin systems are linked using intra-residue NOE and HNCA correlations. The protocol can readily accommodate additional NMR experiments not included in the minimal dataset if more correlations are required.
Table 1.
The ABACUS dataset
| Experiment | TD(F1), SW1 (Hz) | TD(F2), SW2 (Hz) | Number of scans | Acquisition times (h) |
|---|---|---|---|---|
| 1H-15N HSQC* | 256, 2200 | 8 | 0.63 | |
| CT-HNCA* | 68, 4800 | 92, 2200 | 16 | 9.9 |
| CBCA(CO)NH* | 132, 10000 | 92, 2200 | 32 | 12 |
| HNCO* | 96, 1800 | 92, 2200 | 8 | 7.2 |
| HBHA(CBCACO)NH* | 200, 4800 | 92, 2200 | 16 | 30 |
| 15N-NOESY-HSQC | 600, 12000 | 80, 2200 | 8 | 43 |
| H(C)CH-TOCSY (aliphatic) | 204, 6400 | 204, 14500 | 4 | 22 |
| (H)CCH-TOCSY (aliphatic) | 204, 14500 | 204, 14500 | 4 | 22 |
| 13C-NOESY-HSQC | 600, 12000 | 204, 14500 | 4 | 50 |
| CT-1H-13C-HSQC | 640, 14500 | 16 | 3 | |
| H(C)CH-TOCSY (aromatic) | 112, 3200 | 72, 5200 | 8 | 24 |
| (H)CCH-TOCSY (aromatic) | 72, 5200 | 72, 5200 | 8 | 17 |
| 13C-NOESY-HSQC (aromatic) | 600, 12000 | 72, 5200 | 8 | 38 |
Specific scalar-coupled experiments were chosen based on a balance between those with optimal information content and those that do not suffer from low sensitivity due to excessive relaxation losses and/or correlate weaker scalar couplings. Acquisition times are based on sparse data (30%) and a protein concentration of 0.75 mM−1 mM. All TOCSY- and NOESY-based experiments were collected at 298 K at 800 MHz while those labeled with asterisks were collected at 600 MHz. CT refers to constant-time. TD and SW refer to the time domain and sweep width, respectively. The average total acquisition time for the minimal dataset is approximately 13 days, which includes spectra sufficient for both sequential and side chain assignment as well as complete structure determination using ABACUS. Approximately four days of NMR data acquisition is required to collect the N-rooted experiments for semi-automated assignment of main chain resonances using FAWN
Fig. 1.
Definition of a PB spin system. The PB (peptide bond) spin system (circled) is the basic structural unit in the ABACUS protocol
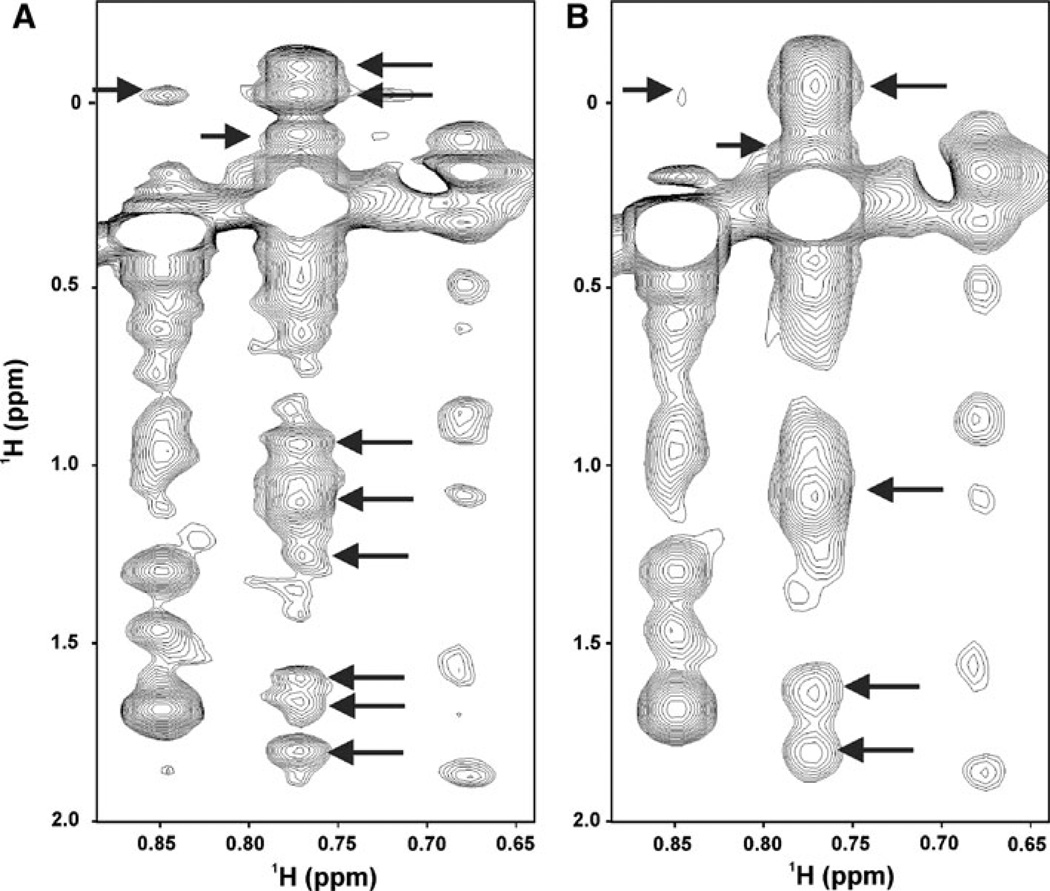
In this context, non-uniform sampling (NUS) was used for two different purposes. For less crowded triple resonance experiments for which there was sufficient sensitivity, NUS was used to reduce the number of data points collected in the indirect dimensions, as in the HNCA and HNCO, thereby decreasing data collection times without compromising spectral resolution. For more crowded spectra such as 3D-HCCH-TOCSY and 15N- and 13C-edited NOESY experiments, NUS was used to increase the digital resolution in the indirect dimensions without an increase in data collection time or loss of sensitivity (relative to conventional uniformly sampled/Fourier transformed spectra; Fig. 2; (Gutmanas et al. 2002; Luan et al. 2005; Orekhov et al. 2003)). A practical guide for setting up an existing conventional multidimensional pulse sequence in NUS mode (on Varian and Bruker spectrometers) is provided on the NMRwiki website of the Northeast Structural Genomics (NESG) consortium at: http://www.nmr2.buffalo.edu/nesg.wiki.
Fig. 2.
NUS and MDD increase the resolution of multidimensional NMR data. 13C-edited NOESY spectrum collected for a 121 residue protein, Atu0922, from Agrobacterium tumefaciens with a 100 ms mixing time at 800 MHz. (a) 300 complex points in 1H indirect dimension without spectral folding whereas and processed with MDD. (b) The spectrum in A was reprocessed with half the number of indirect complex points, employing parameters commonly used in the collection of fully sampled/Fourier Transformed data. Arrows in (a) indicate peaks that are not clearly resolved in a conventional NOESY spectrum
Following NUS data collection, reconstruction of a multidimensional spectrum with conventional appearance is achieved with MDDGUI (accessible through the NESG NMRwiki), a graphical user interface and processing tool that guides the user through the processing stages of NUS data using NMRPipe and MDDNMR protocols (Delaglio et al. 1995; Gutmanas et al. 2002; Orekhov et al. 2003), phasing and apodization of the first plane, FT of the acquisition dimension, and reconstruction and processing of the indirect dimensions of the dataset. The integration with NMRPipe allows for straightforward implementation of user-specific processing protocols. In this application, the use of NMRPipe simplifies the handling of the reconstructed dataset, which can be readily converted to most spectral visualization programs (e.g. SPARKY (Goddard)) for viewing and subsequent peak picking, and identification of spin systems for subsequent analysis in FMCGUI.
Assignment strategy
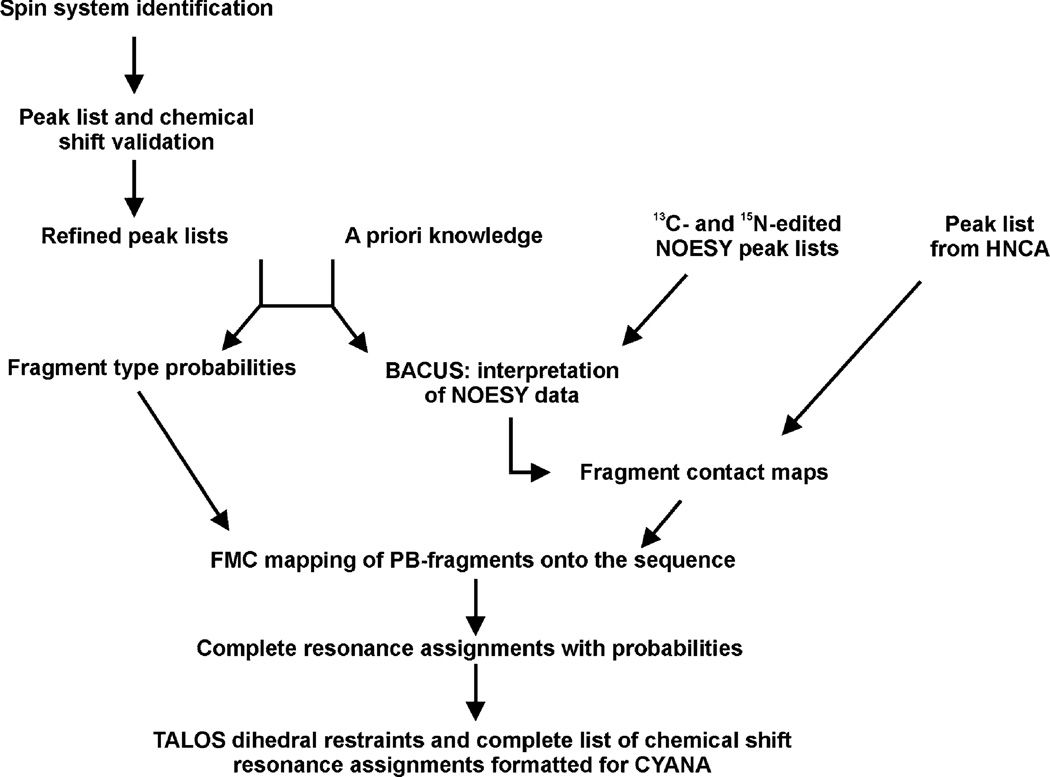
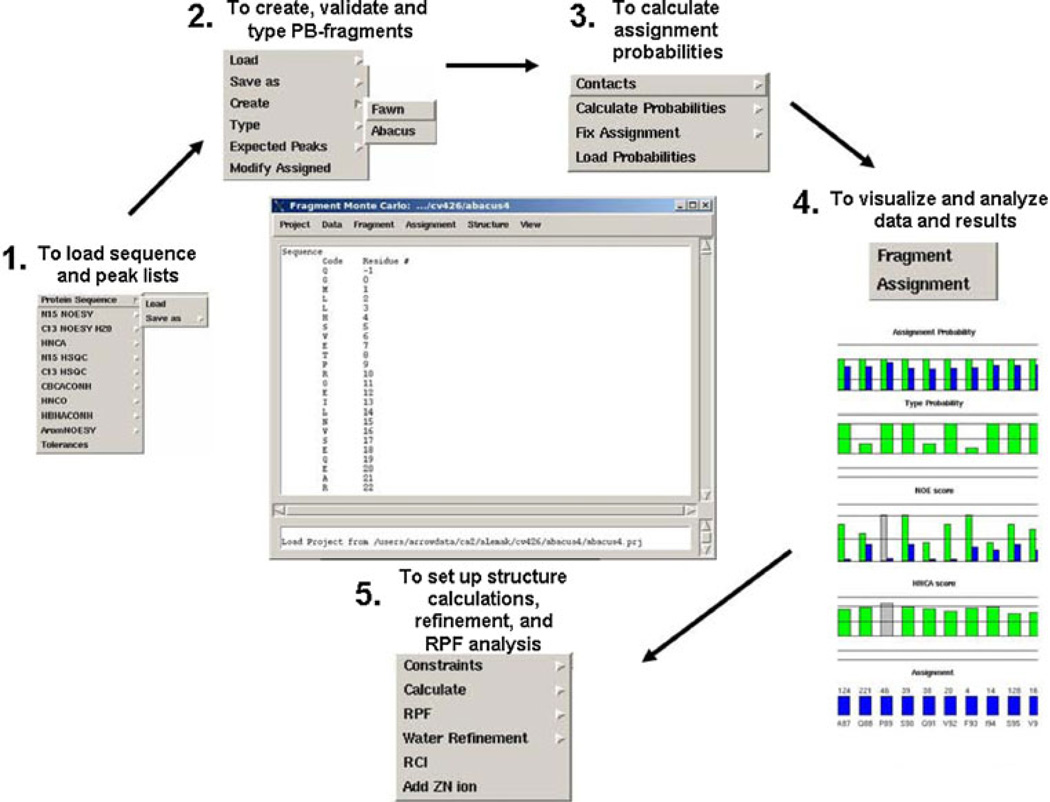
An overview of the assignment strategy used by the ABACUS/FMCGUI approach is presented in Fig. 3. The spin system identification step consists of a few iterations of manual peak picking of the NMR spectra and analysis of the peak lists and chemical shifts with FMCGUI. The peak picking strategy is illustrated with spectral “strips” in Fig. 4. Peak picking of the N-rooted spectra such as the 15N-1H HSQC, HNCA, HNCO, CBCA(CO)NH, and the HBHA(CBCACO)NH is straightforward, in particular since ABACUS/FMCGUI requires peak labeling only in the 15N-1H HSQC (Fig. 4a). The side chain analysis is extended beyond the Cβ/Hβ using the complementary three-dimensional (H)CCH- and H(C)CH-TOCSY spectra (Fig. 4b). Using software such as SPARKY (Goddard), the user manually maps the backbone and side chain scalar correlations to specific peaks in the 2D 13C-1H HSQC, thereby defining all of the PB spin systems in the protein. Of note, the user is not required to label any side chain γ-, δ-, or ε- carbons or hydrogens; rather, ABACUS automatically assigns all of these 13C and 1H frequencies to corresponding side chain positions. At any stage of spin system compilation, FMCGUI can generate corresponding expected peak lists for visual inspection of the spin systems as guides for subsequent peak picking of additional NOE cross peaks in the 13C- and 15N-edited NOESY spectra. This semi-automated spin system identification step is greatly facilitated by an iterative analysis of the evolving spin systems and their chemical shifts by FMCGUI. The analysis and validation of the spin systems utilizes a goodness of fit model that compares the resonances identified in each spin-system to those reported in the Biological Magnetic Resonance Bank (BMRB) database for each amino acid type (Ulrich et al. 2008). FMCGUI allows the user to address any discrepancies or ambiguities encountered during peak picking to improve the outcomes of the ABACUS procedure.
Fig. 3.
Overview of ABACUS and structure calculation workflow. Peaks are manually picked in the HNCO, HNCA, CBCA(CO)NH, HBHA (CBCACO)NH, H(C)CH- and (H)CCH-TOCSY experiments, along with those obtained from the HNCA and 13C- and 15N-edited NOESY. These peak lists are prerequisites for the ABACUS protocol. A reference BMRB chemical shift list identifies potential mismatches or deviations between experimental spin systems and those of standard amino acids. The amino acid sequence and the peak lists from the scalar-coupled experiments are used to match individual spin systems with corresponding amino acid types. NOESY and HNCA data are used to establish connectivities (e.g. contact maps). Fragment type probabilities and contact maps are utilized in FMC simulations to map corresponding spin systems to specific positions in the amino acid sequence. The result is a complete chemical shift list that is used to generate angular and distance constraints for subsequent use in CYANA
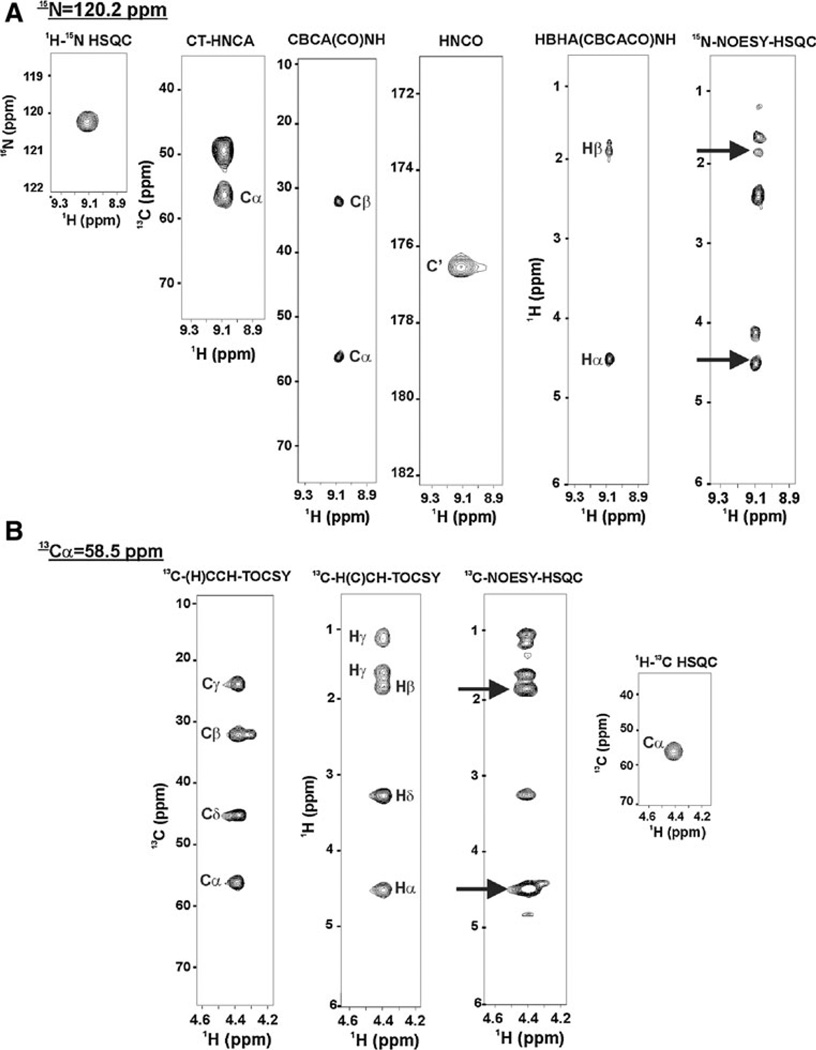
Fig. 4.
Summary of the algorithm used by FMCGUI and ABACUS to define spin systems and sequential connectivities based on peak lists from the minimal dataset. The procedure begins by identifying spins for the PB fragment highlighted in Fig. 1: in (a) The 15N-1H HSQC is used as a reference spectrum to define the 1H-15N correlation for the aspartate residue, the CBCACONH and HNCA identify Cα and Cβ for the arginine, the HNCO is used to define C’ for later use in TALOS and to identify overlapping spin systems. The HBHA(CBCACO)NH confirms the Hα and Hβ in the 15N-edited NOESY (arrows identify through-space NOE correlations of protons). In (b) Complementary (H)CCH-TOCSY and H(C)CH-TOCSY experiments allow for facile assignment of side chain resonances beyond β carbon that can be easily mapped to the corresponding strip in the 13C-edited NOESY and correlation peak in the constant time 1H-13C HSQC. Peak lists are generated for these experiments and are loaded into FMCGUI for implementation of the FAWN and/or ABACUS protocols
The input data required for this approach consists of the amino acid sequence, peak lists from the N-rooted experiments, 2D 13C-1H HSQC, and the 13C- and 15N-edited NOESYs. In principle, if there were no problems encountered during the spin system identification step (e.g. due to overlap or missing peaks), the peak lists should be sufficient for complete automated assignment using the ABACUS assignment protocol in FMCGUI. However since this is rarely the case, FMCGUI facilitates the implementation of the ABACUS assignment protocol in an interactive, semi-automated manner. The peak lists loaded into FMCGUI (Fig. 5, panel 1) are first validated and screened for potential chemical shift and formatting errors so as to generate a refined peak list for subsequent use in the assignment procedure. These data are used to create individual PB fragments with the corresponding amino acid type probabilities. The second step of the procedure evaluates and scores a possible link between individual PB fragments through the creation of PB contact maps using the HNCA and 13C- and 15N-edited NOESY experiments (Fig. 5, panel 2). PB fragment types and contact maps are used in fragment Monte Carlo simulations to map individual PB spin systems onto the amino acid sequence (Fig. 5, panel 3). The result from the simulation is reported in terms of assignment probabilities, which are analyzed using visualization tools in FMCGUI (Fig. 5, panel 4). The quality of each assignment is measured in terms of assignment probabilities and a goodness of fit test of the assignment to the NOE and HNCA data. Those assignments with low probabilities alert the user to which spectral strips should be inspected manually for potential errors in peak picking. Corrections can then be made to the peak lists resulting in improved assignment probabilities. Importantly, this approach rapidly identifies the intra-residue and sequential NOEs, which ensures a good match between the scalar coupled peak assignments (e.g. the chemical shift list) and the corresponding NOEs. Furthermore, the agreement between these two data sets leads to better and more rapid convergence of protein structures calculations.
Fig. 5.
FMCGUI overview. The process begins by loading in the amino acid sequence and respective peak lists from corresponding experiments defined in the minimal dataset (Panel 1). Tolerances are set to improve the reliability of the automated assignment process. The Fragment menu item helps to organize the information for each PB spin system into individual fragments (Panel 2). Once all of the fragments (spin systems) are assembled, FMCGUI has a built-in chemical shift databank that quickly searches for errors in the peak picking process. In addition, the user-friendly interface identifies potential errors in the peak lists which may produce errors in the assignment list. Assignment probabilities are determined using a Fragment Monte Carlo routine with ABACUS and/or FAWN approaches (Panel 3). Confidence in the final chemical shift assignment list is highly dependent on the NOE and HNCA scores (Panel 4). These data are easily manipulated and can be readily exported into CYANA and TALOS formats (Panel 5). Structural ensembles are read back into FMCGUI to assist in the calculation of recall and precision scores and structure refinement in CNS
Our original report of the ABACUS approach (Lemak et al. 2008) threaded individual PB spin systems by making exclusive use of information-rich NOE patterns to determine sequential connectivities. We have since optimized this protocol by including inter- and intra-residue correlations from the HNCA spectrum to improve the probabilistic assignment of each spin system within the amino acid sequence. However, the success of the ABACUS assignment does not rely on uninterrupted HNCA connectivities along the backbone. Furthermore, incorporating the HNCA data contributed to the development of FAWN (Fragment Assignment with NOEs). FAWN is an additional application housed in FMCGUI that uses the same search algorithms as ABACUS, with the exception that it does not make use of the complementary three-dimensional H(C)CH- and (H)CCH-TOC-SY and 13C-edited NOESY experiments. The lack of side chain spin information in FAWN necessitates more manual intervention in the assignment process as compared to that of ABACUS. Nevertheless, this application has been successfully used to rapidly obtain backbone assignments for NMR studies in which complete resonance assignment of the protein is not required such as in NMR-based titration experiments, 15N-based backbone relaxation experiments and CS-Rosetta (Shen et al. 2008; Shen et al. 2009b).
Refinement strategies in FMCGUI
The nature of the graphical interface simplifies the manner in which assignment data and probabilities are visualized and analyzed (Fig. 5, panel 4). This allows the user to quickly focus attention on problematic (e.g. low probability) assignments for manual inspection of spectra when necessary. In addition to the inclusion of FAWN and ABACUS in FMCGUI, other features include: (a) efficient detection of chemical shift assignment errors for each PB spin system, (b) straightforward export of chemical shift and peak lists into TALOS, CYANA and AutoStructure (Huang et al. 2006) formats (Fig. 5, panel 5), (c) facile guided setup of CYANA structure calculation protocols with or without hydrogen bond restraints, and d) incorporation of Zn2+-coordination constraints and residual dipolar coupling (RDC) data for refinement in CNS (Brünger et al. 1998) with explicit water (Fig. 5, panel 5).
Additionally, FMCGUI calculates structure quality coefficients such as recall and precision scores for given structural ensembles (Huang et al. 2005). These values reflect how well the structural ensemble agrees with the NOESY data: the recall score is the percentage of peaks that are consistent with the structure, whereas the precision score is the percentage of expected peaks from the structure that are already included in the NOESY peak list. Based on this analysis, FMCGUI generates expected and nonexpected NOESY peak lists to be used as a tool to improve the structure by weeding out artifacts and in picking NOE peaks that may have been missed or otherwise left out. An ensemble of CYANA structures is chosen for refinement once their recall and precision scores are greater than 0.9. Upon completion of the calculation, FMCGUI creates a separate directory that includes a single text file detailing all of the distance and dihedral violations, along with the refined structures and experimental inputs (e.g. NOESY peak lists, dihedral angle and hydrogen bond restraints). The files are formatted to enable immediate submission to the Protein Structure Validation Suite (PSVS) (Bhattacharya et al. 2007) and deposition into the protein databank database (PDB) and biological magnetic resonance bank (BMRB).
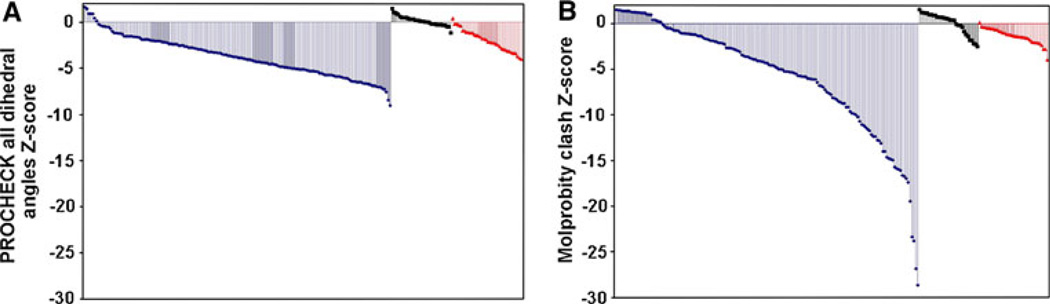
The NUS/MDD/ABACUS/FMCGUI approach has been applied to over 36 proteins with a wide range of secondary structures, and degrees of difficulty (Table 2). In order to ensure that the overall method described above yields high quality structures, we analyzed these structures using the PSVS structure quality package and compared the results to those of similarly sized solution structures and high resolution crystal structures deposited in the PDB during the same time period. All our structures have good backbone conformations as reported by the PROCHECK backbone dihedral angle Z scores, which are better than −3.86. However, we focused our analysis on the side chain geometry (all dihedral angles Z score; Fig. 6a) and packing (Molprobity clash score; Fig. 6b), which are more sensitive to small errors in chemical shift and NOE assignments. In this regard, we find that the quality factors of our ABACUS-derived structures are comparable to those in the “better half” of the solution structures from non-Structural Genomics groups, and are approaching those of high resolution crystal structures.
Table 2.
Representative list of protein structures determined by the NUS/MDD/ABACUS/FMCGUI approach
| Protein name | Taxonomy | PDB code | Nres | %α | %β | Qreson (%) |
|---|---|---|---|---|---|---|
| ATC1776 | A. tumefaciens | 2JYA | 101 | 16.3 | 31.3 | 93.5 |
| ATC2521 | A. tumefaciens | 2JQ4 | 83 | 84.3 | 0 | 94.4 |
| ATC0727 | A. tumefaciens | 2K54 | 123 | 35.8 | 46.3 | 96 |
| ATC1183 | A. tumefaciens | 2K2P | 64 | 45.3 | 23.4 | 89.2 |
| DnaK suppressor protein | A. tumefaciens | 2KQ9 | 112 | 51.8 | 0 | 96 |
| ATC0223 | A. tumefaciens | 2K7I* | 124 | 29.0 | 27.4 | 87.4 |
| ATC0905 | A. tumefaciens | 2KNR | 118 | 32.3 | 27.1 | 97.2 |
| ATC0852 | A. tumefaciens | 2KJZ* | 122 | 23.8 | 33.6 | 89.2 |
| AF2351 | A. fulgidus | 2KKU | 139 | 22.3 | 29.5 | 87.7 |
| CV0237 | C. violaceum | 2KP6 | 79 | 64.6 | 0 | 90.9 |
| CV0863 | C. violaceum | 2×8N | 109 | 27.5 | 22.9 | 98.4 |
| Zn finger protein YBIL | E. coli | 2KGO | 88 | 0 | 45.5 | 97.8 |
| C-terminal oxidized NleG2-3 | E. coli | 2KKY | 102 | 33.3 | 15.7 | 96.3 |
| EC0640 | E. coli | 2K8E | 116 | 26.7 | 31.0 | 92.7 |
| C-terminal reduced NleG2-3 | E. coli | 2KKX | 102 | 28.4 | 24.5 | 96.3 |
| HP0488 | H. pylori | 2JOQ | 86 | 34.1 | 24.2 | 96.2 |
| Cbx4 | H. sapiens | 2K28 | 58 | 36.7 | 23.3 | 94.8 |
| Cbx7 | H. sapiens | 2K1B | 55 | 27.3 | 32.7 | 94.4 |
| Cul7-CPH | H. sapiens | 2JNG | 102 | 18.8 | 34.4 | 94.8 |
| N-terminal Pirh2 | H. sapiens | 2K2C | 137 | 6.6 | 24.1 | 92.4 |
| Cytochrome-b5-like domain HERC2 E3 ligase | H. sapiens | 2KEO | 92 | 57.6 | 16.3 | 93.5 |
| Ub-like domain of NFATC2IP | H. sapiens | 2JXX | 78 | 24.4 | 34.6 | 93.6 |
| Ub-like domain of ubiquilin 1 | H. sapiens | 2KLC | 79 | 27.9 | 32.9 | 90.8 |
| Ub-like domain of Herpud2 | H. sapiens | 2KDB | 77 | 24.7 | 29.9 | 95.6 |
| Ub-protein ligase E3A | H. sapiens | 2KR1 | 64 | 51.6 | 0 | 89.3 |
| Usp7 Ub-like domain | H. sapiens | 2KVR | 128 | 37.5 | 14.8 | 93.9 |
| FK506-binding protein | H. sapiens | 2KFV | 73 | 68.5 | 0 | 95.7 |
| PA1076 | P. aeruginosa | 2K4V | 125 | 34.4 | 32.8 | 92.1 |
| RP4601 | R. palustris | 2JN4 | 66 | 0 | 42.4 | 94.5 |
| RPA3114 | R. palustris | 2JQ5 | 128 | 39.8 | 36.7 | 94.7 |
| RPA1320 | R. palustris | 2IDA | 102 | 23.5 | 13.7 | 95.6 |
| RP3384 | R. palustris | 2JTV | 65 | 58.5 | 23.1 | 90.4 |
| YST0336 | S. cerevisiae | 2JYN | 146 | 56.9 | 5.5 | 92.1 |
| SF3929 | S. flexneri | 2KO6 | 89 | 61.8 | 0 | 93.9 |
| SSO0164 | S. solfataricus | 2KCO | 133 | 12.8 | 24.8 | 86 |
| 30S ribosomal protein S27A | T. acidophilum | 2K4X | 55 | 0 | 12.7 | 90 |
Secondary structure composition of each protein was calculated using PYMOL. Ub refers to ubiquitin; N refers to the total number of residues in the protein; Qassn is the percent completeness of assigned resonances (backbone and side chain 13C, 15N, 1H resonances). Structures that were calculated as dimers are represented by an asterisk
Fig. 6.
Approach yields high quality protein structures. As one measure of structure quality, we compare the PROCHECK all dihedral angle (a) and Molprobity clash Z-scores (b) for NUS/MDD/ ABACUS derived structures (red; PDB accession codes 2JTV, 2KP6, 2KEO, 2KFV, 2KQ9, 2K4X, 2JYN, 2×8N, 2K8E, 2K2P, 2K28, 2JOQ, 2KKX, 2KR1, 2KO6, 2KKY, 2JXX, 2JQ4, 2JYA, 2K4V, 2KDB, 2JQ5, 2K54, 2K1B, 2KLC, 2K7I, 2KNR, 2KCO, 2K2C, 2KGO, 2IDA, 2KVR, 2JUF, 2KJZ, 2JN4, 2KKU), high-resolution X-ray crystal structures of similar sized proteins deposited in April 2009 with a resolution of < 2Å (black), and other similarly sized proteins determined by conventional NMR methods from non-Structural Genomics groups (blue), deposited in the PDB from January 1st, 2008–June 30th, 2009
Discussion
Here, we report a semi-automated method for the rapid assignment and high quality structure determination of small to medium sized proteins in solution by NMR. Our method has several attractive aspects. First, NUS/MDD facilitates spectral analysis by improving the resolution of TOCSY and NOESY data (Gutmanas et al. 2002; Orekhov et al. 2003), thus expediting the assignment of resonances beyond the β carbon. This, in turn, generates a more thorough and complete chemical shift reference list (Table 2), and aids in the assignment of NOE distance restraints involving side chain protons for structure calculation. This, in conjunction with the analysis performed by AutoStructure (Huang et al. 2006), significantly improves the precision in describing the structure of the folded core. Among other features, the graphical interface houses the platforms for semi-automated assignment of backbone (FAWN and ABACUS) and side chain resonances (ABACUS). The inclusion of a reference chemical shift databank (Ulrich et al. 2008) within the software allows one to cross-validate tabulated PB fragment chemical shifts and ensure their accuracy before the calculation of assignment probabilities. This provides an efficient means for the identification of manual errors in entering raw data and/or incompleteness in the peak lists. Moreover, the use of NOEs at the front-end of the procedure saves time by minimizing discrepancies between the resonance assignments and NOE peak lists, with the added bonus that all spectral information required for a full structure determination is already collected as part of the ABACUS dataset. Finally, this semi-automated platform is a self-contained system that allows for facile generation of resonance and NOE assignment lists for rapid structure determination in CYANA, further refinement using other programs and additional data, evaluation and validation of the structure ensemble, and deposition of data into public databases.
Our approach facilitates the entire process of determining NMR structures and is flexible enough for application to a wide range of proteins with varying amounts and type of regular secondary structure and levels of experimental difficulty. For example, proteins exhibiting conformational heterogeneity arising from exchanges between oxidized and reduced states (Wu et al. 2010) as well as symmetric homodimers (PDB accession codes 2K7I and 2KJZ) have been determined using this method. Similarly, the higher resolution afforded by NUS and MDD, together with the ABACUS assignment tools, has enabled rapid and complete resonance assignment of natively disordered regions within our target proteins (PDB accession codes 2GPF, 2KCO).
While the method was developed and validated on proteins less than 160 residues, we believe this strategy is highly adaptable to fractionally deuterated, or other specifically labeled and deuterated proteins of higher molecular weight, as well as for larger multi-domain proteins in which the effective relaxation times are equal to those of the individual sub-domains. Furthermore, we envision that incorporation of a CS-Rosetta module into the method may allow even more rapid convergence of very high quality structures with excellent side chain packing that is often lacking in many de novo experimental NMR structures. Experiments along these lines are underway. FMCGUI is written in python and will be made available upon request as open access software that can be modified and customized by individual users.
Acknowledgments
The authors would like to thank Doung-uen (Kevin) Lee for help in the initial design of FMCGUI, members of the Arrowsmith Lab for their input, and Dr. Binchen Mao for assistance in running PSVS analysis. This work was supported by the US National Institute of Health Protein Structure Initiative (P50-GM62413-01 and GM67965) through the Northeast Structural Genomics Consortium; the Natural Sciences and Engineering Research Council of Canada; the Canadian Institutes of Health Research (CIHR), and the Ontario Ministry of Health and Long Term Care (OMOHLTC). The views expressed do not necessarily reflect those of the OMOHLTC. SC is the recipient of a CIHR post-doctoral fellowship and CHA holds a Canada Research Chair in Structural Proteomics. MS holds a Vinnmer fellowship from VINNOVA (The Swedish Governmental Agency for Innovation Systems).
Contributor Information
Alexander Lemak, Ontario Cancer Institute and The Campbell Family Cancer Research Institute, Department of Medical Biophysics, University of Toronto, 101 College Street, Toronto, ON M5G 1L7, Canada; The Northeast Structural Genomics Consortium, University of Toronto, 101 College Street, Toronto, ON M5G 1L7, Canada.
Aleksandras Gutmanas, Ontario Cancer Institute and The Campbell Family Cancer Research Institute, Department of Medical Biophysics, University of Toronto, 101 College Street, Toronto, ON M5G 1L7, Canada; The Northeast Structural Genomics Consortium, University of Toronto, 101 College Street, Toronto, ON M5G 1L7, Canada.
Seth Chitayat, Ontario Cancer Institute and The Campbell Family Cancer Research Institute, Department of Medical Biophysics, University of Toronto, 101 College Street, Toronto, ON M5G 1L7, Canada.
Murthy Karra, Ontario Cancer Institute and The Campbell Family Cancer Research Institute, Department of Medical Biophysics, University of Toronto, 101 College Street, Toronto, ON M5G 1L7, Canada.
Christophe Farès, Ontario Cancer Institute and The Campbell Family Cancer Research Institute, Department of Medical Biophysics, University of Toronto, 101 College Street, Toronto, ON M5G 1L7, Canada; The Northeast Structural Genomics Consortium, University of Toronto, 101 College Street, Toronto, ON M5G 1L7, Canada.
Maria Sunnerhagen, Division of Molecular Biotechnology, Department of Physics, Chemistry and Biology, Linköping University, 58183 Linköping, Sweden.
Cheryl H. Arrowsmith, Ontario Cancer Institute and The Campbell Family Cancer Research Institute, Department of Medical Biophysics, University of Toronto, 101 College Street, Toronto, ON M5G 1L7, Canada The Northeast Structural Genomics Consortium, University of Toronto, 101 College Street, Toronto, ON M5G 1L7, Canada, carrow@uhnres.utoronto.ca.
References
- Atreya H, Sahu SC, Chary KV, Govil G. A tracked approach for automated NMR assignments in proteins (TATAPRO) J Biomol NMR. 2000;17:125–136. doi: 10.1023/a:1008315111278. [DOI] [PubMed] [Google Scholar]
- Bahrami A, Assadi AH, Markley JL, Eghbalnia HR. Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput Biol. 2009;5:1–12. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna J, Laue ED. Conventional and exponential sampling for 2D NMR experiments with application to a 2D NMR spectrum of a protein. J Magn Reson. 1987;75:387–389. [Google Scholar]
- Bax A, Clore GM, Gronenborn AM. 1H-1H correlation via isotropic mixing of 13C magnetization: a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J Magn Reson B. 1990;88:425–431. [Google Scholar]
- Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures deterined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- Billeter M, Wagner G, Wu¨thrich K. Solution NMR structure determination of proteins revisited. J Biomol NMR. 2008;42:155–158. doi: 10.1007/s10858-008-9277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Christendat D, Yee A, Dharamsi A, Kluger Y, Savchenko A, Cort JR, Booth V, Mackereth CD, Saridakis V, Ekiel I, Kozlov G, Maxwell KL, Wu N, McIntosh LP, Gehring K, Kennedy MA, Davidson AR, Pai EF, Gerstein M, Edwards AM, Arrowsmith CH. Structural proteomics of an archaeon. Nat Struct Mol Biol. 2000;7:903–909. doi: 10.1038/82823. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Freeman R, Kupče E. New methods for fast multidimensional NMR. J Biomol NMR. 2003;27:101–113. doi: 10.1023/a:1024960302926. [DOI] [PubMed] [Google Scholar]
- Goddard T, Kneller DG. Sparky 3. San Francisco: University of California; [Google Scholar]
- Grzesiek S, Bax A. Correlating backbone amide and side-chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc. 1992a;114:6291–6293. [Google Scholar]
- Grzesiek S, Bax A. An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J Magn Reson. 1992b;99:201–207. [Google Scholar]
- Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. J Magn Reson. 1992c;96:432–440. [Google Scholar]
- Grzesiek S, Bax A. Amino-acid type determination in the sequential procedure of uniformly C-13/N-15-enriched proteins. J Biomol NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- Gutmanas A, Jarvoll P, Orekhov VY, Billeter M. Three-way decomposition of a complete 3D 15 N-NOESY-HSQC. J Biomol NMR. 2002;24:191–201. doi: 10.1023/a:1021609314308. [DOI] [PubMed] [Google Scholar]
- Helgstrand M, Kraulis P, Allard P, Härd T. ANSIG for Windows: an interactive computer program for semiautomatic assignment of protein NMR spectra. J Biomol NMR. 2000;18:329–336. doi: 10.1023/a:1026729404698. [DOI] [PubMed] [Google Scholar]
- Huang Y, Powers R, Montelione GT. Protein NMR recall, precision and F-measure scores (RPF scores): structure quality assessment measures based in information retrieval statistics. J Am Chem Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- Huang Y, Tejero R, Powers R, Montelione GT. A topology-constrained distance network algorithm for protein structure determination from NOESY data. Proteins. 2006;62:587–603. doi: 10.1002/prot.20820. [DOI] [PubMed] [Google Scholar]
- Ikura M, Kay LE, Bax A. A novel approach for sequential assignment of 1H, 13C, and 15 N spectra of proteins: hetero-nuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry. 1990a;29:4659–4667. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- Ikura M, Krinks M, Torchia DA, Bax A. An efficient NMR approach for obtaining sequence-specific resonance assignments of larger proteins based on multiple isotopic labeling. FEBS Lett. 1990b;266:155–158. doi: 10.1016/0014-5793(90)81528-v. [DOI] [PubMed] [Google Scholar]
- Ikura M, Marion D, Kay LE, Shih H, Krinks M, Klee CB, Bax A. Heteronuclear 3D NMR and isotopic labeling of calmodulin. Towards the complete assignment of the 1H NMR spectrum. Biochem Pharmacol. 1990c;40:153–160. doi: 10.1016/0006-2952(90)90190-v. [DOI] [PubMed] [Google Scholar]
- Ikura M, Kay LE, Bax A. Improved three-dimensional 1H-13C-1H correlation spectroscopy of a 13C-labeled protein using constant-time evolution. J Biomol NMR. 1991a;1:299–304. doi: 10.1007/BF01875522. [DOI] [PubMed] [Google Scholar]
- Ikura M, Kay LE, Krinks M, Bax A. Triple-resonance multidimensional NMR study of calmodulin complexed with the binding domain of skeletal muscle myosin light-chain kinase: indication of a conformational change in the central helix. Biochemistry. 1991b;30:5498–5504. doi: 10.1021/bi00236a024. [DOI] [PubMed] [Google Scholar]
- Ikura M, Spera S, Barbato G, Kay LE, Krinks M, Bax A. Secondary structure and side-chain 1H and 13C resonance assignments of calmodulin in solution by heteronuclear multidimensional NMR spectroscopy. Biochemistry. 1991c;30:9216–9228. doi: 10.1021/bi00102a013. [DOI] [PubMed] [Google Scholar]
- Kay L, Clore GM, Bax A, Gronenborn AM. Four-dimensional heteronuclear triple-resonance NMR spectroscopy of interleu-kin-1 beta in solution. Science. 1990a;249:411–414. doi: 10.1126/science.2377896. [DOI] [PubMed] [Google Scholar]
- Kay L, Ikura M, Tschudin R, Bax A. Three-dimensional triple resonance NMR spectroscopy of isotopically enriched proteins. J Magn Reson. 1990b;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Kim S, Szyperski T. GFT NMR, a new approach to rapidly obtain precise high-dimensional NMR spectral information. J Am Chem Soc. 2003;125:1385–1393. doi: 10.1021/ja028197d. [DOI] [PubMed] [Google Scholar]
- Kobayahi N, Iwahara J, Koshiba S, Tomizawa T, Tochio N, Guntert P, Kigawa T, Yokoyama S. KUJIRA, a package of integrated modules for systemic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J Biomol NMR. 2007;39:31–52. doi: 10.1007/s10858-007-9175-5. [DOI] [PubMed] [Google Scholar]
- Lee W, Westler WM, Bahrami A, Eghbalnia HR, Markley JL. PINE-SPARKY: graphical interface for evaluating automated probabilistic peak assignments in protein NMR spectroscopy. Bioinformatics. 2009;25:2085–2087. doi: 10.1093/bioinformatics/btp345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemak A, Steren CA, Arrowsmith CH, Llinas M. Sequence specific resonance assignment via multicanonical Monte Carlo search using an ABACUS approach. J Biomol NMR. 2008;41(1):29–41. doi: 10.1007/s10858-008-9238-2. [DOI] [PubMed] [Google Scholar]
- Linge J, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- Logan T, Olejniczak ET, Zi RX, Fesik SW. Side chain and backbone assignments in isotropically labeled proteins from two heteronuclear triple resonance experiments. FEBS Lett. 1992;314:413–418. doi: 10.1016/0014-5793(92)81517-p. [DOI] [PubMed] [Google Scholar]
- Luan T, Jaravine V, Yee A, Arrowsmith CH, Orekhov VY. Optimization of resolution and sensitivity of 4D NOESY using multi-dimensional decomposition. J Biomol NMR. 2005;33:1–14. doi: 10.1007/s10858-005-1363-6. [DOI] [PubMed] [Google Scholar]
- Marion D, Driscoll PC, Kay LE, Wingfield PT, Bax A, Gronenborn AM, Clore GM. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15 N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1 beta. Biochemistry. 1989a;28:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- Marion D, Kay LE, Sparks SW, Torchia DA, Bax A. Three-dimensional heteronuclear NMR of nitrogen-15 labeled proteins. J Am Chem Soc. 1989b;111:1514–1515. [Google Scholar]
- Montelione G, Zheng D, Huang YJ, Gunsalus KC, Szyperski T. Protein NMR spectroscopy in structural genomics. Nat Struct Mol Biol. 2000;7:982–985. doi: 10.1038/80768. [DOI] [PubMed] [Google Scholar]
- Muhandiram D, Kay LE. Graident-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J Magn Reson B 103. 1994 [Google Scholar]
- Orekhov V, Ibraghimov I, Billeter M. Optimizing resolution in multidimensional NMR by three-way decomposition. J Biomol NMR. 2003;27:165–173. doi: 10.1023/a:1024944720653. [DOI] [PubMed] [Google Scholar]
- Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS + : a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009a;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. J Biomol NMR. 2009b;43:63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupsky C, Boyko RF, Booth VK, Sykes BD. Smartnotebook: a semi-automated approach to protein sequential NMR resonance assignments. J Biomol NMR. 2003;27:313–321. doi: 10.1023/a:1025870122182. [DOI] [PubMed] [Google Scholar]
- Snyder D, Chen Y, Denissova NG, Acton T, Aramini JM, Ciano M, Karlin R, Liu J, Manor P, Rajan PA, Rossi P, Swapna GV, Xiao R, Rost B, Hunt J, Montelione GT. Comparisons of NMR spectral quality and success in crystallization demonstrate that NMR and X-ray crystallography are complementary methods for small protein structure determination. J Am Chem Soc. 2005;127:16505–16511. doi: 10.1021/ja053564h. [DOI] [PubMed] [Google Scholar]
- Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A. Use of dipolar 1H-15 N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution. Nat Struct Mol Biol. 1997;4:732–738. doi: 10.1038/nsb0997-732. [DOI] [PubMed] [Google Scholar]
- Ulrich E, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte DF, Tolmie DE, KentWenger R, Yao H, Markley JL. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valafar H, Mayer KL, Bougault CM, LeBlond PD, Jenney FE, Jr., Bereton PS, Adams MW, Prestegard JH. Backbone solution structures of proteins using residual dipolar couplings: application to a novel structural genomics target. J Struct Funct Genomics. 2004;5:241–254. doi: 10.1007/s10969-005-4899-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuister G, Bax A. Resolution enhancement and spectral editing of uniformly 13C-enriched proteins by homonuclear broadband 13C decoupling. J Magn Reson. 1992;98:428–435. [Google Scholar]
- Wong L, Masse JE, Jaravine V, Orekhov V, Pervushin K. Automatic assignment of protein backbone resonances by direct spectrum inspection in targeted acquisition of NMR data. J Biomol NMR. 2008;42:77–86. doi: 10.1007/s10858-008-9269-8. [DOI] [PubMed] [Google Scholar]
- Wu B, Skarian T, Yee A, Jobin MC, Dileo R, Semesi A, Fares C, Lemak A, Coombes BK, Arrowsmith CH, Singer AU, Savchenko A. NleG type 3 effectors from enteroharmorrhagicEscherichia coli are U-box E3 ubiquitin ligases. PLoS Pathog 6. 2010 doi: 10.1371/journal.ppat.1000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich K. NMR of proteins and nucleic acids. New York: Willey; 1986. [Google Scholar]
- Yee A, Chang X, Pineda-Lucena A, Wu B, Semesi A, Le B, Ramelot T, Lee GM, Bhattacharyya S, Gutierrez P, Denisov A, Lee CH, Cort JR, Kozlov G, Liao J, Finak G, Chen L, Wishart D, Lee W, McIntosh LP, Gehring K, Kennedy MA, Edwards AM, Arrow-smith CH. An NMR approach to structural proteomics. Proc Natl Acad Sci USA. 2002;99(4):1825–1830. doi: 10.1073/pnas.042684599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee A, Savchenko A, Ignachenko A, Lukin J, Xu X, Skarina T, Evdokimova E, Liu CS, Semesi A, Guido V, Edwards AM, Arrowsmith CH. NMR and X-ray crystallography, complementary tools in structural proteomics of small proteins. J Am Chem Soc. 2005;127:16512–16517. doi: 10.1021/ja053565+. [DOI] [PubMed] [Google Scholar]
- Zheng D, Huang YJ, Moseley HN, Xiao R, Aramini J, Swapna GV, Montelione GT. Automated protein fold determination using a minimal NMR constraint strategy. Protein Sci. 2003;12:1232–1246. doi: 10.1110/ps.0300203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman D, Kulikowski CA, Huang Y, Feng W, Tashiro M, Shimotakahara S, Chien C, Powers R, Montelione GT. Automated analysis of protein NMR assignments using methods from artificial intelligence. J Mol Biol. 1997;269:592–610. doi: 10.1006/jmbi.1997.1052. [DOI] [PubMed] [Google Scholar]
- Zuiderweg E. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]